Abstract
Genes for σ-like factors of bacterial-type RNA polymerase have not been characterized from any multicellular eukaryotes, although they probably play a crucial role in the expression of plastid photosynthesis genes. We have cloned three distinct cDNAs, designated SIG1, SIG2, and SIG3, for polypeptides possessing amino acid sequences for domains conserved in σ70 factors of bacterial RNA polymerases from the higher plant Arabidopsis thaliana. Each gene is present as one copy per haploid genome without any additional sequences hybridized in the genome. Transient expression assays using green fluorescent protein demonstrated that N-terminal regions of the SIG2 and SIG3 ORFs could function as transit peptides for import into chloroplasts. Transcripts for all three SIG genes were detected in leaves but not in roots, and were induced in leaves of dark-adapted plants in rapid response to light illumination. Together with results of our previous analysis of tissue-specific regulation of transcription of plastid photosynthesis genes, these results indicate that expressed levels of the genes may influence transcription by regulating RNA polymerase activity in a green tissue-specific manner.
Keywords: tissue specificity, photosynthesis, transit peptides, green fluorescent protein, cDNA
The chloroplast is a semi-autonomous organelle whose genetic information is encoded in the nuclear and plastid genomes. The plastid genome encodes genes for photosynthesis, as well as genes for housekeeping functions such as protein synthesis. There is evidence that photosynthesis genes are transcribed by a multimeric Escherichia coli-type RNA polymerase (RNAP), and that housekeeping genes are transcribed by a monomeric T7 or T3 bacteriophage-type RNAP (1). The −10 and −35 sequences, 5′-TATAAT-3′ and 5′-TTGACA-3′, respectively, in promoters of many plastid genes (2) are recognized by the E. coli-type RNAP (3). The E. coli RNAP is composed of a core complex of α, β, and β′ subunits and one of a variety of σ factors, the principal one being σ70, which is capable of binding to the −10 and −35 sequences (4–6). Determination of the complete nucleotide sequences of plastid genomes from liverwort (7), tobacco (8), rice (9), and other plants has resulted in finding of genes, rpoA, rpoB, and rpoC, probably encoding α, β, and β′ subunits, respectively, of a plastid RNAP. In higher plants, rpoC is duplicated, rpoC1 for β′ subunit and rpoC2 for β" subunit. Amino acid sequences deduced from maize plastid genes, rpoC2, rpoB, rpoC1, and rpoA, have proved to correspond to those determined chemically of 180-, 120-, 78-, and 38-kDa polypeptides, respectively, of highly purified maize plastid RNAP (10, 11).
No homolog for the bacterial-like σ factor of RNAP has been detected in the plastid genomes so far sequenced. However, some reports indicate that plastid σ-like factors (SLFs) exist in higher plants. Antibodies against an E. coli σ70 homolog from the cyanobacterium Anabaena sp. PCC7120 have cross-reacted with polypeptides in purified plastid RNAPs from maize and rice (12). In spinach 90- and 33-kDa polypeptides have been identified in a plastid RNAP that are immunologically related to the σ70 factor of E. coli RNAP, and these proteins bound to the promoter of gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (rbcL) and correctly initiated transcription in association with RNAP core enzyme from spinach (13). Functionally distinct 67-, 52-, and 29-kDa SLFs have been identified in mustard, and none of them alone bind to DNA, but they confer enhanced binding to promoters and transcriptional activity on RNAP core enzyme (14, 15). These experiments suggest that genes for homologs of the E. coli σ70 factor exist and are encoded in the nuclear genome. Nuclear genes for such σ factors have recently been reported in the red alga Cyanidium caldarium (16, 17) but not in any multicellular eukaryotes.
