Abstract
Among the seven tyrosine autophosphorylation sites identified in the intracellular domain of tyrosine kinase fibroblast growth factor receptor-1 (FGFR1), five of them are dispensable for FGFR1-mediated mitogenic signaling. The possibility of dissociating the mitogenic activity of basic FGF (FGF2) from its urokinase-type plasminogen activator (uPA)-inducing capacity both at pharmacological and structural levels prompted us to evaluate the role of these autophosphorylation sites in transducing FGF2-mediated uPA upregulation. To this purpose, L6 myoblasts transfected with either wild-type (wt) or various FGFR1 mutants were evaluated for the capacity to upregulate uPA production by FGF2. uPA was induced in cells transfected with wt-FGFR1, FGFR1-Y463F, -Y585F, -Y730F, -Y766F, or -Y583/585F mutants. In contrast, uPA upregulation was prevented in L6 cells transfected with FGFR1-Y463/583/585/730F mutant (FGFR1–4F) or with FGFR1-Y463/583/585/730/766F mutant (FGFR1–5F) that retained instead a full mitogenic response to FGF2; however, preservation of residue Y730 in FGFR1-Y463/583/585F mutant (FGFR1–3F) and FGFR1-Y463/583/585/766F mutant (FGFR1–4Fbis) allows the receptor to transduce uPA upregulation. Wild-type FGFR1, FGFR1–3F, and FGFR1–4F similarly bind to a 90-kDa tyrosine-phosphorylated protein and activate Shc, extracellular signal-regulated kinase (ERK)2, and JunD after stimulation with FGF2. These data, together with the capacity of the ERK kinase inhibitor PD 098059 to prevent ERK2 activation and uPA upregulation in wt-FGFR1 cells, suggest that signaling through the Ras/Raf-1/ERK kinase/ERK/JunD pathway is necessary but not sufficient for uPA induction in L6 transfectants. Accordingly, FGF2 was able to stimulate ERK1/2 phosphorylation and cell proliferation, but not uPA upregulation, in L6 cells transfected with the FGFR1-Y463/730F mutant, whereas the FGFR1-Y583/585/730F mutant was fully active. We conclude that different tyrosine autophosphorylation requirements in FGFR1 mediate cell proliferation and uPA upregulation induced by FGF2 in L6 cells. In particular, phosphorylation of either Y463 or Y730, dispensable for mitogenic signaling, represents an absolute requirement for FGF2-mediated uPA induction.
INTRODUCTION
Fibroblast growth factors (FGFs) comprise a family of at least 18 related heparin-binding polypeptides that play an essential role in the regulation of embryogen esis, angiogenesis, differentiation, and cell proliferation (Basilico and Moscatelli, 1992). In particular, basic FGF (FGF2),1 one of the prototypes of the family, induces, among other responses, cell proliferation and upregulation of urokinase-type plasminogen activator (uPA) production in different cell types, including endothelial cells and tumor cell lines (Presta et al., 1988, 1989). uPA upregulation is involved in different processes in which extracellular proteolysis is required, including cell migration, developmental tissue reorganization, angiogenesis, and invasive growth in normal and pathologic conditions (Dano et al., 1985; Blasi and Verde, 1990; Tienari et al., 1991; Kwaan, 1992).
FGFs interact on the cell surface with membrane-spanning tyrosine kinase FGF receptors (TK-FGFRs) classified as subclass IV (Ullrich and Schlessinger, 1990; Fantl et al., 1993). The first FGFR to be characterized (FGFR1/flg [Lee et al., 1989]) is a single membrane-spanning molecule with three extracellular immunoglobulin (Ig)-like domains, an acidic box located between the first and the second Ig-like loop, a transmembrane domain, a juxtamembrane (JM) region, and an intracellular catalytic TK domain split by a 14 amino acid insertion. Since then, three other genes encoding TK-FGFRs have been discovered: FGFR2/bek (Dionne et al., 1990), FGFR3 (Keegan et al., 1991), and FGFR4 (Partanen et al., 1991). Several RNA alternative spliced variants that structurally differ in the number of Ig-like loops and/or in the absence of the intracellular domain (soluble forms) were also described for FGFR1 and FGFR2 (Johnson and Williams, 1993). The growth factor binding site of FGFR appears to be located in the second half of the third Ig-like loop; three variants of this region, encoded by different exons, have been described: IIIa, IIIb, and IIIc. The IIIa sequence seems to be unique for the FGFR1-soluble receptor form (Johnson et al., 1991), whereas IIIb and IIIc are found in FGFR1, FGFR2 (Johnson and Williams, 1993), and FGFR3 (Chellaiah et al., 1994) membrane-spanning molecules. Binding studies from several laboratories indicate that IIIc variants show a broad spectrum of ligands (Johnson and Williams, 1993; Ornitz et al., 1996). Indeed, we have shown that the IIIc variants of FGFR1, -2, -3, and -4 are all able to transduce uPA upregulation by FGF-1, FGF2, and FGF-4 in Chinese hamster ovary transfectants (Rusnati et al., 1996).
As a general mechanism, binding of growth factors to cognate TK receptors and subsequent conformational alteration of the extracellular domain leads to receptor oligomerization (Ullrich and Schlessinger, 1990). The interactions between adjacent cytoplasmic domains lead to receptor autophosphorylation and activation of kinase function by allosteric mechanisms (Ullrich and Schlessinger, 1990). Autophosphorylation of TK receptors normally occurs at a conserved tyrosine residue located in the kinase domain that allosterically regulates the Vmax of the receptor and at various tyrosine residues distributed along the intracellular portion (Ullrich and Schlessinger, 1990). Phosphorylated tyrosines serve as docking sites for downstream signal transduction molecules containing either Src-homology 2 or phosphotyrosine-binding domains (Cantley et al., 1991; Koch et al., 1991; Margolis, 1992; Pawson and Schlessinger, 1993; Pawson, 1995; Blaikie et al., 1994; Kavanaugh and Williams, 1994; Kavanaugh et al., 1995; Bork and Margolis, 1995). Thus, the capacity of growth factors to exert a complex array of biological responses on the same cell type is thought to reflect the capacity of different docking transducer proteins to associate with the activated TK receptor, leading to the switch of multiple intracellular signals (Pawson and Schlessinger, 1993).
The multiple signal transduction pathways activated by FGFR1 are not fully elucidated. Recent observations have demonstrated that activation of FGFR1 induces tyrosine phosphorylation of the lipid-anchored docking protein FRS2 that forms a direct complex, as well as an Shp-2 tyrosine phosphatase-mediated complex, with Grb2/Sos, thus linking FGFR1 activation to the Ras/MAPK kinase (also referred to as MEK)/MAPK (also referred to as ERK) pathway (Wang et al., 1996; Kouhara et al., 1997). Moreover, FGFR1 activation leads to tyrosine phosphorylation of 80K-H protein, distinct from FRS2, that also binds to Grb2 (Goh et al., 1996). Furthermore, a novel membrane-associated adapter protein, named SNT-2, has been shown to interact with FGFR1 in vitro and to be tyrosine-phosphorylated after FGFR1 stimulation in vivo (Xu et al., 1998).
