Abstract
Gp180, a duck protein that was proposed to be a cell surface receptor for duck hepatitis B virus, is the homolog of metallocarboxypeptidase D, a mammalian protein thought to function in the trans-Golgi network (TGN) in the processing of proteins that transit the secretory pathway. Both gp180 and mammalian metallocarboxypeptidase D are type I integral membrane proteins that contain a 58-residue cytosolic C-terminal tail that is highly conserved between duck and rat. To investigate the regions of the gp180 tail involved with TGN retention and intracellular trafficking, gp180 and various deletion and point mutations were expressed in the AtT-20 mouse pituitary corticotroph cell line. Full length gp180 is enriched in the TGN and also cycles to the cell surface. Truncation of the C-terminal 56 residues of the cytosolic tail eliminates the enrichment in the TGN and the retrieval from the cell surface. Truncation of 12–43 residues of the tail reduced retention in the TGN and greatly accelerated the turnover of the protein. In contrast, deletion of the C-terminal 45 residues, which truncates a potential YxxL-like sequence (FxxL), reduced the protein turnover and caused accumulation of the protein on the cell surface. A point mutation of the FxxL sequence to AxxL slowed internalization, showing that this element is important for retrieval from the cell surface. Mutation of a pair of casein kinase II sites within an acidic cluster showed that they are also important for trafficking. The present study demonstrates that multiple sequence elements within the cytoplasmic tail of gp180 participate in TGN localization.
INTRODUCTION
Duck hepatitis B virus is a member of the hepadnavirus group that infects liver cells, causing acute and chronic hepatitis (Ganem and Varmus, 1987). A candidate receptor for duck hepatitis B virus was previously described by its ability to bind to viral particles (Kuroki et al., 1994, 1995). This 180-kDa protein, named gp180, was shown to bind to the PreS envelope protein of the virus (Kuroki et al., 1994). The gp180-binding region within PreS overlaps key areas that have been established as important for viral infectivity (Kuroki et al., 1994). Cloning of the cDNA for gp180 revealed that it is a member of the metallocarboxypeptidase gene family with approximately 50% amino acid identity to carboxypeptidase E (CPE), a neuropeptide-processing enzyme (Kuroki et al., 1995).
CPE was originally identified as an enkephalin-producing carboxypeptidase in secretory vesicles isolated from various neuroendocrine tissues (Fricker, 1988, 1991). CPE removes the basic amino acids that remain on the C terminus of peptide precursors after endopeptidase processing (Fricker, 1988, 1991). For many years, CPE was thought to be the only carboxypeptidase involved in the production of numerous neuroendocrine peptides; however, the recent finding that peptide processing is substantially reduced, but not eliminated, in mice that lack CPE activity attributable to a point mutation (Cpefat/Cpefat mice) suggests that another carboxypeptidase is able to partially compensate for CPE (Naggert et al., 1995). A search for additional CPE-like enzymes turned up a 180-kDa enzyme named metallocarboxypeptidase D (CPD) (Song and Fricker, 1995).
Partial amino acid sequence analysis of bovine and rat CPD matched the N terminus of gp180 that was deduced from the cDNA clone (Kuroki et al., 1995; Song and Fricker, 1995, 1996). Furthermore, both CPD and gp180 have similar tissue distributions (Kuroki et al., 1995; Song and Fricker, 1996; Xin et al., 1997) and enzymatic properties (Song and Fricker, 1995; Eng et al., 1998). Finally, cloning and sequence analysis of cDNA for rat and human CPD revealed this enzyme to be a homolog of the duck gp180 (Tan et al., 1997; Xin et al., 1997). Both mammalian CPD and gp180 have three metallocarboxypeptidase domains followed by a transmembrane domain and a cytosolic tail. Antisera to various domains of CPD were used to confirm that the C-terminal tail of the protein is cytosolic (Varlamov and Fricker, 1998). In addition to the avian and mammalian proteins, the Silver gene product of Drosophila presumably also represents a homolog, although it appears to contain only two carboxypeptidase domains and no transmembrane domain (Settle et al., 1995). The overall amino acid identity between duck gp180 and rat CPD is 75%. The highest homology between the two proteins is the cytosolic tail; of the 58 amino acids within this region, there is only one conservative change between the duck and rat/human sequences (Figure 1). This high degree of conservation implies that this region performs an essential function.