We have demonstrated that expression of plastid photosynthesis genes is reduced predominantly at the level of transcriptional initiation in nonphotosynthetic tissues of Arabidopsis thaliana, indicating the importance of regulation through RNAP activity in plastids (18). Therefore, we thought that cloning of genes for SLFs from multicellular eukaryotes was needed for complete understanding of the regulatory mechanism. We report here three cDNAs for putative σ factors from the higher plant A. thaliana. Amino acid sequences deduced from the cloned cDNAs have high homology to conserved regions of the bacterial σ70 factor family.
MATERIALS AND METHODS
Plant Materials.
A. thaliana ecotype Columbia was grown on vermiculite for 4 weeks at 22°C under 16-hr light/8-hr dark. A. thaliana was also grown on a Murashige–Skoog (MS) agar medium (19) without sugar at 22°C in continuous light at 3,000 lux for 3 weeks until the growth stage of generation of 8 rosette leaves, and employed for reverse transcription (RT)–PCR. Tobacco (Nicotiana tabacum cv. Petit Havana) was grown on a MS agar medium (19) containing 3% sucrose at 28°C under continuous light at 4,000 lux.
Preparation of Nucleic Acids.
Total cellular DNA was prepared from leaves with cetyltriethylammonium bromide (CTAB) (20), and RNA was isolated from leaves and roots with a Total RNA Separator Kit (CLONTECH) according to the manufacturer’s instructions. Poly(A)+ RNA fraction was recovered from the total RNA by using Oligotex-dT30 (Takara, Otsu, Japan). Total cellular RNA was also prepared by Isogen (NipponGene, Toyama, Japan) and treated with RQ1 (RNase-free DNase, Promega) following the suppliers’ instructions for employment for RT-PCR.
Screening of cDNAs for σ70 Homologs.
The amino acid sequence GYKFSTYAMWWIRQAITRSIAD, which is responsible for DNA melting and recognition of the −10 sequence in bacterial promoters, was highly conserved in σ70 factors in bacteria. The sequence was subjected to homology search in the database of A. thaliana expressed sequence tags (ESTs). Three EST clones were found with accession numbers [stock numbers at the Arabidopsis Biological Resource Center (ABRC), number of nucleotides reported, including unidentified ones], N65838 [240C23T7, 538 bp], T88387 [155H23T7, 389 bp], and N97044 [242P3T7, 535 bp]. DNA fragments corresponding to the first two of these clones were amplified by PCR from total cellular DNA from A. thaliana with primers designed on the basis of their nucleotide sequences in the EST database. The PCR products were inserted into pT7Blue-T (Novagen) and sequenced to confirm the inserts. These two inserts, as well as a SalI-XhoI fragment from 242P3T7A, were labeled with [α-32P]dCTP by using a Random Primer DNA Labeling Kit (Takara), and used to screen a λ ZAP cDNA library (4 × 105 plaque-forming units) of A. thaliana leaves, which was constructed by using the ZAP-cDNA Synthesis Kit (Stratagene). The inserts of positively hybridizing clones were sequenced by an Applied Biosystems 373A DNA sequencer.
Hybridization of Nucleic Acids.
Total cellular DNA digested with PstI or EcoRI was electrophoresed in 1% agarose gels, and poly(A)+ RNA was electrophoresed in 1.2% agarose gels containing 0.66 M formaldehyde (21). The nucleic acids were transferred to nylon membranes (Hybond-N+, Amersham) with 0.4 M NaOH for DNA and 20× SSC for RNA (1× SSC = 0.15 M NaCl/0.015 M sodium citrate, pH 7.0). The prehybridization and hybridization were performed at 65°C (21). Gene-specific DNA probes were labeled with [α-32P]dCTP by using a Random Primer DNA Labeling Kit (Takara). Membranes were washed with 2× SSC containing 0.1% SDS for 30 min at 65°C, and with 0.5× SSC plus 0.1% SDS under the same conditions. Radioactivity on the membranes was detected by BAS2000 Bio-Imaging Analyzer (Fujix, Tokyo).
RT-PCR.
