Abstract
Introduction: Genetic factors influence circulating thyroid hormone levels, but the common gene variants involved have not been conclusively identified. The genes encoding the iodothyronine deiodinases are good candidates because they alter the balance of thyroid hormones. We aimed to thoroughly examine the role of common variation across the three deiodinase genes in relation to thyroid hormones.
Methods: We used HapMap data to select single-nucleotide polymorphisms (SNPs) that captured a large proportion of the common genetic variation across the three deiodinase genes. We analyzed these initially in a cohort of 552 people on T4 replacement. Suggestive findings were taken forward into three additional studies in people not on T4 (total n = 2513) and metaanalyzed for confirmation.
Results: A SNP in the DIO1 gene, rs2235544, was associated with the free T3 to free T4 ratio with genome-wide levels of significance (P = 3.6 × 10−13). The C-allele of this SNP was associated with increased deiodinase 1 (D1) function with resulting increase in free T3/T4 ratio and free T3 and decrease in free T4 and rT3. There was no effect on serum TSH levels. None of the SNPs in the genes coding for D2 or D3 had any influence on hormone levels.
Conclusions: This study provides convincing evidence that common genetic variation in DIO1 alters deiodinase function, resulting in an alteration in the balance of circulating free T3 to free T4. This should prove a valuable tool to assess the relative effects of circulating free T3 vs. free T4 on a wide range of biological parameters.
There is a strong association between a single nucleotide polymorphism in the type 1 iodothyronine deiodinase (DIO1) gene and serum levels of free T4 and free T3, but not TSH, demonstrating the importance of genetic factors in determining thyroid hormone levels.
There is known to be a genetic component to circulating thyroid hormone levels (1,2), but the common gene variants involved have not been conclusively identified.
The three deiodinase enzymes [deiodinase type 1 (D1), D2, and D3] play a vital role in maintaining euthyroidism both at a serum and local tissue level (3). Their action is important in maintaining different levels of thyroid hormone activity in different tissues, in different disease states, and at different stages of development. They achieve this by altering the balance of the thyroid hormones between the active hormone T3, the inactive prohormone T4, which can be activated by conversion to T3, and the inactivated metabolites rT3 and diiodothyronine (T2).
D1 and D2 are predominantly activating enzymes; both convert T4 to T3 (and rT3 to T2) by outer ring deiodination. D1 is found in liver, kidney, thyroid, and pituitary in humans, and D2 in skeletal muscle, central nervous system, pituitary, thyroid, heart, and brown adipose tissue (3). Predictions based on isolated cell deiodinase activity and reported tissue activities in humans suggest that both are responsible for maintaining serum levels of T3, although D2 predominates in hypothyroidism and D1 in hyperthyroidism (4). D3 inactivates thyroid hormones by inner ring deiodination, converting T3 to T2 and T4 to rT3. It has been found in the central nervous system and placenta in adults and in many additional tissues in the fetal state (3). A more detailed description of deiodinase action can be found in recent reviews (3,5).
Previous studies have suggested that genetic variation in the deiodinase enzyme-encoding genes may influence thyroid hormone levels and ratios (6,7,8). The most comprehensive of these studies, of four single-nucleotide polymorphisms (SNPs) in 995 elderly Dutch individuals, found the strongest association between rs11206244 in the DIO1 gene and T3/rT3 ratio (P < 0.001) and weaker associations with T3 (P = 0.004), free T4 (P = 0.04), and rT3 (P = 0.03) (6). Additional evidence is required to assess more comprehensively the role of common variation in the DIO1 genes. It is now widely accepted that genetic association studies need to cover a large proportion of variation across a gene and that levels of statistical confidence need to exceed P < 5 × 10−7. None of these variants has been shown to influence TSH levels, although TSH secretion is controlled by negative feedback of T4 and T3 on the hypothalamus and pituitary. It should also be noted that unlike thyroid hormone receptors, there are no known individuals with inactivating mutations of the deiodinase genes and that deiodinase knockout mice display quite mild phenotypes (9,10). This means our understanding of the importance of genetic variation in the deiodinase genes in humans is limited.
In this study, we aimed to thoroughly assess the role of common variation in the deiodinase genes in relation to thyroid hormone levels and the relationship of circulating concentrations of free T3 (fT3) to fT4. We selected SNPs that covered a large proportion of common variation across the three DIO genes. We initially examined the role of this variation in a study of individuals with little or no thyroidal production of fT3 on T4 replacement therapy because we would expect functional variants to have the greatest effect in this population. We then examined associations in three additional studies of individuals without known thyroid disease. We show that a common variant in intron 3 of the DIO1 gene rs2235544 is a clear genetic marker associated with changes in the conversion of fT4 to fT3 but without altering TSH levels.
Subjects and Methods
Details of the studies used are given in Table 1. The initial analysis was performed on the Weston Area T3/T4 Study (WATTS) cohort of 552 individuals with thyroid disease being treated with T4. We followed up SNPs reaching suggestive levels of significance (P < 0.05) in three more studies consisting of 3424 individuals (including 911 pregnant females): two population studies [Exeter Family Study of Childhood Health (EFSOCH) and Invecchiare in Chianti (InCHIANTI)] and a study of people being tested for abnormal thyroid function on the basis of clinical indication by their general practitioners (GPs) [Depression and Thyroid Disease (DEPTH)]. In all cohorts except WATTS, individuals with any history of thyroid disease or a TSH outside the range of 0.05–10.0 mU/liter were excluded, as were individuals taking medications known to alter thyroid hormone levels (amiodarone, lithium, carbimazole, methimazole, or propylthiouracil). All individuals were of white European ancestry.