Figure 1.
Sequences of the cytoplasmic tail of human/rat CPD, duck gp180, and gp180 mutants. Numbers correspond to position from the C terminus. The YxxL-like sequence (FHRL) and the CKII sites are indicated in bold type. Point mutations are indicated by asterisks.
The cytosolic regions of many integral membrane proteins are involved in the compartmentalization and trafficking of the proteins (Sandoval and Bakke, 1994; Marks et al., 1997). For example, the C-terminal cytosolic tails of resident TGN proteins such as TGN38, furin (an endopeptidase), and PAM (a neuropeptide-processing enzyme) are important for their TGN localization and their trafficking to and from the cell surface (Luzio et al., 1990; Tausk et al., 1992; Bos et al., 1993; Humphrey et al., 1993; Wong and Hong, 1993; Bosshart et al., 1994; Molloy et al., 1994; Schafer et al., 1995). Recently, endogenous CPD in AtT-20 cells was shown to be localized in the TGN and to cycle to and from the cell surface (Varlamov and Fricker, 1998). In the present study, we examined the role of the cytosolic tail of CPD/gp180 using gp180 constructs expressed in AtT-20 cells. Mutations within the gp180 tail identified regions and sequence elements that are involved in TGN retention and retrieval. Some of these sequence elements are similar to those found in furin, PAM, and TGN38; however, the activity of these elements differs between the proteins. Important elements for the TGN retention and retrieval of gp180 from the cell surface include an FxxL sequence, which resembles a YxxL motif, and casein kinase II (CKII) sites within an acidic cluster of residues.
MATERIALS AND METHODS
Recombinant DNA Procedures
For the construction of a gp180-expressing vector, a 4.2-kb NcoI fragment encoding the gp180 cDNA was first inserted into a modified version of the plasmid pVL1392L (PharMingen, San Diego, CA) to introduce a consensus Kozak sequence that includes the initiating ATG of gp180. pVL1392L was created by inserting the double-stranded linker GGCCGCGCCATGGCTTAAGTGAGGTAATCTAG into the NotI and XbaI sites of its polylinker. The linker contains an NcoI site (underlined). The initiating ATG of gp180 is encoded within one end of the 4.2-kb NcoI fragment. Incorporation of the fragment in the correct orientation introduces the Kozak sequence and generated the plasmid pVL180. Gp180 and the Kozak sequence were then excised from pVL180 as a 4.2-kb NotI–XbaI fragment and ligated into the vector pcDNA3 (Invitrogen, San Diego, CA), resulting in the gp180 expression vector p180X.
Deletion mutations indicated in Figure 1 were constructed using PCR. Tailored fragments (60–190 nucleotides) spanning the transmembrane/lumenal border to the deletion site within the cytoplasmic tail were obtained using AmpliTaq polymerase (Perkin Elmer-Cetus, Norwalk, CT) and oligonucleotide primers. Primers corresponding to regions within the cytoplasmic domain incorporated an AflII site followed by a stop codon and an XbaI site. The AflII site would code for an additional two amino acids (LK) at the C terminus of each deletion mutant. The Δ56 mutant has the additional residues SD at its C terminus. The primer corresponding to the transmembrane/lumenal border incorporated a native XhoI site. PCR products were purified using a PCR purification kit (Qiagen, Hilden, Germany) and then digested with XhoI and XbaI for 3 h. Substitution of the XhoI–XbaI fragment of p180X resulted in plasmids encoding gp180 with C-terminal truncations of its cytoplasmic domain.