Total cellular RNA prepared from A. thaliana was treated with RQ1 DNase, following the supplier’s instructions. RT-PCR was performed with ≈1 μg of the total RNA and oligo(dT)12–18 in 21 μl of reaction mixture by using the SuperScript Preamplification System for First Strand cDNA Synthesis (GIBCO/BRL). An aliquot (2 μl for SIG genes, described in Results, or 1 μl for ACT2 encoding actin 2) from the reaction mixture was further subjected to PCR with AmpliTaq (Perkin–Elmer). Primers for PCR of SIG1, SIG2, and SIG3 (see Results) were designed to amplify 395-, 254-, and 342-bp DNA fragments, respectively, on the basis of nucleotide sequences of cDNAs determined in this investigation: 5′-CCTCCGAGGTCGCTAGACCAG-3′ and 5′-GCTCAGGTGGGTGGGTTCTATGC-3′ for SIG1, 5′-GGTCTCCCTGGAGAAACTCATC-3′ and 5′-CTCCAGCGCCACAAGCCCTACC-3′ for SIG2, and 5′-GATTCGAGCAGCTAACCAATGCC-3′ and 5′-GAGGTCTTCGGCGTTGGGTTGG-3′ for SIG3. ACT2 (22) was also examined as an internal standard with primers, 5′-GAAGATTAAGGTCGTTGCACCACCTG-3′ and 5′-ATTAACATTGCAAAGAGTTTCAAGGT-3′, to amplify the 477-bp DNA fragment with cDNA or the 563-bp one with genomic DNA, if any, contaminating the RNA fractions.
Transient Expression of Chimeric Genes.
DNA fragments encoding the first 83 and 89 amino acid residues of the ORFs of SIG2 and SIG3 cDNAs, respectively, were amplified by PCR with pairs of synthetic oligonucleotides containing new NcoI sites (italicized): 5′-TTCCATGGCTACTGCAGCTG-3′ and 5′-TTCCATGGTAGAAGCAACATCATC-3′ for SIG2, and 5′-TTCCATGGCTTCCTTCA-3′ and 5′-TTCCATGGATAGAAACGACC-3′ for SIG3. Two PCR products were digested with NcoI and inserted into the NcoI site of CaMV35S-sGFP(S65T)-nos [pUC18, harboring a synthetic gene for improved green fluorescent protein sGFP(S65T), hereafter referred to as “GFP”] driven by cauliflower mosaic virus 35S promoter and NOS terminator (23), resulting in two chimeric GFP constructs, SIG2-GFP and SIG3-GFP. The synthetic GFP gave ≈100 times higher fluorescent signal upon excitation with 488-nm light in comparison with that of the native GFP (23). The chimeric constructs were introduced into tobacco leaf cells by using Biolistic PDS-1000/He (Bio-Rad). Gun parameters employed were as follows: pressure rupture disks rated at 1,100 psi (7.58 MPa), a vacuum at 27 inches of Hg (reaching 9.83 kPa), distance to target tissues 6 cm, and gold particles 1.0 μm in diameter. After bombardment, cells were incubated for 20 hr at 28°C under continuous light, and observed by using the Bio-Rad MRC-1024 Confocal Imaging System (480×).
RESULTS
Three Distinct cDNAs for Polypeptides with Conserved Domains in σ Factors.
Three cDNA clones (reported partial sequences of 538 bp, 389 bp, and 535 bp, including unidentified nucleotides) in the EST database of A. thaliana contain animo acid sequences that are highly homologous to regions for melting DNA and recognizing the −10 promoter sequence of bacterial σ factors. The sequences in these clones were employed as probes to screen a leaf cDNA library from A. thaliana, and positively hybridizing clones, 6, 9, or 7, respectively, for these three ESTs, were obtained from the library of 4 × 105 plaque-forming units. Three cDNAs, corresponding to stock numbers 240C23T7, 155H23T7, and 242P3T7, were designated SIG1, SIG2, and SIG3, respectively. The largest ORFs of SIG1, SIG2, and SIG3 coded for polypeptides with 572, 502, and 571 amino acids, respectively.