Table 1.
Baseline characteristics
| Characteristic | WATTS | EFSOCH males | EFSOCH pregnant femalesa | InCHIANTI | DEPTH |
|---|---|---|---|---|---|
| n | 552 | 877 | 911 | 1200 | 436 |
| No. female (%) | 460 (83.3) | 662 (55.2) | 335 (76.8) | ||
| Age (yr) | 57.2 ± 10.8 | 32.7 ± 5.9 | 30.4 ± 5.2 | 68.4 ± 15.5 | 43.7 ± 15.0 |
| BMI (kg/m2) | 29.2 ± 61 | 26.6 ± 3.8 | 24.0 ± 4.6 | 27.1 ± 4.1 | |
| fT4 (pmol/liter) | 21.1 ± 3.7 | 16.5 ± 2.1 | 12.2 ± 1.6 | 18.7 ± 4.6 | 16.5 ± 7.0 |
| fT3 (pmol/liter) | 3.84 ± 0.72 | 5.51 ± 0.6 | 4.2 ± 0.5 | 6.63 ± 1.0 | 4.67 ± 0.72 |
| TSH (mU/liter), median (IQR) | 0.91 (0.29, 2.08) | 1.82 (1.33, 2.53) | 1.85 (1.35, 2.50) | 1.3 (0.84, 2.00) | 1.5 (1.0, 2.4) |
Results are mean ± sd unless otherwise specified. IQR, Interquartile range.
BMI in pregnant females is based on self-reported prepregnancy weight.
Subjects/cohorts
WATTS
WATTS cohort consists of people on T4 replacement, recruited from GP practices in the Bristol and Weston-super-Mare areas in the West of England between March 2000 and June 2002. Entry criteria have been previously published (11). They were involved in a study on the benefits of combined T3/T4 therapy; however, in this study, only baseline data were used (on a stable dose of T4 only). The study was approved by the local research ethics committees in Bristol.
EFSOCH
The EFSOCH study is a prospective study of birth weight and early postnatal growth. Blood was taken from mothers, fathers, and offspring who lived in the central part of Exeter, Devon, UK. All mothers in the study were pregnant at the time of blood sampling. Therefore, the fathers and pregnant mothers were analyzed separately, because pregnancy has well known effects on thyroid hormone levels and may also have effects on deiodinase function. Study design and protocol have been described in detail previously (12).
InCHIANTI
The InCHIANTI study is a representative, population-based sample of residents from two small towns near Florence, Italy, initially recruited to investigate risk factors for the onset of disability in older persons. It included 298 subjects aged less than 65 yr and 1155 aged 65 yr or older, and data collection took place between September 1998 and March 2000. The study design and protocol have been described in detail previously (13). The study was approved by the Instituto Nazionale Riposo e Cura Anziani (INRCA) ethical committee.
DEPTH
The DEPTH study is a prospective study to investigate the relationship between thyroid function and mood in a population with no known thyroid disease, presenting to primary care. Participants comprised patients not on T4 and with no previously documented thyroid abnormality in whom the GP considered that thyroid function testing was clinically indicated. It included five GP practices in Bristol, UK, and data collection took place between September 2005 and July 2007. Participants had a venous blood sample taken for thyroid function analysis and DNA extraction and filled out two questionnaires with mood assessments, the 12-question version of the General Health Questionnaire (GHQ-12) and the Patient Health Questionnaire (PHQ). The study was approved by the local research ethics committees in Bristol.
Biochemical measurements
WATTS
Serum TSH and fT4 were measured from a serum sample by RIA (Diagnostic Products Corp. Los Angeles, CA). Free T3 was measured by chemiluminescence assay (Elecsys system 1010; Roche Diagnostics, Mannheim, Germany). The laboratory reference ranges were TSH 0.3–4.0 mU/liter, fT4 10–24 pmol/liter, and fT3 2.8–7.1 pmol/liter. Coefficients of variation (CV) were TSH 5.5–8.0%, fT4 7.7–10.0%, and fT3 11.7–12.6%. rT3 was measured by an in-house RIA (Erasmus University Medical Centre) (14).
EFSOCH
Serum TSH, fT4, and fT3 were analyzed using the electrochemiluminescent immunoassay, run on the Modular E 170 Analyzer (Roche, Burgess Hill, UK). The manufacturer’s population reference ranges were TSH 0.35–4.5 mU/liter, fT4 11–24 pmol/liter, and fT3 3.9–6.8 pmol/liter. The between-batch CV were TSH 5.4%, fT4 7%, and fT3 4.2%.
InCHIANTI
TSH, fT4, and fT3 were measured in EDTA plasma using chemiluminescent assays (Vitros TSH Reagent; Ortho-Clinical Diagnostics, Johnson & Johnson Medical S.p.A Section, Raritan, NJ). Reference normal ranges were TSH 0.46–4.68 mIU/liter, fT4 10–28 pmol/liter, and fT3 4.3–8.1 pmol/liter. Intraassay CV were TSH less than 5.3%, fT4 less than 5.3%, and fT3 less than 5.1%.