To construct the point mutations indicated in Figure 1, a small 230-nucleotide XhoI–XbaI fragment that encompasses the transmembrane and cytoplasmic domain was first ligated into the vector pAlter (Promega, Madison, WI) at its SmaI/XbaI sites, resulting in the plasmid pAlt-Tail. The XhoI end of the 230-bp fragment had been blunt-ended with Klenow before ligation. Fusion of the blunt-ended XhoI site with the SmaI site regenerates the XhoI site. The pAlt-Tail plasmid was used as the template for mutagenesis. Point mutations were made using Quik-Change Mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer’s instructions. Briefly, complementary oligonucleotide pairs incorporating the respective mutations in the middle of each oligonucleotide were used with Pfu polymerase (Stratagene) to replicate the pAlt-Tail plasmid, introducing the mutation. After replication, the reaction was digested with DpnI, an enzyme that cleaves only the methylated DNA template plasmid. The remaining uncleaved replicated plasmids were then transformed into Escherichia coli. Mutations were screened by DNA sequence analysis. XhoI–XbaI fragments from the mutated pAlt-Tail plasmids were then ligated into the gp180 expression vector p180X, substituting the nonmutated XhoI–XbaI fragment of p180X for the mutant. All plasmids were confirmed by sequence analysis of the modified region.
Cells and Media
AtT-20 cells and stable transfectants were cultivated in DMEM (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum, 100 U penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. To establish stable transfectants, AtT-20 cells (approximately 40% confluent in a 100 mm dish) were transfected with 10 μg of linearized DNA using the calcium phosphate method (Davis et al., 1986). Approximately 16–18 h after transfection, cells were washed three times with phosphate-buffered saline (PBS) and then incubated with DMEM. On the following day, cells were trypsinized, replated into three 100 mm dishes, and cultivated for 24 h. Media (DMEM) containing G418 (650 μg/ml) was then used to cultivate cells and select for stable transfectants. Single clones were screened by Western analysis. Clones selected for further analysis were checked for expression levels by labeling with [35S]Met for 15 min followed by immunoprecipitation with anti-gp180 antiserum. Only those clones that expressed levels of the transfected protein that were within two- to threefold of the level of transfected full-length gp180 were used for subsequent studies. In some studies, cells were treated for the indicated time with 100 μg/ml cycloheximide or with lysosomal protease inhibitors (100 μg/ml leupeptin, 100 μg/ml E-64, 100 μg/ml pepstatin A, and 2 mM methionine methyl ester).
Antibodies
Polyclonal anti-gp180 antiserum was raised in rabbits using only the 170-kDa lumenal domain of gp180, which had been generated from a recombinant protein expressed in the baculovirus system. Polyclonal anti-CPD rabbit antiserum was raised using CPD purified from rat brain, as described previously (Song and Fricker, 1996). Texas Red-labeled anti-rabbit immunoglobulin G (IgG) and fluorescein-labeled anti-mouse IgG were obtained from Vector Laboratories (Burlingame, CA). A monoclonal antibody to syntaxin 6 was obtained from Dr. Richard Scheller (Stanford University), and a monoclonal antibody to LAMP-1 was obtained from the Developmental Studies Hybridoma Bank (University of Iowa).
Pulse–Chase Analysis
Cells grown to 90% confluence on 12-well plates (Falcon; Becton Dickinson, Lincoln Park, NJ) were washed twice with PBS and once with methionine-free DMEM. The cells were then starved of methionine by incubating in methionine-free DMEM for 1 h at 37°C. Cells were metabolically labeled (pulsed) with DMEM containing [35S]Met (100 μCi/ml) for 15 min at 37°C. The pulse was quenched by placing the plates on ice and washing four times with cold PBS. Pulse-labeled cells in replicate plates were chased for different periods at 37°C in complete DMEM. Cell pellets were solubilized in lysis buffer (50 mM Tris 7.4, 150 mM NaCl, 1% Triton X-100, and 1% deoxycholate). Proteins containing the lumenal domain of gp180 were immunoprecipitated with anti-gp180 antibodies (1:500) and protein A Sepharose (35 μl) overnight at 4°C. The protein A Sepharose beads were then washed three times with lysis buffer. Bound proteins were eluted from the beads by heating at 95°C for 5 min in gel loading buffer containing 0.1% SDS and then subjected to denaturing PAGE (SDS-PAGE). The gel was fixed in 30% methanol and 10% acetic acid, soaked in Fluoro-hance (Research Products International, Mt. Prospect, IL), dried, and then subjected to autoradiography at -80°C.
