Abstract
Aerobic microorganisms have evolved a variety of siderochromes, special ligands which can dissolve insoluble ferric iron and facilitate its transport into the cell. We have found that enb mutants of Salmonella typhimurium blocked in the biosynthesis of enterobactin (its natural iron carrier) are able to utilize siderochromes of different types made by other microorganisms as iron carriers. The antibiotic albomycin δ2 was used to select mutants defective in ferrichrome-mediated iron uptake. Twelve classes of albomycin-resistant mutants, named sid, were defined on the basis of their growth responses to other siderochromes. Most of these classes have genetic lesions in loci that are cotransduced with panC (represented at 9 min on the genetic map). The locus designated sidJ is cotransduced with enb, whereas sidK and sidL are linked with neither panC nor enb. Genetic and physiological data indicate that S. typhimurium has several transport systems of high specificity for a variety of siderochromes produced by other microorganisms.
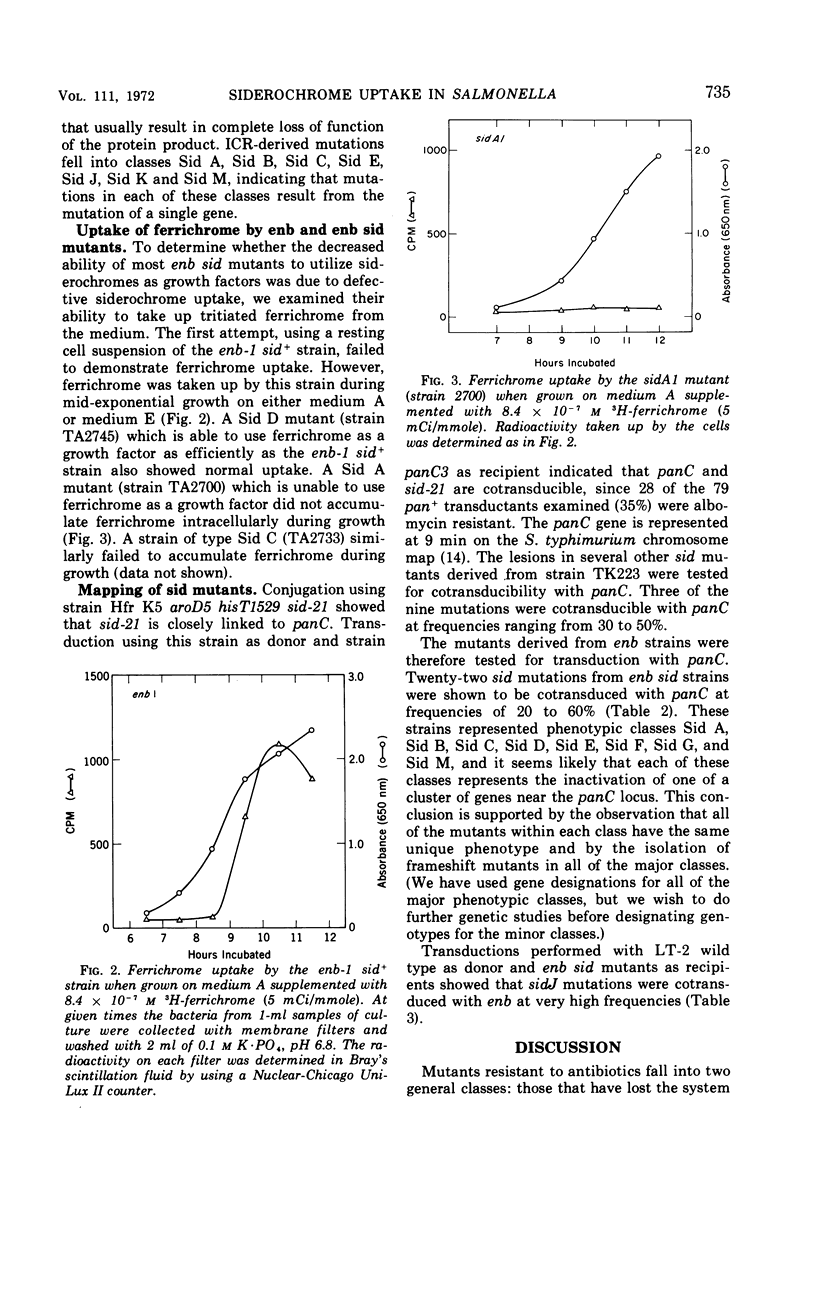
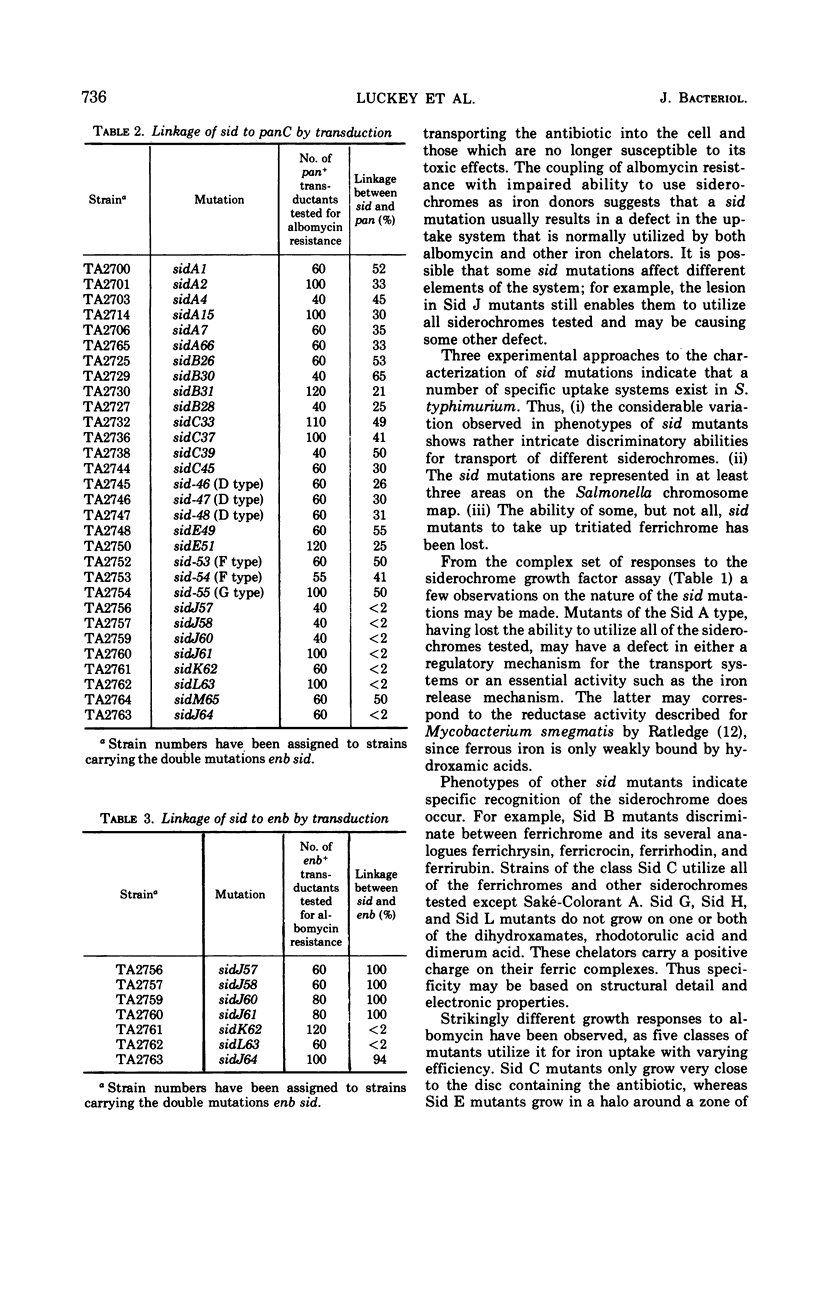
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Whitfield H. J., Jr Frameshift mutagenesis in Salmonella. Cold Spring Harb Symp Quant Biol. 1966;31:221–225. doi: 10.1101/sqb.1966.031.01.030. [DOI] [PubMed] [Google Scholar]
- BRAZHNIKOVA M. G., MIKESH O., LOMAKINA N. N. Issledovanie odnorodnosti al'bomitsina. Biokhimiia. 1957 Jan-Feb;22(1-2):111–117. [PubMed] [Google Scholar]