DEPTH
Serum TSH and free T4 were measured from serum samples by chemiluminescent immunometric assay on the Immulite 2000 (Siemens, Gwynedd, UK). Free T3 was measured by electrochemiluminescence immunoassay on the Elecsys 1010 (Roche Diagnostics, Mannheim, Germany). The laboratory reference ranges were TSH 0.3–4.0 mU/liter, fT4 10–24 pmol/liter, and fT3 2.8–7.1 pmol/liter. Intraassay CV were: TSH less than 6.3%, fT4 less than 7.5%, and fT3 less than 2.2%.
Tag SNP selection and genotyping
We used genotype data from the European individuals in the International Haplotype Mapping Project (http://www.hapmap.org) to select a set of SNPs that capture the majority of common variation across the three deiodinase genes (DIO1, DIO2, and DIO3) including 50 kb either side of the genes. We used a minor allele frequency of at least 10%. The 21, seven, and seven SNPs in the DIO1, DIO2, and DIO3 genes required nine, four, and six SNPs, respectively, to capture all common variants with an r2 greater than 0.8. These were, for D1, rs11206237, rs11206244, rs2235544, rs2268181, rs2294511, rs2294512, rs4926616, rs731828, and rs7527713; for D2, rs12885300, rs225011, rs225014, and rs225015; and for D3, rs1190716, rs17716499, rs7150269, rs8011440, rs945006, and rs1190715. In previous studies, rs11206244 has also been referred to as D1a-C/T, and rs225014 has been referred to as D2-Thr92Ala (6,8).
Genotyping for the WATTS and DEPTH cohorts was performed by KBiosciences (http://www.kbioscience.co.uk) using their own novel fluorescence-based competitive allele-specific PCR (KASPar). Assays were designed for each of the 19 SNPs by KBiosciences. Design of an assay for the SNP rs1190715 was not possible; hence, genotyping was performed on 18 SNPs only. In WATTs, two additional SNPs, rs1190716 and rs12885300, failed quality control because of a high number of duplicate errors (>1%) and deviation from Hardy-Weinberg equilibrium (P < 0.05), respectively. The percentage of duplicate samples included was 20%. Concordance between duplicate samples for the 16 SNPs used was at least 99%. Use of these 16 SNPs resulted in coverage of 100, 85, and 71% of the common (minor allele frequency >10%), HapMap-based variation in DIO1, DIO2, and DIO3, respectively, at r2 greater than 0.8.
Genotyping on the EFSOCH cohort was performed in-house (Peninsula Medical School) using TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. The percentage of duplicate samples included was 12%. Concordance between duplicate samples was at least 99% for rs2235544.
Genotyping for the InCHIANTI cohort was performed as part of a genome-wide association study using the Illumina Infinium HumanHap550 genotyping chip. Samples were initially assessed for genotype success rate (>98%) and concordance of reported and genotype-based gender.
Statistical analysis
We used a log transformation for serum TSH, but fT3, rT3, and fT4 did not require normalization. To detect an association between thyroid parameters and genotypes, linear regression was used, with log TSH, fT4, fT3, or rT3 as the dependent variables and genotype (as zero, one, or two alleles) as the independent. The analysis was performed both unadjusted and using age and sex as covariates. Analyses were performed using STATA version 9.0 (StataCorp, College Station, TX).
Metaanalysis was performed using an inverse variance weighting method as implemented using the metan command of STATA version 9.0. We tested for heterogeneity using the Q statistics and I2 metric as implemented in STATA version 9.0.
Results
Association of deiodinase gene variation in patients on T4 replacement therapy
We first tested deiodinase gene variants in 552 individuals from the WATTS study. These individuals are hypothyroid with little or no endogenous thyroid hormone production and are treated with T4. They produce little or no thyroidal T3 and are therefore an ideal group to assess function of enzymes that convert fT4 to fT3. Sixteen of the 18 SNPs were successfully genotyped and passed quality control criteria in the WATTs study. Two SNPs in the DI01 gene, rs2235544 and rs11206244, were associated with fT3/fT4 ratio at P < 0.01 (Table 2). The same SNPs were also associated with individual fT3, rT3, and fT4 levels but not TSH levels (see Table 3 and supplemental Table 1, A–D, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org).
Table 2.