Immunofluorescence Microscopy
Cells (stable transfectants) were grown on growth-supporting glass coverslips (Fisher Scientific, Houston, TX) in either 6- or 12-well plates. In some studies, cells were treated with cycloheximide (100 μg/ml) for 2 h or with brefeldin A (10 μg/ml) before immunostaining. Before fixation, cells were washed twice with DMEM and once with PBS. Cells were fixed for 10 min in 4% paraformaldehyde in PBS and then washed with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min, washed with PBS, and then blocked with 5% bovine serum albumin (BSA) in PBS for 1 h. For LAMP-1 staining, cells were additionally fixed with ice-cold methanol for 2 min after the paraformaldehyde fixation. In analyses for steady-state distribution, cells were immunostained for 1 h with primary antisera diluted in 5% BSA in PBS (anti-gp180; 1:2500). Unbound antibodies were removed by washing with 0.2% Tween-20 in PBS (3 × 5 min) and PBS (1 × 5 min). To detect bound antibodies, Texas Red-conjugated goat anti-rabbit IgG (1:200, diluted in 5% BSA) was added and incubated in the dark for 1 h. Unbound antibodies were removed by washing as described above, and then the coverslips were mounted on glass slides with Prolong antifade reagent (Molecular Probes, Eugene, OR). Representative single plane of focus images were obtained with a Bio-Rad (Bio-Rad, Hercules, CA) confocal microscope.
Antibody Internalization
Cells were grown on glass coverslips in 12-well plates. For continuous uptake of antisera, cells were incubated for 1 h with anti-gp180 antiserum (1:500) in DMEM supplemented with 20 mM HEPES, pH 7.4, and 2 mg/ml BSA. For experiments examining the time course of antisera uptake, cells were washed three times with ice-cold DMEM and then incubated on ice for 1 h with anti-gp180 antiserum (1:250) in DMEM supplemented with HEPES and BSA. Unbound antiserum was removed by washing four times with cold PBS, and the cells were then incubated for various times at 37°C in DMEM supplemented with HEPES and BSA. At the indicated time points, the plates were placed on ice, and the cells were washed with cold PBS. Cells were fixed, permeabilized with Triton X-100, and stained with fluorescently labeled secondary antibodies.
RESULTS
The Cytoplasmic Domain of Gp180 Contains Essential Information for Both Its Retrieval from the Cell Surface and Its Steady-State Accumulation in the TGN
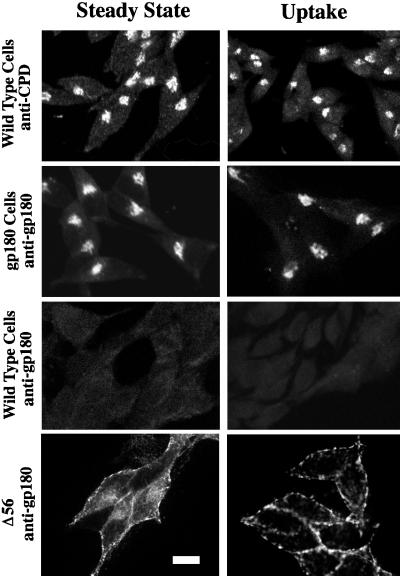
In a previous study, endogenous CPD in AtT-20 cells was found to be localized in the TGN and to traffic between this compartment and the cell surface (Varlamov and Fricker, 1998). To establish an experimental system that could examine the potential role of the cytoplasmic domain in the localization of CPD/gp180 to the TGN, we first assessed whether duck gp180 can be used as a reporter construct in AtT-20 cells. Immunofluorescence labeling of permeabilized AtT-20 cells stably transfected with gp180 cDNA shows a juxtanuclear staining pattern with a gp180-specific antiserum (Figure 2, row 2). This distribution is similar to that observed for endogenous CPD in AtT-20 cells using an antiserum specific for rodent CPD (Figure 2, row 1). The antiserum raised against duck gp180 does not recognize the endogenous mouse CPD or any other protein in the wild-type AtT-20 cells (Figure 2, row 3). This specificity of detection permits gp180 constructs to be analyzed in the presence of endogenous CPD in AtT-20 cells.
Figure 2.