Similarity of Functional Domains in σ Factors.
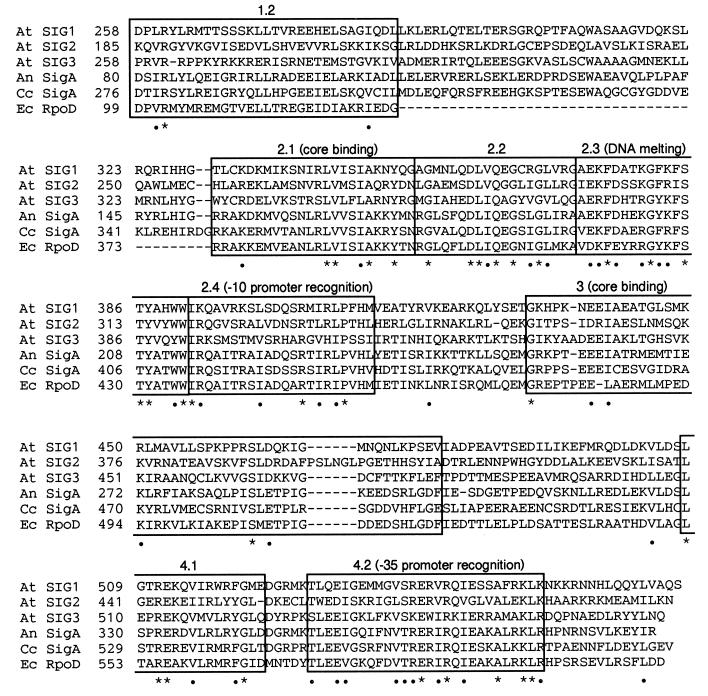
Amino acid sequences deduced from SIG1, SIG2, and SIG3 were compared with σ factors from E. coli (24), Anabaena sp. PCC7120 (25), and Cyanidium caldarium (17) (Fig. 1). The comparison showed that all ORFs of SIG genes are highly conserved in two major regions (designated regions 2 and 4) in bacterial σ factors (26). Subregions 2.1, 2.3, 2.4, and 4.2 are involved in interaction with core enzyme, DNA melting, recognition of −10 promoter sequence, and recognition of −35 promoter sequence, respectively. The subregion 4.1 can be modeled as an amphipathic α-helix. Region 3 may contribute to binding to core RNAP. The functions of subregions 1.2 and 2.2 are unclear. Table 1 shows comparisons of SIG1, SIG2, and SIG3 with the highly conserved subregions 2.1, 2.3, 2.4, and 4.2 from E. coli, Anabaena, and C. caldarium. Among three SIGs, SIG1 has the highest similarity to bacterial σ factors, and SIG3 has the least similarity. Among the subregions of the SIGs, 2.3 is the most conserved, while 2.1 and 2.4 are the least conserved, especially in SIG3 (18–29%). The subregion 4.2 in SIG2 is more diverse from bacterial σ factors, showing 43% in spite of 57–61% in SIG1 and 57–64% in SIG3.
Figure 1.
Alignment of conserved domains in the deduced amino acid sequences of A. thaliana SIG1 (At SIG1), SIG2 (At SIG2), and SIG3 (At SIG3) with those of SigA from Anabaena sp. PCC 7120 (An SigA), SigA from C. caldarium (Cc SigA), and σ70 factor encoded in rpoD from E. coli (Ec RpoD). Regions 1–4 in bacterial σ factors (5, 26) are boxed. Residues marked with an asterisk (∗) or a dot (•) are identical or similar, respectively, in all six sequences. The numbers between the product designations and the amino acid sequences are those of amino acid residues. There are 246 amino acids in a gap between subregion 1.2 and subregion 2.1 in E. coli RpoD.
Table 1.