The fT3/fT4 ratio by genotype in WATTs cohort
| SNP | Common homozygous
|
Heterozygous
|
Minor homozygous
|
P for trend
|
||||
|---|---|---|---|---|---|---|---|---|
| Mean fT3/fT4 (95% CI) | n | Mean fT3/fT4 (95% CI) | n | Mean fT3/fT4 (95% CI) | n | Unadjusted | Adjusteda | |
| DIO1 | ||||||||
| rs11206237 | 0.187 (0.183, 0.192) | 399 | 0.187 (0.180, 0.195) | 130 | 0.192 (0.177, 0.207) | 16 | 0.82 | 0.96 |
| rs11206244 | 0.193 (0.187, 0.199) | 239 | 0.186 (0.181, 0.192) | 238 | 0.175 (0.166, 0.185) | 69 | 0.005 | 0.004 |
| rs2235544 | 0.177 (0.171, 0.184) | 143 | 0.189 (0.184, 0.195) | 288 | 0.196 (0.187, 0.205) | 111 | 0.001 | 0.001 |
| rs2268181 | 0.187 (0.182, 0.192) | 387 | 0.189 (0.182, 0.196) | 140 | 0.192 (0.178, 0.205) | 19 | 0.63 | 0.88 |
| rs2294511 | 0.192 (0.186, 0.197) | 240 | 0.185 (0.179, 0.190) | 248 | 0.183 (0.171, 0.195) | 56 | 0.08 | 0.08 |
| rs2294512 | 0.186 (0.180, 0.191) | 252 | 0.186 (0.180, 0.191) | 245 | 0.205 (0.190, 0.220) | 50 | 0.06 | 0.02 |
| rs4926616 | 0.189 (0.183, 0.194) | 245 | 0.188 (0.182, 0.194) | 236 | 0.186 (0.174, 0.197) | 56 | 0.67 | 0.68 |
| rs731828 | 0.185 (0.179, 0.191) | 183 | 0.189 (0.183, 0.194) | 278 | 0.190 (0.179, 0.201) | 84 | 0.32 | 0.19 |
| rs7527713 | 0.187 (0.182, 0.192) | 362 | 0.189 (0.182, 0.196) | 157 | 0.184 (0.170, 0.198) | 26 | 0.96 | 0.75 |
| DIO2 | ||||||||
| rs225011 | 0.184 (0.177, 0.190) | 172 | 0.189 (0.183, 0.195) | 264 | 0.190 (0.182, 0.198) | 107 | 0.22 | 0.24 |
| rs225014 | 0.186 (0.180, 0.192) | 223 | 0.187 (0.181, 0.193) | 236 | 0.193 (0.184, 0.203) | 87 | 0.25 | 0.28 |
| rs225015 | 0.187 (0.181, 0.193) | 237 | 0.187 (0.181, 0.193) | 236 | 0.193 (0.183, 0.204) | 71 | 0.42 | 0.46 |
| DIO3 | ||||||||
| rs17716499 | 0.191 (0.185, 0.196) | 202 | 0.184 (0.178, 0.189) | 248 | 0.193 (0.183, 0.202) | 95 | 0.89 | 0.97 |
| rs7150269 | 0.191 (0.184, 0.197) | 169 | 0.186 (0.180, 0.192) | 254 | 0.187 (0.179, 0.196) | 121 | 0.48 | 0.50 |
| rs8011440 | 0.190 (0.184, 0.195) | 215 | 0.186 (0.180, 0.192) | 246 | 0.188 (0.178, 0.199) | 85 | 0.65 | 0.54 |
| rs945006 | 0.187 (0.183, 0.192) | 438 | 0.187 (0.178, 0.196) | 92 | 0.200 (0.156, 0.243) | 10 | 0.69 | 0.55 |
CI, Confidence interval.
Adjusted for age and sex.
Table 3.
Relationships between rs2235544 and thyroid function in all studies
| Measure | AA
|
AC
|
CC
|
B coefficient | se. | P for trenda | Metaanalysis P valueb | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (95%CI) | n | Mean (95%CI) | n | Mean (95%CI) | n | |||||
| fT3/fT4 | ||||||||||
| WATTS | 0.18 (0.17, 0.18) | 143 | 0.19 (0.18, 0.19) | 288 | 0.20 (0.19, 0.21) | 111 | 0.009 | 0.003 | 0.001 | |
| EFS-M | 0.32 (0.32, 0.33) | 224 | 0.34 (0.33, 0.34) | 430 | 0.35 (0.34, 0.36) | 201 | 0.011 | 0.002 | 1.3 × 10−6 | |
| InChianti | 0.35 (0.34, 0.36) | 244 | 0.37 (0.36, 0.37) | 533 | 0.38 (0.37, 0.39) | 320 | 0.015 | 0.003 | 1.5 × 10−7 | |
| DEPTH | 0.29 (0.28, 0.30) | 116 | 0.30 (0.29, 0.31) | 192 | 0.30 (0.29, 0.31) | 115 | 0.005 | 0.004 | 0.244 | |
| 3.6 × 10−13 | ||||||||||
| EFS-F | 0.34 (0.34, 0.35) | 236 | 0.35 (0.35, 0.36) | 450 | 0.35 (0.34, 0.36) | 210 | 0.005 | 0.003 | 0.072 | |
| fT3 | ||||||||||
| WATTS | 3.75 (3.65, 3.85) | 143 | 3.88 (3.80, 3.96) | 288 | 3.91 (3.76, 4.07) | 111 | 0.076 | 0.045 | 0.092 | |
| EFS-M | 5.43 (5.36, 5.49) | 225 | 5.53 (5.47, 5.58) | 431 | 5.55 (5.46, 5.64) | 201 | 0.053 | 0.027 | 0.05 | |