- Gibson F., Magrath D. I. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim Biophys Acta. 1969 Nov 18;192(2):175–184. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- Maehr H., Pitcher R. G. Identity of albomycin 2 and antibiotic Ro 5-2667. J Antibiot (Tokyo) 1971 Dec;24(12):830–834. doi: 10.7164/antibiotics.24.830. [DOI] [PubMed] [Google Scholar]
- O'Brien I. G., Gibson F. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta. 1970 Aug 14;215(2):393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- Pollack J. R., Ames B. N., Neilands J. B. Iron transport in Salmonella typhimurium: mutants blocked in the biosynthesis of enterobactin. J Bacteriol. 1970 Nov;104(2):635–639. doi: 10.1128/jb.104.2.635-639.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J. R., Neilands J. B. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970 Mar 12;38(5):989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- Ratledge C. Transport of iron by mycobactin in Mycobacterium smegmatis. Biochem Biophys Res Commun. 1971 Nov;45(4):856–862. doi: 10.1016/0006-291x(71)90417-7. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Current linkage map of Salmonella typhimurium. Bacteriol Rev. 1970 Jun;34(2):176–193. doi: 10.1128/br.34.2.176-193.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967 Feb;31(2):207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Zimmermann W., Knüsel F. Permeability of Staphylococcus aureus to the Sideromycin antibiotic A 22,765. Arch Mikrobiol. 1969 Oct;68(2):107–112. doi: 10.1007/BF00413870. [DOI] [PubMed] [Google Scholar]