Identity of A. thaliana SIG1, SIG2, and SIG3 to other factors in the σ70 family in major conservative regions
| σ factor subregion | Identity, %
|
||
|---|---|---|---|
| SIG1 | SIG2 | SIG3 | |
| Escherichia coli | |||
| 2.1 | 50 | 42 | 25 |
| 2.3 | 61 | 44 | 56 |
| 2.4 | 55 | 45 | 27 |
| 4.2 | 57 | 43 | 57 |
| Anabaena | |||
| 2.1 | 58 | 50 | 29 |
| 2.3 | 78 | 61 | 67 |
| 2.4 | 55 | 64 | 18 |
| 4.2 | 61 | 43 | 57 |
| Cyanidium caldarium | |||
| 2.1 | 46 | 50 | 21 |
| 2.3 | 78 | 67 | 56 |
| 2.4 | 50 | 55 | 27 |
| 4.2 | 57 | 43 | 64 |
See Fig. 1 for sequences of the σ factors.
Numbers and Expression of the SIG Genes.
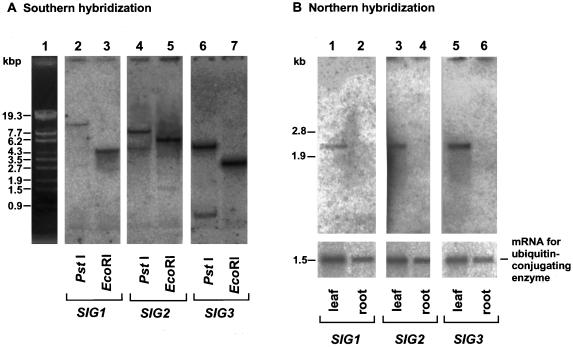
Total cellular DNA from A. thaliana was digested with PstI or EcoRI and subjected to Southern hybridization using SIG1, SIG2, and SIG3 as probes (Fig. 2A). One band was generated in each case except for SIG2 and SIG3 digested with PstI (lanes 4 and 6). Multiple bands in these lanes resulted from the presence of PstI sites in these two SIG genes. Therefore, SIG1, SIG2, and SIG3 are probably single-copy genes in A. thaliana.
Figure 2.
Southern and Northern blot analyses of SIG genes. (A) Southern hybridization with SIG gene probes of total cellular DNA (8 μg per lane) digested with PstI (lanes 2, 4, and 6) and EcoRI (lanes 3, 5, and 7). DNA in lanes 2 and 3, lanes 4 and 5, and lanes 6 and 7, was hybridized with 1.7-kb EcoRI fragment from SIG1 cDNA, 1.6-kb XbaI fragment from SIG2 cDNA, or 1.2-kb XhoI fragment from SIG3 cDNA, respectively. (B) Northern hybridization with SIG probes of total poly(A)+ RNA (0.5–1.0 μg per lane) from leaves (lanes 1, 3, and 5) and roots (lanes 2, 4, and 6). The cDNA for ubiquitin-conjugating enzyme, which is constitutively expressed (M.S., Y.N., and H.K., unpublished results), was used as a probe as an internal control for equivalence of amounts of RNA loaded into lanes. Each of the SIG genes gives rise to ≈2.3-kb mRNA.
We have already demonstrated that suppressed expression of the plastid genome-encoded photosynthesis genes in roots of A. thaliana is due predominantly to transcriptional regulation (18). To examine the possibility that there is an involvement of SIGs in the transcriptional regulation, levels of transcripts for the SIG genes in RNAs from leaves and roots were investigated by Northern hybridization. All transcripts for SIG1, SIG2, and SIG3 were detected in leaves, but barely in roots (Fig. 2B). This finding suggests that expressed levels of SIG genes correlate with transcriptional activities of plastid photosynthesis genes evaluated by run-on transcription (18).