| InChianti | 6.51 (6.42, 6.60) | 244 | 6.58 (6.52, 6.64) | 533 | 6.57 (6.49, 6.65) | 320 | 0.034 | 0.030 | 0.261 | |
| DEPTH | 4.66 (4.52, 4.79) | 116 | 4.71 (4.61, 4.81) | 193 | 4.65 (4.53, 4.78) | 115 | −0.015 | 0.045 | 0.750 | |
| 0.058 | ||||||||||
| EFS-F | 4.15 (4.08, 4.22) | 237 | 4.20 (4.15, 4.24) | 449 | 4.20 (4.13, 4.27) | 209 | 0.029 | 0.024 | 0.226 | |
| fT4 | ||||||||||
| WATTS | 21.7 (21.1, 22.3) | 143 | 21.1 (20.6, 21.5) | 288 | 20.4 (19.7, 21.0) | 111 | −0.631 | 0.231 | 0.007 | |
| EFS-M | 16.9 (16.6, 17.2) | 224 | 16.5 (16.4, 16.7) | 431 | 16.1 (15.9, 16.4) | 201 | −0.408 | 0.101 | 5.8 × 10−5 | |
| InChianti | 19.4 (18.8, 20.0) | 244 | 18.5 (18.2, 18.9) | 533 | 17.9 (17.5, 18.3) | 324 | −0.772 | 0.162 | 2.1 × 10−6 | |
| DEPTH | 16.5 (15.9, 17.1) | 116 | 16.8 (15.4, 18.2) | 193 | 15.9 (15.4, 16.4) | 115 | −0.361 | 0.464 | 0.440 | |
| 2.1 × 10−9 | ||||||||||
| EFS-F | 12.2 (12.0, 12.5) | 236 | 12.1 (12.0, 12.3) | 450 | 12.1 (11.9, 12.4) | 209 | −0.044 | 0.076 | 0.561 | |
| TSH | ||||||||||
| WATTS | 0.79 (0.61, 1.03) | 143 | 0.65 (0.55, 0.78) | 288 | 0.67 (0.53, 0.85) | 111 | 0.032 | 0.040 | 0.510 | |
| EFS-M | 1.31 (1.27, 1.36) | 220 | 1.29 (1.26, 1.31) | 427 | 1.31 (1.27, 1.35) | 200 | −0.0008 | 0.011 | 0.938 | |
| InChianti | 1.24 (1.13, 1.35) | 234 | 1.25 (1.18, 1.33) | 516 | 1.35 (1.25, 1.46) | 311 | 0.046 | 0.030 | 0.127 | |
| DEPTH | 2.81 (2.10, 3.70) | 116 | 2.77 (2.25, 3.42) | 193 | 2.15 (1.67, 2.77) | 115 | −0.056 | 0.042 | 0.180 | |
| 0.90 | ||||||||||
| EFS-F | 1.28 (1.24, 1.33) | 236 | 1.29 (1.26, 1.32) | 448 | 1.31 (1.28, 1.35) | 208 | 0.010 | 0.011 | 0.357 | |
EFS-F, EFSOCH females; EFS-M, EFSOCH males.
Adjusted for sex and age.
Excludes EFSOCH females and WATTS.
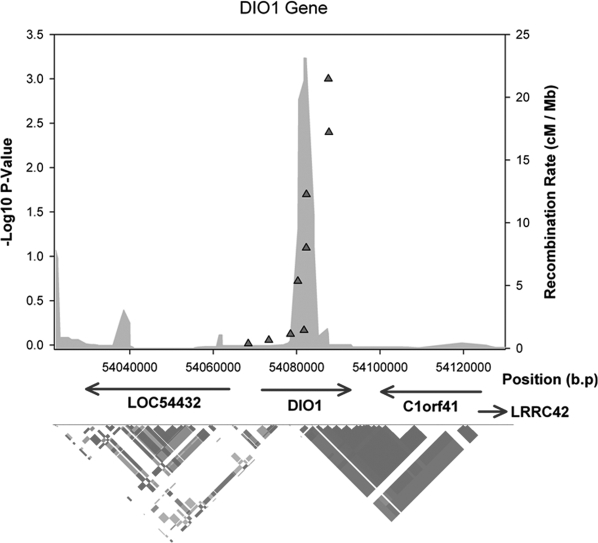
There were no associations at P < 0.01 in the DIO2 or DIO3 genes with any of the ratio or individual thyroid hormone measures. There was evidence of linkage disequilibrium (LD) between these SNPs within our data, strongest between rs2294511 and rs4926616 (r2 = 0.49), and rs2235544 and rs11206244 (r2 = 0.41). All other pairwise LD correlations within the DIO1 gene were r2 less than 0.16. The LD structure of the DIO1 gene and associated statistics are shown in Fig. 1.
Figure 1.
LD structure and associations in WATTS diagram of the DIO1 gene.
Associations in population-based samples
To provide more robust evidence, and to assess whether associations were specific to patients with thyroid disease or were present in the general population, we followed up SNPs associated at P < 0.01 in males and pregnant females from a second study. The associations between rs2235544 and rs11206244 and fT3/fT4 ratio were confirmed in the EFSOCH males (P = 1.3 × 10−6) with the same allele A, being associated with decreased ratio (supplemental Table 2). There was no evidence of association in the pregnant females (P = 0.07), in whom thyroid hormone levels were measured at 28 wk gestation.
Given the LD (r2 = 0.41) between rs2235544 and rs11206244, we performed conditional analysis including both SNPs in the regression model for fT3/fT4 ratio in both WATTs and EFSOCH males and metaanalyzed the two results. This revealed that rs2235544 was driving the association (P = 6.8 × 10−4 conditioning on rs11206244) rather than rs11206244 (P = 0.46 conditioning on rs2235544).