Distribution and uptake from the cell surface of endogenous CPD in AtT-20 cells, and full-length and C-terminally truncated gp180 in transfected AtT-20 cells. Wild-type AtT-20 cells and stable transfectants expressing similar levels of either gp180 or the gp180 deletion mutant (Δ56) were analyzed by confocal microscopy. Cells were fixed and permeabilized at steady state or after 1 h of antiserum uptake at 37°C (see MATERIALS AND METHODS). Antisera used for staining at steady state or uptake are indicated as anti-CPD or anti-gp180. Staining was detected using Texas Red-conjugated anti-rabbit IgGs from goat. At least two single clones were analyzed with comparable results. Bar, 10 μm.
To determine whether the juxtanuclear staining of gp180 in AtT-20 cells reflects localization to the TGN, the cells were treated with brefeldin A before staining with an antiserum to gp180. Brefeldin A causes a redistribution of Golgi but not TGN proteins from the juxtanuclear compartment and releases β-COP from Golgi membranes to the cytoplasm (Fujiwara et al., 1988; Lippincott-Schwartz et al., 1989; Reaves and Banting, 1992). Treatment of AtT-20 cells expressing full-length gp180 with brefeldin A for 30 min had little effect on the juxtanuclear staining of gp180 (our unpublished results), as previously found for endogenous CPD (Varlamov and Fricker, 1998) and furin (Molloy et al., 1994). This result suggests that the juxtanuclear staining of gp180 in transfected AtT-20 cells is not Golgi but a post-Golgi compartment such as the TGN.
To determine whether gp180 was expressed on the cell surface and could be recycled to the TGN, gp180-expressing AtT-20 cells were incubated with antiserum at 4°C for 1 h, washed with cold PBS to remove unbound antiserum, and then incubated at 37°C for 1 h. Cells were subsequently permeabilized and then labeled with fluorescently tagged secondary antiserum. Antiserum to gp180 was internalized to a juxtanuclear compartment (Figure 2, row 2). The distribution of the internalized antiserum to gp180 was generally similar to the distribution of internalized antiserum to rodent CPD in wild-type AtT-20 cells (Figure 2, row 1). In contrast, only background staining was observed with the anti-gp180 antiserum in wild-type AtT-20 cells (Figure 2, row 3), which further demonstrates the specificity of this antiserum for the duck protein.
To determine whether the cytoplasmic domain contains information for TGN localization and trafficking of gp180, a deletion mutant was constructed removing 56 of the 58 residues of the cytoplasmic domain (Figure 1). Upon expression in AtT-20 cells, the Δ56 mutant shows a diffuse distribution throughout the cell, with some accumulation near the cell perimeter that is suggestive of cell surface localization (Figure 2, row 4). In antiserum uptake experiments, cells expressing the Δ56 mutant show a pattern that resembles cell surface staining, suggesting that this mutant enzyme is not retrieved from the cell surface. Thus, the cytoplasmic domain is important for this enzyme’s enrichment in the TGN and retrieval from the cell surface.
Effect of Cycloheximide on Cytoplasmic Domain Deletion Mutants
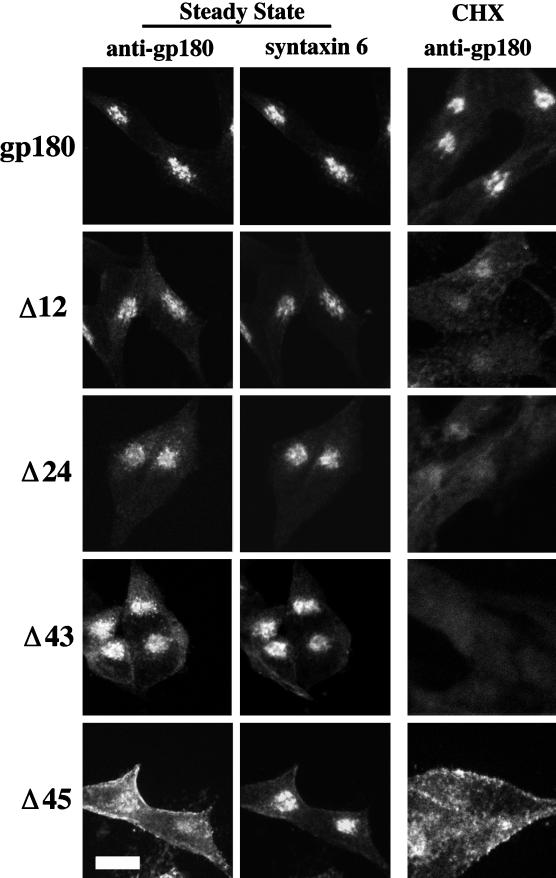
A series of deletion mutations that results in the successive shortening of the cytoplasmic domain from its C terminus were incorporated into the gp180 construct (Figure 1). Stable transfectants expressing similar levels of these mutant constructs were analyzed. The steady-state staining pattern for all of the deletion mutants showed some juxtanuclear staining (Figure 3, and our unpublished results). Costaining of the cells with an antibody to syntaxin 6, which has been previously localized to the TGN (Bock et al., 1997), showed substantial colocalization with gp180. The Δ12 to Δ43 deletion mutants also showed colocalization with syntaxin 6, with some additional distribution beyond the syntaxin 6 distribution (Figure 3). The Δ45 deletion mutant showed a broad distribution that had partial overlap with syntaxin 6 (Figure 3).