Seven autophosphorylation sites in the intracellular domain of FGFR1 have been identified: Y463, Y583, Y585, Y653, Y654, Y730, and Y766 (Mohammadi et al., 1991, 1996a). Among them, phosphorylated tyrosine 766 has been shown to bind phospholipase C (PLC)γ (Mohammadi et al., 1991), although PLCγ activation appears to be dispensable for cell proliferation, differentiation, and uPA upregulation induced by FGFs (Mohammadi et al., 1991, 1992; Peters et al., 1992; Spivak-Kroizman et al., 1994; Huang et al., 1995; Roghani et al., 1996). Tyrosines 653 and 654 are crucial instead for receptor kinase activity, and their neutralization hampers receptor autophosphorylation (Mohammadi et al., 1996a). Simultaneous multiple mutations of the autophosphorylation sites Y463, Y583, Y585, and Y730 do not perturb the ability of FGFR1 to mediate cell proliferation and differentiation (Mohammadi et al., 1996a). The biological function(s) of these sites and their relationship with FGFR1-activated signal transduction pathways therefore remain undefined.
Previous results in our laboratory had shown that the mitogenic activity and the uPA-inducing capacity of FGF2 are mediated by different signal transduction pathways in cultured endothelial cells (Presta et al., 1989). We observed also that various FGF2 mutants devoid of the capacity to upregulate uPA production still retain receptor-binding activity and full mitogenic capacity (Isacchi et al., 1991; Presta et al., 1992, 1993). Accordingly, we demonstrated that the interaction of FGF2 with FGFR1 is quantitatively and qualitatively different in mediating mitogenicity and uPA upregulation in endothelial cells. Nevertheless, TK activity of FGFR1 is essential for both cellular responses (Rusnati et al., 1996).
uPA gene expression induced by FGF2 in 3T3 fibroblasts requires the activation of the Ras/Raf-1/MEK/ERK2/JunD pathway (Besser et al., 1995). Also, uPA upregulation is impaired in TK-deficient FGFR1-Y653/654F L6 transfectants (Roghani et al., 1996). In the present work, we investigated the role played by different FGFR1 autophosphorylation sites in mediating FGF2-induced uPA upregulation. To this purpose, L6 myoblasts transfected with various FGFR1 mutants were evaluated for the capacity to upregulate uPA production after stimulation by FGF2. The results demonstrate that the uPA-inducing activity of FGF2 depends on tyrosine phosphorylation events in FGFR1 that include residues Y463 and Y730. Simultaneous mutagenesis of these residues abolishes FGF2-mediated uPA upregulation without affecting the mitogenic activity of the growth factor. Our findings shed a new light on the mechanism(s) responsible for the dissociation of the mitogenic activity of FGF2 from its uPA-inducing capacity.
MATERIALS AND METHODS
Site-directed Mutagenesis and Generation of Stable Cell Lines
Site-directed mutagenesis of FGFR1 cDNA was performed according to the protocol of the manufacturer (Amersham, Arlington Heights, IL). Oligonucleotide sequences and generation of single and multiple mutations on FGFR1 have already been described (Mohammadi et al., 1996a). FGFR1-Y463/730F and FGFR1-Y583/585/730F mutants were generated by replacing the wild-type BstEII fragment in FGFR1-Y730F with the corresponding mutagenized fragments from FGFR1-Y463F and FGFR1-Y583/585F mutants, respectively. All of the cDNAs were subcloned in pMJ30 and used together with pSV2neo to transfect L6 myoblasts by the calcium phosphate precipitation method (Chen and Okayama, 1987). G418-resistant colonies were screened for expression of FGFR1 by binding assay with 125I-labeled FGF2 as a specific probe. Cell lines expressing similar levels of FGF receptors were used for further analysis. L6 parental and transfected cells were grown in DMEM supplemented with 10% FCS.
[3H]thymidine Incorporation and Cell Proliferation Assays
Cells were seeded in 48-well plates at 20,000 cells/cm2 in DMEM plus 10% FCS. After 48 h of serum starvation in DMEM plus 0.1% FCS, either FGF2 (30–100 ng/ml) or 10% FCS as a control was added to the wells. Twenty hours later, cells were incubated with [3H]thymidine (1 μCi/ml), and after an additional period of 6 h samples were directly precipitated in 5% trichloroacetic acid and incubated at 4°C for 1 h. Then cells were lysed in 0.5 M sodium hydroxide, and after neutralization with 1:10 (vol/vol) of 5 M hydrochloric acid and addition of liquid scintillator, they were analyzed for the amount of [3H]thymidine incorporated in a counting device (Beckman, Fullerton, CA). For the cell proliferation assay, cells were seeded in 96-well plates at 20,000 cells/cm2 in DMEM plus 10% FCS. After 24 h of serum starvation in DMEM plus 0.1% FCS, cells were stimulated for 2 d with increasing concentrations of FGF2. Then cell number was estimated by a colorimetric assay after staining with crystal violet. Briefly, cells were fixed for 20 min at room temperature with 2.5% glutaraldehyde, stained with 0.1% crystal violet in 20% methanol, and solubilized with 10% acetic acid. Then wells were read at 595 nm in a microplate reader against a calibration curve set up with a known number of cells.
Plasminogen Activator and Northern Blot Assays
To evaluate the uPA-inducing activity of FGF2, L6 transfectants were seeded at 50,000 cells/cm2 in DMEM containing 10% FCS. Twenty-four hours later, medium was replaced with DMEM plus 0.5% FCS in the absence or presence of FGF2. The day after, cell-associated uPA activity was measured using the plasmin chromogenic substrate H-d-norleucyl-hexahydrotyrosil-lysine-p-nitroanilide-acetate (American Diagnostica, Greenwich, CT) as described (Presta et al., 1989). Steady-state levels of uPA mRNA were evaluated by Northern blot analysis of total RNA (20 μg/sample) according to standard procedures (Chomczynski and Sacchi, 1987; Sambrook et al., 1989) using a murine uPA probe (kindly provided by P. Mignatti, New York University).