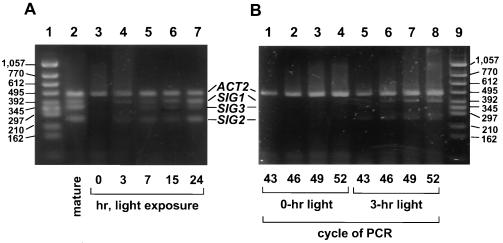
Light induction of SIG transcripts in leaves of dark-adapted plants was examined by RT-PCR. Transcripts for ACT2 (encoding actin 2), which is reported to be constitutively expressed (22), were determined as internal standards both for proving no contamination with genomic DNA in the RNA fractions and estimating the amounts of SIG transcripts. Expression of all three SIG genes was obviously under light control (Fig. 3). SIG1 and SIG2 transcripts were induced during 3-hr illumination (Fig. 3B) and seemed to reach a plateau between 7 and 15 hr in the light induction (Fig. 3A).
Figure 3.
Time-sequential changes of transcripts for SIG genes in dark-adapted plants after exposure to light. (A) A. thaliana grown for 3 weeks was incubated in the dark for 3 days or maintained under the same light conditions for plants indicated as “mature.” Leaves of individual dark-adapted plants were harvested at 0, 3, 7, 15, and 24 hr after initiation of the exposure to light at 3,000 lux. RT-PCR products were subjected to electrophoresis after PCR cycles when intensities of signals for ACT2 were unsaturated and comparable among RNA fractions harvested at different illumination times. RT-PCR products (5 μl from 50 μl of reaction mixture) were electrophoresed in 3% agarose gel (Agarose H, NipponGene), stained with SYBR green I (FMC BioProducts), and observed by FluorImager SI (Molecular Dynamics). (B) RT-PCR products of 0- and 3-hr illumination after indicated PCR cycles were electrophoresed and stained with SYBR green I.
Destination of SIG Gene Products for Chloroplasts.
Transit peptides can be predicted by psort, a program for protein sorting (http://psort.nibb.ac.jp/index.html) (27). The N-terminal regions of SIG2 and SIG3 (Fig. 4) were predicted to be transit peptides at probability higher than 90%. These N-terminal sequences are enriched with Ser and Thr, a property of transit peptides (28). However, the N-terminal region of SIG1 ORF (Fig. 4) was less likely to be a transit peptide.
Figure 4.
N-terminal sequences of SIG1, SIG2, and SIG3. The first 90 amino acids in the ORFs of SIG1 (SIG1), SIG2 (SIG2), and SIG3 (SIG3) cDNAs are represented. Ser and Thr residues are indicated in boldface.
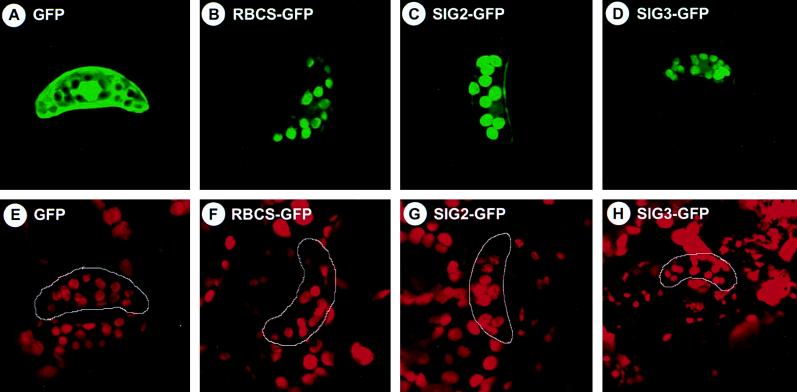
We have tested whether N-terminal regions of SIGs can function as transit peptides for plastid-targeting by transient expression assays with an improved GFP. We made two constructs in which peptides composed of the first 83 amino acids of SIG2 ORF and of the first 89 residues of SIG3 ORF (Fig. 4) were fused to the N terminus of GFP (SIG2-GFP and SIG3-GFP) and placed under the control of cauliflower mosaic virus 35S promoter. Tobacco leaves were bombarded with one of these constructs. The GFP alone (23) and a chimeric construct of GFP with the transit peptide of small subunit of Rubisco [RBCS-GFP, as a positive control (23)] were used. Fig. 5 shows results with tobacco guard cells, in which it is easier to visualize GFP accumulated in chloroplasts. Green fluorescence was observed only in chloroplasts of the guard cells (Fig. 5 B, C, and D), where chloroplasts were identified by red fluorescence of chlorophyll (Fig. 5 F, G, and H). Without these N-terminal sequences, GFP alone was not localized in chloroplasts (Fig. 5 A and E). We obtained similar results with leaves of A. thaliana (data not shown). These data suggest that the N-terminal regions of SIG2 and SIG3 ORFs can function as transit peptides for import into chloroplasts. We could not confirm the function of N-terminal region of SIG1 as a transit peptide by transient expression with GFP (data not shown).