Figure 3.
Steady-state localization of gp180 deletion mutants. Stable transfectants expressing similar levels of gp180 or gp180 deletion constructs (depicted in Figure 1) were either treated with cycloheximide (CHX) for 2 h at 37°C (right column) or were not treated (left and middle columns). Cells were fixed, permeabilized, and costained with antiserum to gp180 and antibodies to syntaxin 6. Left and right, gp180 antiserum followed by Texas Red-conjugated goat anti-rabbit IgGs; middle, syntaxin 6 antibodies followed by fluorescein-conjugated anti-mouse IgGs. At least two separate clones were analyzed with comparable results. Bar, 10 μm.
To determine whether the juxtanuclear staining represented only newly synthesized proteins, a blockade on protein synthesis was introduced by treating cells with cycloheximide for 2 h. The juxtanuclear staining of full-length gp180 remains relatively unchanged as compared with untreated cells (Figure 3, CHX). In contrast, cells expressing the Δ12 mutant show a reduction in juxtanuclear staining after the cycloheximide treatment. The Δ24 mutant shows an even greater reduction of staining after cycloheximide treatment, whereas the Δ33, Δ36, and Δ43 mutants show virtually no staining above background after this treatment (Figure 3, and our unpublished results). After CHX, the Δ45 deletion mutant shows diffuse staining (Figure 3). These results suggest that newly synthesized mutant proteins are reduced in their capacity to be retained in a juxtanuclear compartment and further suggests that the cellular half-lives of the Δ12 to Δ43 mutants, but not the Δ45 mutant, are decreased.
Pulse–Chase Analysis with [35S]Met
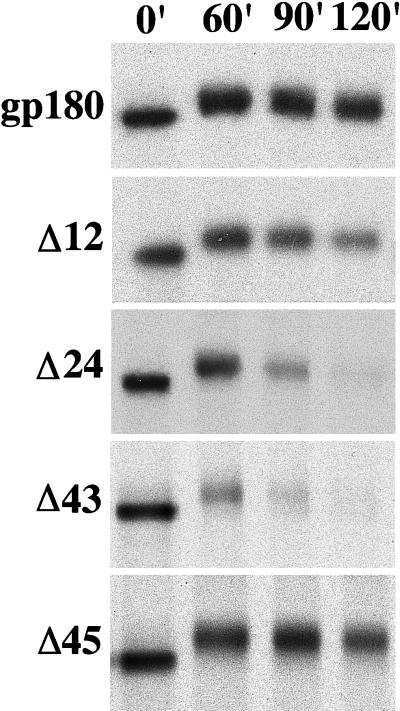
To examine the half-lives of gp180 and the mutant proteins, a pulse–chase analysis was performed (Figure 4). Stable transfectants expressing the various proteins were pulsed in media containing [35S]Met for 15 min and then chased in basal media for the indicated time (Figure 4). Proteins were isolated by immunoprecipitation. The amount of full-length gp180 detected after the 2-h chase period remained relatively unchanged. The molecular weight increase of the gp180 protein detected between pulse and chase times has been observed for endogenous CPD in AtT-20 cells (Varlamov and Fricker, unpublished observations). In comparison to full-length gp180, the Δ12 mutant exhibits some reduction in half-life, and the Δ24, Δ33, Δ36, and Δ43 mutants show an even greater reduction in half-life (Figure 4). The Δ45 mutant was more stable than the Δ43 mutant (Figure 4), consistent with the studies of CHX-treated cells (Figure 3). Quantitation of the results from two separate experiments showed that the half-life of the Δ24, Δ33, Δ36, and Δ43 mutants was 70–90 min, the Δ12 mutant was 120 min, the Δ45 mutant was approximately 6 h, and full-length gp180 had a half-life >7 h. Inspection of the media from the 2 h chase for the Δ33 and Δ43 mutants revealed no immunoreactive material, indicating that the half-life reduction is not caused by secretion or cell surface shedding.