Immunoprecipitation and Immunoblot Analysis
Cells expressing wild-type or mutant receptors were grown to 80–90% confluency in 10-cm-diameter dishes, treated with FGF2 without changing the medium, and maintained at 37°C for the indicated periods of time. At the end of the incubation, cells were washed briefly with ice-cold PBS and lysed in 1 ml of lysis buffer (20 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mM phenylmethylsulfonylfluoride, 2 mM sodium orthovanadate, 1 mM sodium pyrophosphate). Cell lysates were centrifuged for 10 min at 15,000 × g, and protein concentration was determined. For immunoprecipitation, 1-mg aliquots of each sample were incubated overnight with the appropriate antibody and protein A-Sepharose. Immunocomplexes were washed three times with HNTG (20 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton X-100) and then resuspended in 4× SDS-PAGE sample buffer and boiled for 5 min. For ERK2 gel-shift analysis, 50-μg aliquots of each sample were directly boiled for 5 min in 4× SDS-PAGE sample buffer. All of the samples were then subjected to SDS-PAGE under reducing conditions. The gel was prepared according to Sambrook et al. (1989) except for ERK2 shift analysis in which the pH of the separating gel was changed from 8.8 to 8.3 (Besser et al., 1995). Proteins were transferred electrophoretically onto a PolyScreen polyvinylidene difluoride transfer membrane (New England Nuclear Life Science, Boston, MA) and immunoblotted with different antibodies. Membranes were incubated sequentially with horseradish peroxidase-conjugated secondary antibodies and with Renaissance chemiluminescence reagents (New England Nuclear Life Science) according to manufacturer’s instructions and then exposed to Reflection films (New England Nuclear Life Science). The following antibodies were used in this study: anti-FGFR1 (C-15), anti–Jun-D, and antiphosphorylated ERK1/2 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-Shc antibody was a gift from P.G. Pelicci (European Institute of Oncology, Milan, Italy); anti-ERK2 antibody was a gift of Y. Nagamine (Friedrick Miescher Institute, Basel, Switzerland); and antiphosphotyrosine antibody (4G10) was from Upstate Biotechnology (Lake Placid, NY).
ERK2 In Vitro Kinase Assay
Cells expressing wild-type or mutant receptors were plated in a 60-mm dish (1.4 × 106 cells/dish), and 6 h later medium was changed to DMEM containing 1 mg/ml bovine serum albumin and 1 mg/ml transferrin. The day after, cells were treated for 20 min with 10 ng/ml FGF2 at 37°C and then lysed in ERK lysis buffer (50 mM β-Na glycerophosphate, 1.5 mM EGTA, pH 8.5, 2 mM sodium orthovanadate, 1 μM dithiothreitol, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μM benzamidine, 1% Nonidet P-40). Lysates were centrifuged for 10 min at 15,000 × g, and protein concentration was determined. Cell lysates (200 μg) were incubated with anti-ERK2 antiserum and protein A-Sepharose for 2 h at 4°C. The immunoprecipitates were subjected to an in vitro kinase assay. The reaction was performed for 30 min at 37°C in 30 mM Tris, pH 8.0, 20 mM MgCl2, 2 mM MnCl2, 10 μM ATP, 15 μg per assay of myelin basic protein, and 0.1 μCi/sample of [32P]-labeled γATP. Samples were then boiled for 5 min in 4× SDS-PAGE sample buffer and subjected to SDS-PAGE (15%). The gel was dried and exposed to Reflection film.
RESULTS
ERK1/2 Activation and FGF2-mediated uPA Upregulation in FGFR1-transfected L6 Cells
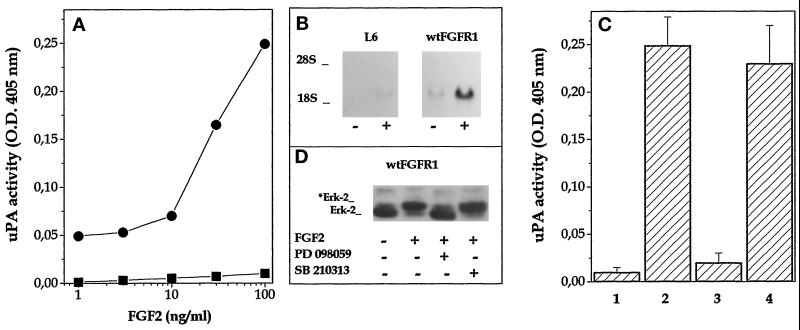
In agreement with previous observations (Roghani et al., 1996), FGF2 induces a dose-dependent increase of cell-associated uPA activity in L6 cells transfected with wild-type FGFR1 cDNA (wt-FGFR1 cells) with an ED50 equal to 20 ng/ml (Figure 1A). Accordingly, an increase in steady-state levels of uPA mRNA was observed in wt-FGFR1 transfectants after 24 h of treatment with 10 ng/ml FGF2 (Figure 1B). uPA upregulation by FGF2 was prevented in L6 transfectants incubated with 100 μM of the TK inhibitor 23 tyrphostin. On the contrary, the uPA-inducing activity exerted by FGF2 in wt-FGFR1 cells was not affected by downregulation of PKC induced by a 16-h pretreatment with 500 ng/ml 12-O-tetradecanoyl phorbol 13-acetate or by incubation with 50 μM of the protein kinase inhibitor N-[2-(methylamino)ethyl]-5-isoquinoline-sulfonamide H-8 (Ido et al., 1991) (our unpublished results).
Figure 1.
FGF2-mediated uPA upregulation in FGFR1-transfected L6 cells requires ERK2 activation. (A) Parental (▪) and wt-FGFR1-transfected cells (•) were stimulated with different amounts of FGF2. Cell-associated uPA activity was evaluated after 24 h. For both cell lines, uPA activity measured in nonstimulated cells (ranging from 0.1 to 0.2 OD at 405 nm) was subtracted from all the values. (B) Northern blot analysis was performed using a murine uPA probe on total RNA extracted from parallel cultures incubated overnight with 10 ng/ml FGF2. Uniform loading of samples was judged by methylene blue staining of the filter. (C) wt-FGFR1-transfected cells were preincubated for 30 min at 37°C with no addition (1, 2) or with 100 μM PD 098059 (3) or 10 μM SB 210313 (4) before addition of 100 ng/ml FGF2 (2–4). After 24 h, cell-associated uPA activity was measured. uPA activity in control nontransfected L6 cells incubated under the same experimental conditions was subtracted from all the values. (D) Parallel cultures were lysed 20 min after FGF2 addition, and cell lysates were probed with anti-ERK2 antibody in a Western blot. *Erk-2 indicates the phosphorylated protein.