Figure 5.
Localization of GFPs fused to N-terminal regions of SIG2 and SIG3. GFP fusion constructs with the N-terminal regions of SIG2 ORF (SIG2-GFP, C and G) and SIG3 ORF (SIG3-GFP, D and H), and the transit peptide of the small subunit of Rubisco (23) (RBCS-GFP, B and F), as well as GFP alone (23) (GFP, A and E), were introduced into tobacco leaves by particle bombardment. Guard cells were observed by using the MRC-1024 Confocal Imaging System (480×) with excitation at 488 nm and emission at 520 nm (A–D), as well as excitation at 647 nm and emission at 666 nm (E–H). The same objects are shown in each pair of upper and lower panels.
DISCUSSION
When the complete nucleotide sequence of tobacco plastid genome was determined in 1986 (8), genes for all subunits of E. coli-type RNAP except for SLFs were found in the plastid genome. In spite of thorough effort to identify genes for SLFs in plants, they were not cloned until 1996, when the genes were isolated from the unicellular red alga Cyanidium caldarium by two research groups (16, 17). In the investigation reported here, putative genes for σ factors from multicellular eukaryotes have now been characterized. It took over a decade to obtain clones for SLFs from higher plants for the following reasons: (i) low contents of mRNA, one in 40,000–70,000 molecules of total poly(A)+ RNA in leaves of A. thaliana as estimated from the frequency of cloning in this investigation; (ii) homology conserved but at degree nonhybridizable to bacterial ones; and (iii) possible interference by the gene products with the growth of the E. coli used for cloning (data not shown).
The ORFs of SIG1, SIG2, and SIG3 encode polypeptides consisting of 572, 502, and 571 amino acids, respectively, with calculated molecular masses of 64, 56, and 65 kDa. Although these are precursors with transit peptides, they are close to 67 kDa (SLF67) and 52 kDa (SLF52) of mustard SLFs associated with the activity of σ factors (16, 29) and to 64-kDa peptides from maize and rice that immunochemically cross-reacted with antibodies against a σ factor from Anabaena (12). SIG1 among the three SIGs most resembles bacterial σ factors and has more than 50% identity to four major conservative subregions—2.1, 2.3, 2.4, and 4.2—in each of the σ factors from E. coli, Anabaena, and C. caldarium except for the subregion 2.1 in C. caldarium. SIG2 has lower identity in its subregion 4.2 for −35 promoter recognition, and SIG3 has significantly low identity in both its subregions 2.1 and 2.4 for DNA melting and −10 promoter recognition, respectively. The differences in amino acid sequences among the subregions of the three SIGs may contribute to fine regulation of transcription of distinct target genes.