Figure 4.
Pulse–chase analysis of gp180 and gp180 deletion mutants. Stable transfectants expressing similar levels of gp180 or gp180 deletion constructs (depicted in Figure 1) were metabolically labeled for 15 min with [35S]Met. Cells on replicate plates were collected either immediately after the pulse (0′) or after the chase times indicated in minutes (60′, 90′, and 120′). Proteins were isolated from detergent extracts using the anti-gp180 antiserum and resolved by SDS-PAGE under reducing conditions on 7% polyacrylamide gels. This analysis was performed with two single clones for each construct, with comparable results.
Point Mutations in the CKII Sites
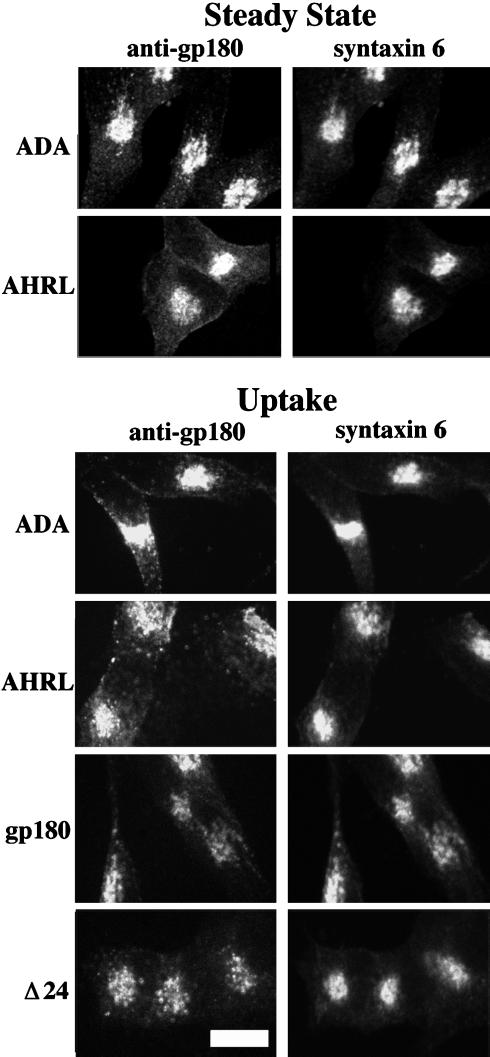
The deletion analysis indicated that a sequence present within the C-terminal 12 residues is important for the relatively long half-life of gp180. This region of gp180 contains an acidic cluster with potential CKII phosphorylation sites in positions -11 and -13 (Figure 1). These CKII sites were mutated (TDT to ADA), destroying the potential for CKII-dependent phosphorylation. The intracellular localization of the ADA mutant shows strong juxtanuclear staining in addition to punctate staining throughout the cell body (Figure 5, top). The juxtanuclear staining showed partial overlap with the distribution of the TGN marker syntaxin 6 (Figure 5). When cells expressing the ADA mutant were exposed to gp180 antiserum continuously for 1 h, the internalized antiserum showed a juxtanuclear distribution that overlapped with the distribution of syntaxin 6 (Figure 5). In addition to this juxtanuclear staining, there was also diffuse punctate staining throughout the cell body that did not overlap with syntaxin 6. Similar results were found with the Δ24 mutant, whereas the full-length gp180 showed more complete overlap with syntaxin 6 (Figure 5). To examine the time course of the internalization, cells were incubated with antiserum at 4°C, the antiserum was removed, and then the cells were incubated at 37°C for the indicated time (Figure 6). These experiments showed that the ADA mutant is internalized from the cell surface within 5 min; however, after 120 min of uptake the antiserum is not detected above background levels (Figure 6), which is similar to the results with the Δ12, Δ24, and Δ43 deletions (Figure 6, and our unpublished results).