The requirement for the activation of the Ras/Raf/MEK/ERK2 pathway had been demonstrated for FGF2-mediated uPA upregulation in 3T3 fibroblasts (Besser et al., 1995). To assess whether this holds true also for wt-FGFR1 transfectants, cells were incubated with FGF2 in the absence or presence of the MEK1 inhibitor PD 098059 (Alessi et al., 1995). PD 098059 treatment prevents both ERK2 phosphorylation and uPA upregulation in L6 cells (Figure 1, C and D). Specificity of the inhibition was demonstrated by the lack of effect of the p38 MAPK inhibitor SB 210313 (Cuenda et al., 1995). In conclusion, FGF2-mediated uPA induction in L6 transfectants depends on TK-FGFR activity, requires ERK2 phosphorylation, and is not mediated by PKC activation.
uPA Upregulation Is Abolished by Mutagenesis of Distinct FGFR1 Autophosphorylation Sites
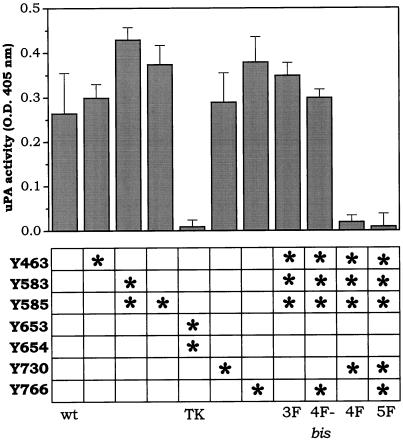
To assess the role of the different tyrosine residues of FGFR1 in uPA upregulation, L6 cells were independently transfected with different receptor mutants in which one or more of the seven identified autophosphorylation sites were mutagenized to phenylalanine (Mohammadi et al., 1996a). After G418 selection and 125I-FGF2 binding analysis, two independent clones for each transfection, bearing 40,000–100,000 receptors/cell, were incubated for 24 h with 100 ng/ml FGF2. At the end of the incubation, cell monolayers were lysed, and cell-associated uPA activity was measured. The results are summarized in Figure 2. No uPA upregulation was observed in cells transfected with the TK-defective mutant FGFR1-Y653/654F, whereas single point mutations in autophosphorylation sites Y463, Y585, Y730, or Y766 did not affect the uPA-inducing capacity of FGF2. FGF2 was able to induce a significant increase of cell-associated uPA activity also in cells transfected with FGFR1-Y583/585F mutant, FGFR1-Y463/583/585F mutant (FGFR1–3F cells), or FGFR1-Y463/583/585/766F mutant (FGFR1–4Fbis cells); however, uPA upregulation by FGF2 was hampered in cells transfected with FGFR1-Y463/583/585/730F mutant (FGFR1–4F cells) or with FGFR1-Y463/583/585/730/766F mutant (FGFR1–5F cells).
Figure 2.
uPA-inducing capacity of different FGFR1 mutants. L6 myoblasts were independently transfected with wt-FGFR1 or with the FGFR1 mutants described in the bottom panel. In each column, an asterisk or asterisks mark the tyrosine autophosphorylation site(s) mutagenized to phenylalanine in the different mutants. Each cell line was stimulated with 100 ng/ml FGF2, and 24 h later cell-associated uPA activity was evaluated (top panel). For each cell line, uPA activity measured in the corresponding nonstimulated cells (ranging from 0.1 to 0.2 OD at 405 nm) was subtracted. Results represent the mean ± SD of two independent experiments in triplicate. For each mutant, similar results were obtained for two independent clones.
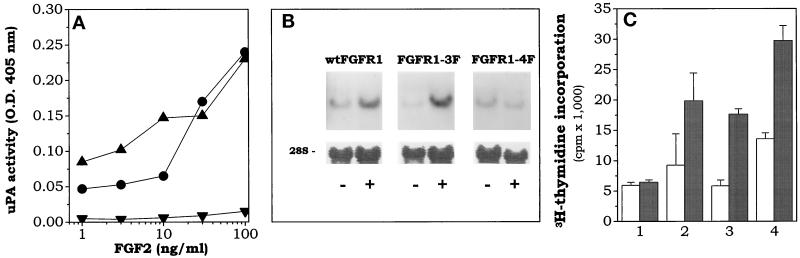
On this basis, FGFR1–3F and FGFR1–4F clones, which differ for the absence or presence of the Y730F mutation, were analyzed further. FGF-1 induces [3H]thymidine incorporation in L6 myoblasts transfected with wt-FGFR1 or FGFR1–4F and FGFR1–5F mutants (Mohammadi et al., 1996a). When [3H]thymidine incorporation and cell-associated uPA activity were measured in parallel cultures of wt-FGFR1, FGFR1–3F, and FGFR1–4F cells exposed to FGF2, the results shown in Figure 3 were obtained. As anticipated, FGF2 stimulates DNA synthesis in all cell lines, but only wt-FGFR1 and FGFR1–3F cells respond to the growth factor with an increase in uPA activity. This latter observation was confirmed by Northern blot analysis of total RNA isolated from L6 transfectants that demonstrated a significant increase in the steady-state levels of uPA mRNA in wt-FGFR1 and FGFR1–3F cells incubated with 10 ng/ml FGF2 for 24 h, but not in FGFR1–4F transfectants (Figure 3B). Interestingly, we did not observe any significant differences in the rate of receptor downregulation and FGF2 internalization among wt-FGFR1, FGFR1–3F, and FGFR1–4F transfectants (Dell’Era, unpublished observations). Finally, control experiments demonstrated that FGFR1–4Fbis and FGFR1–5F cells behave similarly to FGFR1–3F and FGFR1–4F transfectants, respectively (our unpublished results).
Figure 3.
Dissociation of uPA upregulation from mitogenicity in L6 transfectants. (A) wt-FGFR1 (•), FGFR1–3F (▴), and FGFR1–4F (▾) cell lines were stimulated with FGF2. Cell-associated uPA activity was evaluated after 24 h. For each cell line, uPA activity measured in corresponding nonstimulated cells (ranging from 0.1 to 0.2 OD at 405 nm) was subtracted from all the values. (B) Parallel cultures were incubated overnight in the absence or presence of 10 ng/ml FGF2. Then, total RNA was extracted and analyzed by Northern blot using a murine uPA probe. Uniform loading of the gel was judged by methylene blue staining of the filter (28S). (C) Parental L6 myoblasts (1), wt-FGFR1 (2), FGFR1–3F (3), and FGFR1–4F (4) cell lines were seeded in 48-well plates, starved for 48 h in the presence of 0.1% FCS, and treated with 30 ng/ml FGF2 (gray bar) or vehicle (open bar) for 20 h. Then [3H]thymidine was added, and 6 h later samples were collected and [3H]thymidine incorporation was evaluated. Data are the mean ± SD of three determinations.