We have already demonstrated that expression of plastid photosynthesis genes, rbcL for large subunit of Rubisco, psbA for D1 protein in photosystem II reaction center, and atpB/E for β and ɛ subunits of coupling factor 1, is remarkably suppressed by transcriptional regulation in roots in A. thaliana (18). Northern analysis of SIG1, SIG2, and SIG3 gene expression showed that their transcripts were few in roots but were present in leaves (Fig. 2B). This observation suggests that changes of SIG levels could regulate the expression of plastid photosynthesis genes in a tissue-specific manner. The light induction of transcripts for SIG genes (Fig. 3) probably precedes induction of transcripts of plastid photosynthesis genes such as rbcL (data not shown). Therefore, it is very probable that generation of all SIG gene products results in the initiation of transcription of photosynthesis genes after their transport into plastids and association with RNAP core enzyme in plastids. We think that the present investigation has opened a way to study not only the biological significance of each of the three distinct SIGs but also the regulation of their tissue-specific expression.
Acknowledgments
We are indebted to Thomas Newman and the Arabidopsis Biological Resource Center for the EST database and clones and to Jen Sheen for GFP. We thank Steven R. Rodermel for critical reading of the manuscript. The research was supported by Grants-in-Aid from the Ministry of Education, Science and Culture of Japan (Monbusho) and by grants from the New Energy and Industrial Technology Development Organization (NEDO)/the Research Institute of Innovative Technology for the Earth (RITE), and the Toray Science Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNAP, RNA polymerase; SLF, σ-like factor; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; RT, reverse transcription; EST, expressed sequence tag; GFP, green fluorescent protein.
References
- 1.Allison L A, Simon L D, Maliga P. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- 2.Kung S D, Lin C M. Nucleic Acids Res. 1985;13:7543–7549. doi: 10.1093/nar/13.21.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igloi G L, Kel H. Crit Rev Plant Sci. 1992;10:525–558. [Google Scholar]
- 4.Burgess R, Travers A, Dunn J, Bautz E. Nature (London) 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 5.Helmann J D, Chamberlin M J. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 6.Busby S, Ebright R H. Cell. 1992;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 7.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S, Inokuchi H, Ozeki H. Nature (London) 1986;322:572–574. [Google Scholar]
- 8.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng B, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun C-R, Meng B-Y, Li Y-Q, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, Bogorad L. Proc Natl Acad Sci USA. 1990;87:1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, Troxler R F, Bogorad L. Nucleic Acids Res. 1991;19:3431–3434. doi: 10.1093/nar/19.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troxler R F, Zhang F, Hu J, Bogorad L. Plant Physiol. 1994;104:753–759. doi: 10.1104/pp.104.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerbs S, Bräutigam E, Mache R. Mol Gen Genet. 1988;211:459–464. [Google Scholar]
- 14.Bülow S, Link G. Plant Mol Biol. 1988;10:349–357. doi: 10.1007/BF00029885. [DOI] [PubMed] [Google Scholar]
- 15.Tiller K, Eisermann A, Link G. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Troxler R F. Proc Natl Acad Sci USA. 1996;93:3313–3318. doi: 10.1073/pnas.93.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka K, Oikawa K, Kuroiwa H, Kuroiwa T, Takahashi H. Science. 1996;272:1932–1933. doi: 10.1126/science.272.5270.1932. [DOI] [PubMed] [Google Scholar]
- 18.Isono K, Niwa Y, Satoh K, Kobayashi H. Plant Physiol. 1997;114:623–630. doi: 10.1104/pp.114.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 20.Rogers S O, Bendich A J. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.An Y Q, Mcdowell J M, Huang S R, Mckinney E C, Chambliss S, Meagher R B. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- 23.Chiu W L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 24.Burton Z, Burgess R R, Lin J, Moore D, Holder S, Gross C A. Nucleic Acids Res. 1981;9:2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brahamsha B, Haselkorn R. J Bacteriol. 1991;173:2442–2450. doi: 10.1128/jb.173.8.2442-2450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonetto M, Gribskov M, Gross C. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keegstra K, Olsen L J. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- 29.Tiller K, Link G. Plant Mol Biol. 1993;21:503–513. doi: 10.1007/BF00028807. [DOI] [PubMed] [Google Scholar]