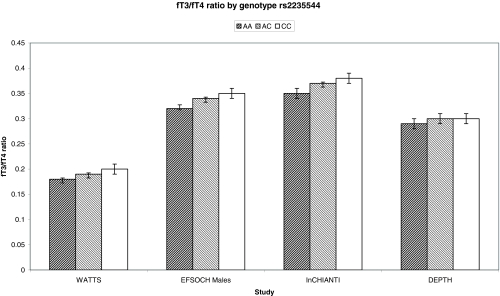
The association between the A allele of rs2235544 and reduced fT3/fT4 ratio was further confirmed in the InCHIANTI study of healthy older males and females (n = 1200; P = 1.5 × 10−7) and was consistent in the smaller DEPTH study of predominantly females (n = 436; P = 0.24). The association of rs2235544 with fT3/fT4 ratio and individual hormone levels in all studies is summarized in Table 3. The fT3/fT4 ratio in the WATTS population is significantly lower than in the populations with normal thyroid function, which is consistent with previous findings in patients on T4 therapy who have a higher fT4 and lower fT3 for the same TSH due to lack of thyroid T3 production (15,16,17). A metaanalysis of the population-based studies, excluding pregnant females, showed strong evidence for an association in the general population (P = 3.6 × 10−13), with no evidence of heterogeneity (P = 0.13; I2 = 50%). Each copy of the C-allele was associated with an increase in the fT3/fT4 ratio of approximately 0.2 sd. The fT3/fT4 ratios by genotype are shown in Fig. 2. By contrast, there was no association of this allele with TSH, either in any individual study or in a metaanalysis of all studies (P = 0.90, Table 3). There was no difference in the effect size between nonpregnant females and males (P > 0.05).
Figure 2.
The fT3/fT4 ratio by genotype of rs2235544. The graph shows the plot of the mean and 95% confidence intervals of fT3/fT4 ratio in the four studies.
Discussion
Our results show that common variation in the DIO1 gene, best tagged by rs2235544, is associated with circulating fT3/fT4 ratio both in a population of people on thyroid hormone replacement and in the general population. In contrast to previous studies, we have assessed a large proportion of the common variation across the DIO genes, and importantly, the statistical confidence of our finding (P = 3.6 × 10−13) exceeds that necessary for multimarker genetic association studies (18). Our study therefore provides robust evidence that a common variant can affect serum thyroid hormone levels.
The effect of rs2235544 on thyroid hormone levels is consistent with the C-allele being associated with higher deiodinase enzymatic activity (increased fT3/fT4 ratio and fT3 and decreased fT4 and rT3). This association is mainly driven by the strong association with decreased fT4 (see Table 3). This association was present both in subjects on T4 replacement, with no endogenous T3 production, and those with normal thyroid function, suggesting that a significant amount of serum T3 is derived from peripheral deiodination of T4 in both populations. These findings are consistent with those from the D1 knockout mouse, in which absent D1 activity results in raised fT4 and rT3 serum levels with a decreased fT3/fT4 ratio and no effect on fT3 or TSH (10) (resembling the effect of the A-allele if this is associated with decreased D1 activity). Previous studies identified putative associations between rs11206244 and fT4, fT3, and rT3 levels but at much lower levels of statistical confidence (6,8). This SNP is partially correlated with rs2235544, but conditional analyses clearly showed rs2235544 is driving the association. The rs2235544 occurs in intron 3 of the DIO1 gene. There are no SNPs in HapMap in the coding region of DIO1 that are strongly correlated with rs2235544. There are also no SNPs within 100 kb of the DIO1 gene that are associated with altered DIO1 expression levels in the recently published genome-wide study of expression levels in lymphocytes (19). This means the mechanism by which this SNP or one in LD with it affects deiodinase function remains unknown. Taking into account the heritability estimates of twin studies for serum fT4 (39–65%) and fT3 (23–64%) (1,2), we estimate, using the regression model, that rs2235544 is responsible for approximately 2% of genetic variance of fT4 and 1.5% of fT3.
No association was observed between any of the SNPs in DIO2 or DIO3 and circulating concentrations of thyroid hormone levels in our cohort of patients on T4, and this is consistent with several previous studies in patients not on T4 (6,20,21). This implies either that none of the common SNPs we have studied in these genes are linked to changes in enzyme function or that these enzymes (D2 in particular) have a smaller role in determining serum (as opposed to intracellular) thyroid hormone levels than was previously thought (3,4,22). One of the three DIO2 SNPs we studied, rs225014, has previously been reported to be associated with reduced enzyme activity in thyroid and skeletal muscle samples (23), although the effect was not apparent in transfected cell lines (24) and phenotype associations with DIO2 SNPs have been inconsistent in other reports (6,23,25,26). Hence, additional evidence is required of the functional association of common variation in the DIO2 and DIO3 genes before we can interpret the lack of association with serum thyroid hormone levels. Studies of the effects of a common variation in DIO3 in pregnant women (not studied here) would also be of interest in view of the high levels of D3 expression in placental tissue.