Signal Transduction Pathway Analysis of FGFR1–3F and FGFR1–4F Mutants
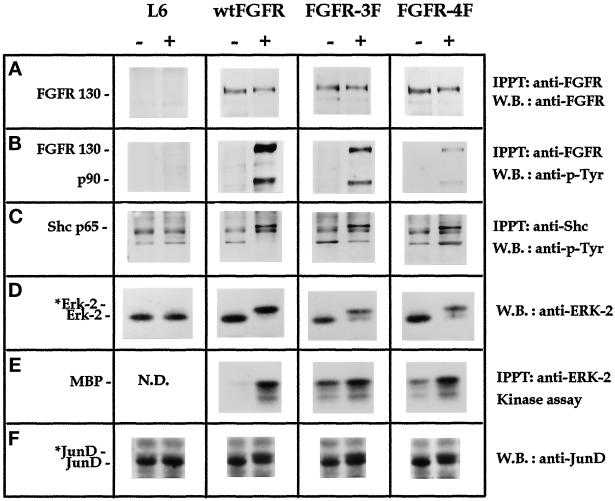
When wt-FGFR1, FGFR1–3F, and FGFR1–4F cells were exposed to FGF2, lysed, immunoprecipitated with anti-FGFR1 antibody, and immunoblotted with antiphosphotyrosine antibodies, mutated receptors underwent tyrosine autophosphorylation, albeit to a lesser extent than the wild-type receptor, because of the neutralization of three or four of the autophosphorylation sites (Figure 4B). Also, tyrosine phosphorylation of a receptor-associated 90-kDa protein was observed in all of the cell lines (Figure 4B). This protein does not cross-react with antibodies directed against Stat3 (Faris et al., 1996), 80K-H (Goh et al., 1996), or FRS2 (Kouhara et al., 1997) (our unpublished results). Reprobing the same membrane with anti-FGFR1 antibody confirmed the presence of similar amounts of the receptor in all the samples (Figure 4A). In agreement with previous observations (Mohammadi et al., 1996a), we found that both FGFR1–3F and FGFR1–4F mutants retain the capacity to tyrosine-phosphorylate Shc (Figure 4C).
Figure 4.
Signal transduction analysis of FGFR1 mutants. Parental L6 myoblasts, wt-FGFR1, FGFR1–3F, and FGFR1–4F cells were nonstimulated (−) or treated (+) either for 5 min (A, B, and C) or 20 min (D, E, and F) with 100 ng/ml FGF2. (A) Anti-FGFR1 immunoprecipitates were analyzed for FGFR1 expression using anti-FGFR1 antibodies. (B) Anti-FGFR1 immunoprecipitates were analyzed for tyrosine phosphorylation using antiphosphotyrosine (P-Tyr) antibodies. (C) Anti-Shc immunoprecipitates were analyzed for tyrosine phosphorylation. (D) Cell lysates (50 μg of total protein) were analyzed with anti-ERK2 antibodies. *Erk-2 indicates the phosphorylated protein. (E) Anti-ERK2 immunoprecipitates were analyzed for kinase activity using myelin basic protein (MBP) as substrate. (F) Cell lysates (50 μg of total protein) were analyzed with anti-JunD antibodies. *JunD indicates the phosphorylated protein. N.D., not determined.
Activation of the Ras/Raf-1/MEK/ERK2/JunD pathway is required for uPA upregulation by FGF2 (see above and Besser et al., 1995). On this basis, the capacity of receptor mutants to activate the last two steps of this pathway was investigated after 20 min of incubation of wt-FGFR1, FGFR1–3F, and FGFR1–4F cells with 10 ng/ml FGF2. Activation of ERK2 was evident in all of the cell lines as a mobility shift of the protein by immunoblotting with anti-ERK2 antibody (Figure 4D) and as an increased enzymatic activity in a kinase assay (Figure 4E). Control experiments confirmed that FGF2-activated FGFR1–4Fbis and FGFR1–5F also undergo autophosphorylation, associate with and phosphorylate the 90-kDa protein, and activate ERK2 (our unpublished results). Furthermore, FGF2 treatment modifies JunD protein in wt-FGFR1, FGFR1–3F, and FGFR1–4F cell lines (Figure 4F).
Simultaneous Mutagenesis of Residues Y463 and Y730 in FGFR1 Is Sufficient to Dissociate uPA Induction from Mitogenesis
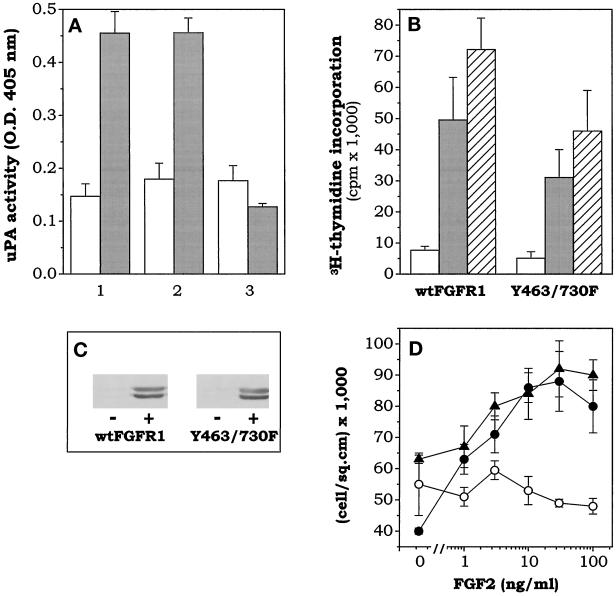
The above data point to a role for Y730 phosphorylation in uPA upregulation, although in a complex context attributable to the simultaneous mutation of residues Y463, Y583, and Y585; however, mutation of Y730 alone is not sufficient to abolish FGF2-mediated uPA induction in L6 transfectants (see Figure 2). On this basis, L6 cells were transfected with FGFR1 mutants bearing the mutation Y730F together with the single mutation Y463F or with the double mutation Y583/585F. As shown in Figure 5, FGF2 was unable to cause a significant increase of uPA activity in FGFR1-Y463/730F cells, whereas FGFR1-Y583/585/730F cells were fully responsive. Nevertheless, in agreement with the results obtained with FGFR1–4F and FGFR1–5F mutants, both FGFR1-Y463/730F and FGFR1-Y583/585/730F mutants were able to stimulate ERK1/2 phosphorylation and DNA synthesis in response to FGF2. Moreover, FGF2 exerted a dose-dependent mitogenic response in FGFR1-Y463/730F transfectants similar to that elicited in wt-FGFR1 cells (Figure 5D). In conclusion, phosphorylation of residues Y463 and Y730 is dispensable for mitogenic signaling but essential for uPA upregulation by FGF2 in FGFR1 transfectants.
Figure 5.
Residues Y463 and Y730 are implicated in uPA upregulation but not mitogenesis by FGF2. (A) wt-FGFR1 (1), FGFR1-Y583/585/730F (2), and FGFR1-Y463/730F (3) transfectants were assayed for cell-associated uPA activity after a 24-h incubation in the absence (open bar) or presence (gray bar) of 100 ng/ml FGF2. (B) wt-FGFR1 and FGFR1-Y463/730F transfectants were seeded in 48-well plates, starved for 48 h in the presence of 0.1% FCS, and then treated with no addition (open bar), 100 ng/ml FGF2 (gray bar), or 10% FCS (dashed bar) for an additional 20 h. Then, [3H]thymidine was added, and 6 h later samples were collected and [3H]thymidine incorporation was evaluated. (C) Parallel cultures were lysed 20 min after FGF2 addition, and cell lysates (30 μg of protein) were probed with antiphosphorylated ERK1/2 antibody in a Western blot. (D) Parental L6 cells (○), wt-FGFR1 (•), and FGFR1-Y463/730F (▴) transfectants were seeded in 96-well plates at 20,000 cells/cm2 in DMEM plus 10% FCS. After 24 h of serum starvation in DMEM plus 0.1% FCS, cells were stimulated with increasing concentrations of FGF2. Cells were counted after 2 d. For each mutant, similar results were obtained for two independent clones in all the assays.