Discovery of a marker that alters the ratio of circulating T3 to T4 may help us answer important questions about thyroid hormone action in humans. T3 is the active hormone that binds to thyroid hormone receptors. However, the local deiodination of T4 to intracellular T3 by the D2 enzyme means that circulating concentrations of both T3 and T4 levels contribute to intracellular T3 levels. Importantly, wide variation in the levels of deiodinase expression (and possibly also of thyroid hormone transporters) (27) between tissues results in wide variation in the relative contribution of the circulating concentrations of these two hormones to thyroid hormone action. In rodent studies, for example, it is estimated that serum T3 contributes 87% of intracellular T3 in the kidney but only 50% in the pituitary and just 20% in the cerebral cortex, the remainder coming from local deiodination of serum T4 (3). Hence, variation in the ratio of T3 to T4 in the circulation is likely to have different effects in different tissues, with potential impact on a wide range of important biological parameters such as body weight, serum lipids, heart rate, bone metabolism, central nervous system development, and psychological well-being. However, very large samples are likely to be required for robust exploration of these effects. To assess the effects of a SNP that alters fT3/fT4 ratio on secondary traits, power calculations need to consider both the correlation between fT3/fT4 ratio and the correlation between fT3/fT4 ratio and the secondary trait (28). For example, rs2235544 is associated with an approximately 0.2 sd difference in fT3/fT4 ratio per allele. The fT3/fT4 ratio is correlated with body mass index (BMI) in published studies with an r of approximately 0.28 (29). This means studies of the effects of rs2235544 on BMI need to include approximately 15,000 individuals to be powered to detect the expected effect of 0.056 (0.2 × 0.28) sd BMI change per allele (at P = 0.01 and 80% power).
The very clear lack of an association between rs2235544 and circulating concentrations of TSH across all our cohorts, despite its effects on the fT3/fT4 ratio, is interesting. It implies that rs2235544 can be used as a tool to explore the effects of changes in the balance of circulating T3 and T4 on different tissues in the absence of an increase in overall thyroid hormone production from the thyroid. It is also consistent with the observation that TSH levels correlate as well if not better with serum fT4 than fT3 in patients on T4 (11) and implies that for the pituitary, the rise in circulating concentrations of fT3 compensates for the fall in fT4 associated with the C-allele of rs2235544. The pituitary itself does express D1 (3), although D2 may predominate in the feedback pathway.
In conclusion, our study provides the most convincing example of common DNA variation that alters circulating thyroid hormone ratios in the general population. This variant may be useful in dissecting causal relationships between thyroid hormone levels and related traits in different tissues.
Supplementary Material
Footnotes
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes on Health, Department of Health and Human Services (Annual Report Number Z01 AG000965-02; http://nidb.nih.gov/search/searchreport.taf?ipid=44815), Endocrine Research Fund, South-West National Health Service Research and Development, and the Special Trustees of United Bristol Healthcare Trust. D.M. is supported in part by National Institutes of Health National Institute on Aging Grant R01 AG24233-01.
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 20, 2008
Abbreviations: BMI, Body mass index; CV, coefficients of variation; D1, deiodinase type 1; DEPTH, Depression and Thyroid Disease; EFSOCH, Exeter Family Study of Childhood Health; fT3, free T3; GP, general practitioner; InCHIANTI, Invecchiare in Chianti; LD, linkage disequilibrium; SNP, single-nucleotide polymorphism; WATTS, Weston Area T3/T4 Study.
References
- Hansen PS, Brix TH, Sorensen TI, Kyvik KO, Hegedus L 2004 Major genetic influence on the regulation of the pituitary-thyroid axis: a study of healthy Danish twins. J Clin Endocrinol Metab 89:1181–1187 [DOI] [PubMed] [Google Scholar]
- Panicker V, Wilson SG, Spector TD, Brown SJ, Falchi M, Richards JB, Surdulescu GL, Lim EM, Fletcher SJ, Walsh JP 2008 Heritability of serum TSH, free T4 and free T3 concentrations: a study of a large UK twin cohort. Clin Endocrinol (Oxf) 68:652–659 [DOI] [PubMed] [Google Scholar]
- Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR 2002 Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev 23:38–89 [DOI] [PubMed] [Google Scholar]
- Maia AL, Kim BW, Huang SA, Harney JW, Larsen PR 2005 Type 2 iodothyronine deiodinase is the major source of plasma T3 in euthyroid humans. J Clin Invest 115:2524–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting A, Yen PM 2007 New insights into thyroid hormone action. Best Pract Res Clin Endocrinol Metab 21:193–208 [DOI] [PubMed] [Google Scholar]
- de Jong FJ, Peeters RP, den Heijer T, van der Deure WM, Hofman A, Uitterlinden AG, Visser TJ, Breteler MM 2007 The association of polymorphisms in the type 1 and 2 deiodinase genes with circulating thyroid hormone parameters and atrophy of the medial temporal lobe. J Clin Endocrinol Metab 92:636–640 [DOI] [PubMed] [Google Scholar]
- Peeters RP, van den Beld AW, Attalki H, Toor H, de Rijke YB, Kuiper GG, Lamberts SW, Janssen JA, Uitterlinden AG, Visser TJ 2005 A new polymorphism in the type II deiodinase gene is associated with circulating thyroid hormone parameters. Am J Physiol Endocrinol Metab 289:E75–E81 [DOI] [PubMed] [Google Scholar]
- Peeters RP, van Toor H, Klootwijk W, de Rijke YB, Kuiper GG, Uitterlinden AG, Visser TJ 2003 Polymorphisms in thyroid hormone pathway genes are associated with plasma TSH and iodothyronine levels in healthy subjects. J Clin Endocrinol Metab 88:2880–2888 [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St Germain DL, Galton VA 2001 Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol 15:2137–2148 [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, Parlow AF, St Germain DL, Galton VA 2006 Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology 147:580–589 [DOI] [PubMed] [Google Scholar]
- Saravanan P, Simmons DJ, Greenwood R, Peters TJ, Dayan CM 2005 Partial substitution of thyroxine (T4) with tri-iodothyronine in patients on T4 replacement therapy: results of a large community-based randomized controlled trial. J Clin Endocrinol Metab 90:805–812 [DOI] [PubMed] [Google Scholar]
- Knight B, Shields BM, Hattersley AT 2006 The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol 20:172–179 [DOI] [PubMed] [Google Scholar]
- Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, Guralnik JM 2000 Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc 48:1618–1625 [DOI] [PubMed] [Google Scholar]
- Visser TJ, Docter R, Hennemann G 1977 Radioimmunoassay of reverse tri-iodothyronine. J Endocrinol 73:395–396 [DOI] [PubMed] [Google Scholar]
- Meier C, Staub JJ, Roth CB, Guglielmetti M, Kunz M, Miserez AR, Drewe J, Huber P, Herzog R, Muller B 2001 TSH-controlled l-thyroxine therapy reduces cholesterol levels and clinical symptoms in subclinical hypothyroidism: a double blind, placebo-controlled trial (Basel Thyroid Study). J Clin Endocrinol Metab 86:4860–4866 [DOI] [PubMed] [Google Scholar]
- Woeber KA 2002 Levothyroxine therapy and serum free thyroxine and free triiodothyronine concentrations. J Endocrinol Invest 25:106–109 [DOI] [PubMed] [Google Scholar]
- Jorde R, Waterloo K, Storhaug H, Nyrnes A, Sundsfjord J, Jenssen TG 2006 Neuropsychological function and symptoms in subjects with subclinical hypothyroidism and the effect of thyroxine treatment. J Clin Endocrinol Metab 91:145–153 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium 2007 Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO 2007 A genome-wide association study of global gene expression. Nat Genet 39:1202–1207 [DOI] [PubMed] [Google Scholar]
- Mentuccia D, Thomas MJ, Coppotelli G, Reinhart LJ, Mitchell BD, Shuldiner AR, Celi FS 2005 The Thr92Ala deiodinase type 2 (DIO2) variant is not associated with type 2 diabetes or indices of insulin resistance in the old order of Amish. Thyroid 15:1223–1227 [DOI] [PubMed] [Google Scholar]
- Gumieniak O, Perlstein TS, Williams JS, Hopkins PN, Brown NJ, Raby BA, Williams GH 2007 Ala92 type 2 deiodinase allele increases risk for the development of hypertension. Hypertension 49:461–466 [DOI] [PubMed] [Google Scholar]
- Pilo A, Iervasi G, Vitek F, Ferdeghini M, Cazzuola F, Bianchi R 1990 Thyroidal and peripheral production of 3,5,3′-triiodothyronine in humans by multicompartmental analysis. Am J Physiol 258:E715–E726 [DOI] [PubMed] [Google Scholar]
- Canani LH, Capp C, Dora JM, Meyer EL, Wagner MS, Harney JW, Larsen PR, Gross JL, Bianco AC, Maia AL 2005 The type 2 deiodinase A/G (Thr92Ala) polymorphism is associated with decreased enzyme velocity and increased insulin resistance in patients with type 2 diabetes mellitus. J Clin Endocrinol Metab 90:3472–3478 [DOI] [PubMed] [Google Scholar]
- Coppotelli G, Summers A, Chidakel A, Ross JM, Celi FS 2006 Functional characterization of the 258 A/G (D2-ORFa-Gly3Asp) human type-2 deiodinase polymorphism: a naturally occurring variant increases the enzymatic activity by removing a putative repressor site in the 5′ UTR of the gene. Thyroid 16:625–632 [DOI] [PubMed] [Google Scholar]
- Guo TW, Zhang FC, Yang MS, Gao XC, Bian L, Duan SW, Zheng ZJ, Gao JJ, Wang H, Li RL, Feng GY, St Clair D, He L 2004 Positive association of the DIO2 (deiodinase type 2) gene with mental retardation in the iodine-deficient areas of China. J Med Genet 41:585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AL, Dupuis J, Manning A, Liu C, Meigs JB, Cupples LA, Larsen PR, Fox CS 2007 The type 2 deiodinase (DIO2) A/G polymorphism is not associated with glycemic traits: the Framingham Heart Study. Thyroid 17:199–202 [DOI] [PubMed] [Google Scholar]
- van der Deure WM, Peeters RP, Visser TJ 2007 Genetic variation in thyroid hormone transporters. Best Pract Res Clin Endocrinol Metab 21:339–350 [DOI] [PubMed] [Google Scholar]
- Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CN, Morris AD, Elliott P, Jarvelin MR, Smith GD, McCarthy MI, Hattersley AT, Frayling TM 2008 Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected, given its effect on BMI. Diabetes 57:1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pergola G, Ciampolillo A, Paolotti S, Trerotoli P, Giorgino R 2007 Free triiodothyronine and thyroid stimulating hormone are directly associated with waist circumference, independently of insulin resistance, metabolic parameters and blood pressure in overweight and obese women. Clin Endocrinol (Oxf) 67:265–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.