DISCUSSION
FGF2 induces, among other responses, cell proliferation and uPA upregulation in different cell types, including endothelial cells (Presta et al., 1989; Rusnati et al., 1996), NIH 3T3 fibroblasts (Besser et al., 1995), and tumor cell lines (Coltrini et al., 1995). Previous results in our laboratory had shown that the mitogenic activity and the uPA-inducing capacity of FGF2 are mediated by distinct signal transduction pathways in cultured endothelial cells (Presta et al., 1989). We observed also that site-directed mutagenesis of FGF2 leads to growth factor mutants that retain full receptor binding and mitogenic activity but are devoid of the capacity to upregulate uPA production (Isacchi et al., 1991; Presta et al., 1992, 1993). Finally, we demonstrated that the interaction of FGF2 with endothelial cell FGFR1 that is able to elicit a mitogenic response is quantitatively and qualitatively different from uPA upregulation requirements (Rusnati et al., 1996).
The results presented here suggest that the observed dissociation between mitogenic activity and uPA-inducing capacity of FGF2 reflects different tyrosine autophosphorylation requirements of FGFR1. Indeed, FGFR1-Y463/730F, FGFR1–4F, and FGFR1–5F mutants retain the capacity to mediate the FGF2 mitogenic signal, although they have lost the ability to stimulate uPA upregulation. It must be pointed out that the inhibition of the uPA-inducing activity of these FGFR1 mutants does not appear to be the mere consequence of a nonspecific decrease of the overall tyrosine autophosphorylation capacity of FGFR1. Indeed, FGFR1 mutants bearing three (FGFR1-Y583/585/730F) or four (FGFR1–4Fbis) tyrosine substitutions are still able to transduce uPA upregulation in L6 cells, whereas the simultaneous mutation of residues Y463 and Y730 is sufficient to abolish this activity.
Residues Y653 and Y654 are essential for the TK activity of FGFR1, whereas Y766 has been shown to bind PLCγ (Mohammadi et al., 1991, 1996a). Previous observations showed that the simultaneous mutation Y653/654F hampers FGF2-mediated uPA induction in L6 transfectants, whereas mutation Y766F is ineffective (Roghani et al., 1996). Our results confirm and extend these observations by showing that also mutations Y585F and Y583/585F do not affect uPA upregulation by FGF2. Residues Y583 and Y585 belong to the kinase insert domain of FGFR1. A FGFR1 chimera in which the kinase insert of FGFR1 was replaced by the kinase insert of PDGF receptor (kindly provided by L. Claesson-Welsh, Uppsala, Sweden) retains the capacity to mediate uPA upregulation in stable Chinese hamster ovary transfectants (our unpublished observations), although no homology in amino acid sequences flanking autophosphorylated tyrosine residues exists between the kinase insert domains of the two receptors. These findings indicate that autophosphorylation sites Y583 and Y585, like Y766, are dispensable for uPA induction by FGF2.
Our data point instead to a role for residues Y463 and Y730 in uPA upregulation. This conclusion is inferred from the observation that among the mutants tested, only those characterized by the simultaneous mutation of both residues (FGFR1-Y463/730F, FGFR1–4F, and FGFR1–5F) were unable to upregulate uPA in response to FGF2. In contrast, the mutation of only one of them, as it occurs in FGFR1-Y463F, FGFR1-Y730F, FGFR1-Y583/585/730F, FGFR1–3F, and FGFR1–4Fbis, as well as FGFR1-Y463/766F (Dell’Era, unpublished observations), does not affect the uPA-inducing capacity of the receptor. Thus, our results suggest a redundancy in FGFR1 tyrosine residues implicated in uPA upregulation, autophosphorylation sites Y463 and Y730 being able to sustain uPA induction despite clear differences in their flanking amino acid sequences. Similar conclusions have been drawn for activation of STAT-5 nuclear import and DNA binding activity mediated by interleukin-2 receptor (Gaffen et al., 1996).
Y463 is located in the JM domain of FGFR1. Autophosphorylation sites in the JM region of insulin, PDGF, and nerve growth factor receptors have been shown to be involved in signal transduction (White et al., 1988; Mori et al., 1993; Obermeier et al., 1994). Recently, the JM domain of FGFR1 has been demonstrated to play an important role in FGF1-mediated neurite outgrowth in PC12 cells transfected with a FGFR1 JM domain-FGFR3 chimera (Lin et al., 1998). This receptor chimera also causes a sustained phosphorylation of ERK2 and FRS2 protein without affecting cell proliferation; however, this capacity appears to be independent of Y463 phosphorylation (Lin et al., 1998). Also, the yeast two-hybrid protein–protein interaction assay has demonstrated the capacity of FRS2 to interact with a JM region of FGFR1 lacking Y463 (Xu et al., 1998). Accordingly, no significant differences in FRS2 phosphorylation was observed in Grb2 immunoprecipitates from lysates of wt-FGFR1 and FGFR1-Y463F transfectants (Dell’Era, unpublished observations).
FGFR1, FGFR2, FGFR3, and FGFR4 are all able to mediate FGF2-induced uPA upregulation in stable Chinese hamster ovary transfectants (Rusnati et al., 1996). Accordingly, Y730 is conserved in the four FGFRs in all of the species cloned so far. This residue has a YMXM motif that resembles the consensus binding site for the SH2 domain of the regulatory subunit of PI 3-kinase; however, neither direct association of PI 3-kinase with FGFR nor its activation after ligand binding has been demonstrated (Wennstrom et al., 1992). It has been proposed that the Y730 counterpart in chicken FGFR (Y728) is a minor site for Shc binding (Ward et al., 1996). Nevertheless, FGF2 causes Shc phosphorylation in FGFR1–4F cells. Finally, crystallography of the kinase domain of FGFR1 has shown that Y730 is buried (Mohammadi et al., 1996b), suggesting that autophosphorylation of this site requires a conformational change of the intracellular portion of FGFR1 after ligand interaction. It is possible that the modality of ligand/receptor interaction may decide the exposure and consequent phosphorylation of Y730, possibly explaining the inability of various mitogenic FGF2 mutants to induce uPA upregulation (Isacchi et al., 1991; Presta et al., 1992, 1993).
The activation of the Ras/Raf/MEK/ERK2/JunD pathway is required for FGF2-mediated uPA upregulation in NIH 3T3 cells that constitutively express FGFR1 (Besser et al., 1995). Accordingly, we have observed that the MEK1 inhibitor PD 098059 (Alessi et al., 1995) prevents ERK2 phosphorylation and uPA upregulation in wt-FGFR1 transfectants, thus indicating that ERK2 activation is required for uPA induction also in L6 cells. The same pathway is activated by FGF2 in FGFR1-Y463/730F, FGFR1–4F, and FGFR1–5F cells; nevertheless, uPA is not induced. This observation suggests that ERK2 activation is necessary but not sufficient for FGF2-induced uPA upregulation and that the activation of a second as yet unidentified signal transduction pathway might be required. This hypothesis is in keeping with the incapacity of FGF2 to induce uPA production, as well as fibrin gel invasion, in murine aortic endothelial cell cultures, although they respond to FGF2 with a rapid activation of ERK2 and cell proliferation (Bastaki et al., 1997). Also, we have found that various mitogenic FGF2 mutants, characterized by the incapacity to induce uPA upregulation in endothelial cells (Isacchi et al., 1991; Presta et al., 1992, 1993), are still able to stimulate ERK1/2 phosphorylation in a manner similar to the wild-type growth factor (Presta and Bastaki, unpublished observations).
In conclusion, we have demonstrated the possibility of dissociating the mitogenic activity of FGF2 from its uPA-inducing capacity by mutational analysis of the seven characterized autophosphorylation sites in FGFR1. Residues Y463 and Y730, dispensable for cell proliferation, are implicated in uPA upregulation by FGF2. To our knowledge, our findings represent the first demonstration for a role of these tyrosine residues in FGFR1 activity.
ACKNOWLEDGMENTS
We are indebted to M.L. Massardi for her skillful technical assistance, and Y. Nagamine and A. Gualandris for helpful discussion. Antibodies raised against 80K-H and FRS2 were generous gifts from Dr. K.C. Goh and Dr. I. Lax, respectively. This work was supported by grants from Ministero Università Ricerca Scientifica e Tecnologica (Cofinanziamento 1997 “Infiammazione: biologia e clinica” and Quota 60%), Associazione Italiana per la Ricerca sul Cancro, Consiglio Nazionale delle Ricerche (Target Project on Biotechnology 97.01186.PF49), and Istituto Superiore di Sanità (AIDS Project) to M.P.
Abbreviations used:
- ERK
extracellular signal-regulated kinase
- FGF2
basic fibroblast growth factor
- FGFR
FGF receptor
- JM
juxtamembrane
- MEK
ERK kinase (also referred to as MAPK kinase)
- TK
tyrosine kinase
- uPA
urokinase-type plasminogen activator
REFERENCES
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- Bastaki M, Nelli EE, Dell’Era P, Rusnati M, Molinari-Tosatti MP, Parolini S, Auerbach R, Ruco LP, Possati L, Presta M. Basic fibroblast growth factor-induced angiogenic phenotype in mouse endothelium. A study of aortic and microvascular endothelial cell lines. Arterioscler Thromb Vasc Biol. 1997;17:454–464. doi: 10.1161/01.atv.17.3.454. [DOI] [PubMed] [Google Scholar]
- Besser D, Presta M, Nagamine Y. Elucidation of a signalling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. [PubMed] [Google Scholar]
- Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- Blasi F, Verde P. Urokinase-dependent cell surface proteolysis and cancer. Semin Cancer Biol. 1990;1:117–126. [PubMed] [Google Scholar]
- Bork P, Margolis B. A phosphotyrosine interaction domain. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–11627. [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coltrini D, Gualandris A, Nelli EE, Parolini S, Molinari-Tosatti MP, Quarto N, Ziche M, Giavazzi R, Presta M. Growth advantage and vascularization induced by basic fibroblast growth factor overexpression in endometrial HEC-1-B cells: an export-dependent mechanism of action. Cancer Res. 1995;55:4729–4738. [PubMed] [Google Scholar]
- Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res. 1985;44:139–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- Dionne CA, Crumley G, Bellot F, Kaplow JM, Searfoss G, Ruta M, Burgess WH, Jaye M, Schlessinger J. Cloning and expression of two distinct high-affinity receptors cross-reacting with acidic and basic fibroblast growth factors. EMBO J. 1990;9:2685–2692. doi: 10.1002/j.1460-2075.1990.tb07454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl WJ, Johnson DE, Williams LT. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- Faris M, Ensoli B, Stahl N, Yancopoulos G, Nguyen A, Wang S, Nel AE. Differential activation of the extracellular signal-regulated kinase, jun kinase and janus kinase-stat pathways by oncostatin M and basic fibroblast growth factor in AIDS-derived Kaposi’s sarcoma cells. AIDS. 1996;10:369–378. doi: 10.1097/00002030-199604000-00004. [DOI] [PubMed] [Google Scholar]
- Gaffen SL, Lai SY, Ha M, Liu X, Hennighausen L, Greene WC, Goldsmith MA. Distinct tyrosine residues within the interleukin-2 receptor β chain drive signal transduction specificity, redundancy, and diversity. J Biol Chem. 1996;271:21381–21390. doi: 10.1074/jbc.271.35.21381. [DOI] [PubMed] [Google Scholar]
- Goh KC, Lim YP, Ong SH, Siak CB, Cao X, Tan YH, Guy GR. Identification of p90, a prominent tyrosine-phosphorylated protein in fibroblast growth factor-stimulated cells, as 80K-H. J Biol Chem. 1996;271:5832–5838. doi: 10.1074/jbc.271.10.5832. [DOI] [PubMed] [Google Scholar]
- Huang J, Mohammadi M, Rodrigues GA, Schlessinger J. Reduced activation of RAF-1 and MAP kinase by a fibroblast growth factor receptor mutant deficient in stimulation of phosphatidylinositol hydrolysis. J Biol Chem. 1995;270:5065–5072. doi: 10.1074/jbc.270.10.5065. [DOI] [PubMed] [Google Scholar]
- Ido M, Nagao Y, Higashigawa M, Shibata T, Taniguchi KX, Hamazaki M, Sakurai M. Differential growth inhibition of isoquinolinesulfonamides H-8 and H-7 towards multidrug-resistant P388 murine leukaemia cells. Br J Cancer. 1991;64:1103–1107. doi: 10.1038/bjc.1991.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isacchi A, Statuto M, Chiesa R, Bergonzoni L, Rusnati M, Sarmientos P, Ragnotti G, Presta M. A six-amino acid deletion in basic fibroblast growth factor dissociates its mitogenic activity from its plasminogen activator-inducing capacity. Proc Natl Acad Sci USA. 1991;88:2628–2632. doi: 10.1073/pnas.88.7.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Lu J, Chen H, Werner S, Williams LT. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. [Review] Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- Kavanaugh WM, Turck CW, Williams LT. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- Kavanaugh WM, Williams LT. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- Kwaan HC. The plasminogen-plasmin system in malignancy. Cancer Metastasis Rev. 1992;11:291–311. doi: 10.1007/BF01307184. [DOI] [PubMed] [Google Scholar]
- Keegan K, Johnson DE, Williams LT, Hayman MJ. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc Natl Acad Sci USA. 1991;88:1095–1099. doi: 10.1073/pnas.88.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Lee PL, Johnson DE, Cousens LS, Fried VA, Williams LT. Purification and complementary DNA cloning of a receptor for basic fibroblast growth factor. Science. 1989;245:57–60. doi: 10.1126/science.2544996. [DOI] [PubMed] [Google Scholar]
- Lin HY, Xu J, Ischenko I, Ornitz DM, Halegoua S, Hayman MJ. Identification of the cytoplasmic regions of fibroblast growth factor (FGF) receptor 1 which play important roles in induction of neurite outgrowth in PC12 cells by FGF-1. Mol Cell Biol. 1998;18:3762–3770. doi: 10.1128/mcb.18.7.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis B. Proteins with SH2 domains: transducers in the tyrosine kinase signaling pathway. Cell Growth Differ. 1992;3:73–80. [PubMed] [Google Scholar]
- Mohammadi M, Dionne CA, Li W, Li N, Spivak T, Honegger AM, Jaye M, Schlessinger J. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358:681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996a;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Honegger AM, Rotin D, Fischer R, Bellot F, Li W, Dionne CA, Jaye M, Rubinstein M, Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (flg) is a binding site for the SH2 domain of phospholipase C-γ1. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996b;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- Mori S, Ronnstrand L, Yokote K, Engstrom A, Courtneidge SA, Claesson-Welsh L, Heldin CH. Identification of two juxtamembrane autophosphorylation sites in the PFGD beta-receptor; involvement in the interaction with Src family tyrosine kinases. EMBO J. 1993;12:2257–2264. doi: 10.1002/j.1460-2075.1993.tb05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeier A, Bradshaw RA, Seedorf K, Choidas A, Schlessinger J, Ullrich A. Neuronal differentiation signals are controlled by nerve growth factor receptor/Trk binding sites for SHC and PLC gamma. EMBO J. 1994;13:1585–1590. doi: 10.1002/j.1460-2075.1994.tb06421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Partanen J, Mäkelä TP, Eerola E, Korhonen J, Hirvonen H, Claesson-Welsh L, Alitalo K. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991;10:1347–1354. doi: 10.1002/j.1460-2075.1991.tb07654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signalling networks. Nature (Lond) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Pawson T, Schlessinger J. SH2 and SH3 domains. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- Peters KG, Marie J, Wilson E, Ives HE, Escobedo J, Del Rosario M, Mirda D, Williams LT. Point mutation of an FGF receptor abolishes phosphatidylinositol turnover and Ca2+ flux but not mitogenesis. Nature. 1992;358:678–681. doi: 10.1038/358678a0. [DOI] [PubMed] [Google Scholar]
- Presta M, Gualandris A, Urbinati C, Rusnati M, Coltrini D, Isacchi A, Caccia P, Bergonzoni L. Subcellular localization and biological activity of M(r) 18,000 basic fibroblast growth factor: site-directed mutagenesis of a putative nuclear translocation sequence. Growth Factors. 1993;9:269–278. doi: 10.3109/08977199308991587. [DOI] [PubMed] [Google Scholar]
- Presta M, Maier JA, Ragnotti G. The mitogenic signaling pathway but not the plasminogen activator-inducing pathway of basic fibroblast growth factor is mediated through protein kinase C in fetal bovine aortic endothelial cells. J Cell Biol. 1989;109:1877–1884. doi: 10.1083/jcb.109.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta M, Maier JA, Rusnati M, Moscatelli D, Ragnotti G. Modulation of plasminogen activator activity in human endometrial adenocarcinoma cells by basic fibroblast growth factor and transforming growth factor beta. Cancer Res. 1988;48:6384–6389. [PubMed] [Google Scholar]
- Presta M, Statuto M, Isacchi A, Caccia P, Pozzi A, Gualandris A, Rusnati M, Bergonzoni L, Sarmientos P. Structure-function relationship of basic fibroblast growth factor: site-directed mutagenesis of a putative heparin-binding and receptor-binding region. Biochem Biophys Res Commun. 1992;185:1098–1107. doi: 10.1016/0006-291x(92)91739-d. [DOI] [PubMed] [Google Scholar]
- Roghani M, Mohammadi M, Schlessinger J, Moscatelli D. Induction of urokinase-type plasminogen activator by fibroblast growth factor (FGF)-2 is dependent on expression of FGF receptors and does not require activation of phospholipase Cγ1. J Biol Chem. 1996;271:31154–31159. doi: 10.1074/jbc.271.49.31154. [DOI] [PubMed] [Google Scholar]
- Rusnati M, Dell’Era P, Urbinati C, Tanghetti E, Massardi ML, Nagamine Y, Monti E, Presta M. A distinct basic fibroblast growth factor (FGF-2)/FGF receptor interaction distinguishes urokinase-type plasminogen activator induction from mitogenicity in endothelial cells. Mol Biol Cell. 1996;7:369–381. doi: 10.1091/mbc.7.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Spivak-Kroizman T, Mohammadi M, Hu P, Jaye M, Schlessinger J, Lax I. Point mutation in the fibroblast growth factor receptor eliminates phosphatidylinositol hydrolysis without affecting neuronal differentiation of PC12 cells. J Biol Chem. 1994;269:14419–14423. [PubMed] [Google Scholar]
- Tienari J, Alanko T, Lehtonen E, Saksela O. The expression and localization of urokinase-type plasminogen activator and its type 1 inhibitor are regulated by retinoic acid and fibroblast growth factor in human teratocarcinoma cells. Cell Regul. 1991;2:285–297. doi: 10.1091/mbc.2.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:205–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Wang J-K, Xu H, Li H-C, Goldfarb M. Broadly expressed SNT-like proteins link FGF receptor stimulation to activators of Ras. Oncogene. 1996;13:721–729. [PubMed] [Google Scholar]
- Ward CW, Gough KH, Rashke M, Wan SS, Tribbick G, Wang J. Systematic mapping of potential binding sites for Shc and Grb2 SH2 domains on insulin receptor substrate-1 and the receptors for insulin, epidermal growth factor, platelet-derived growth factor, and fibroblast growth factor. J Biol Chem. 1996;271:5603–5609. doi: 10.1074/jbc.271.10.5603. [DOI] [PubMed] [Google Scholar]
- Wennstrom S, Landgren E, Blume-Jensen P, Claesson-Welsh L. The platelet-derived growth factor beta-receptor kinase insert confers specific signaling properties to a chimeric fibroblast growth factor receptor. J Biol Chem. 1992;267:13749–13756. [PubMed] [Google Scholar]
- White MF, Livingston JN, Backer JM, Lauris V, Dull TJ, Ullrich A, Kahn CR. Mutation of the insulin receptor at tyrosine 960 inhibits signal transmission but does not affect its tyrosine kinase activity. Cell. 1988;54:641–649. doi: 10.1016/s0092-8674(88)80008-4. [DOI] [PubMed] [Google Scholar]
- Xu H, Lee KW, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]