Figure 5.
Top, steady-state localization of the two point mutations of gp180 (ADA and AHRL). Stable transfectants were fixed, permeabilized, and costained for gp180 and syntaxin 6, as described in Figure 3 legend. Bottom, antiserum uptake for cells expressing full-length gp180, the Δ24 deletion mutant, and the ADA and AHRL point mutants. Cells were incubated in media containing gp180 antiserum (1:500) at 37°C for 1 h. Then, cells were fixed and permeabilized, and antiserum to gp180 was detected with Texas Red-conjugated goat anti-rabbit IgGs. Steady-state levels of syntaxin 6 (i.e., not internalized antibodies) were detected as described in Figure 3 legend. Bar, 10 μm.
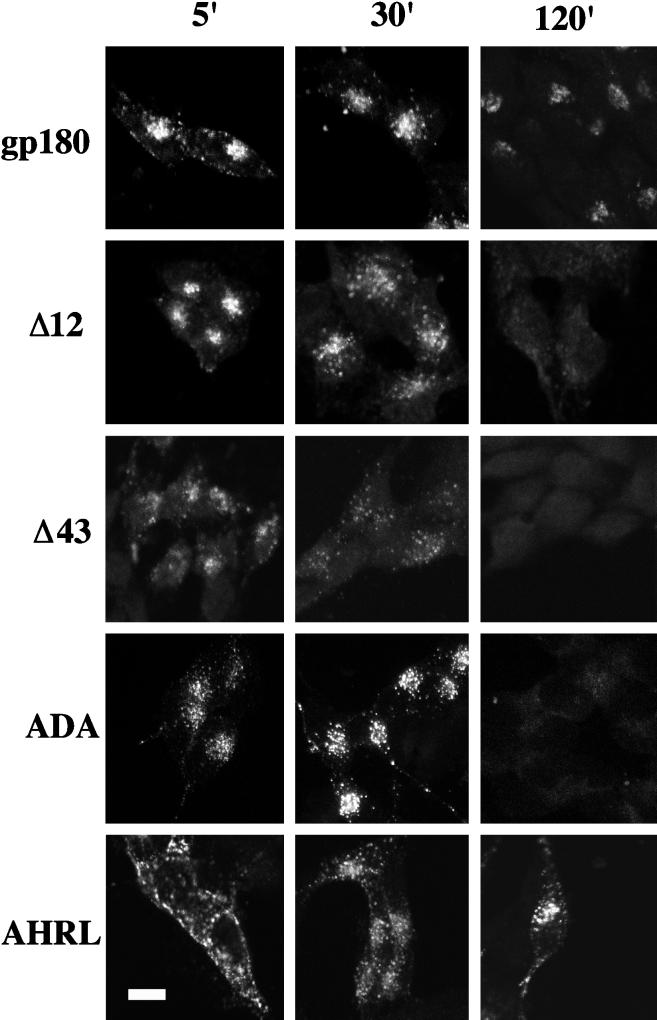
Figure 6.
Time-course analysis of antibody internalization. Stable transfectants expressing similar levels of gp180, the deletion constructs, or the point mutants were incubated with anti-gp180 antiserum at 4°C for 1 h, washed three times with cold PBS, and then incubated at 37°C for the times indicated in minutes (5′, 30′, and 120′). Cells were fixed, permeabilized, and stained with Texas Red-conjugated anti-rabbit IgGs from goat. Shown are the analyses of single clones. Bar, 10 μm.
To test whether the CKII sites are responsible for the shorter half-life of the Δ12 deletion mutant, pulse–chase analysis with [35S]Met was performed. The ADA mutant was more stable (t1/2 = 4 h) than the Δ12 mutant (t1/2 = 2 h), but considerably less stable than the full-length gp180 (t1/2 > 7 h). This finding indicates that the CKII sites within the cytoplasmic tail of gp180 contribute to the routing or retention of the protein and that deletion of this motif leads to increased degradation.
FHRL Is Required for Cell Surface Retrieval and Targeting to the TGN
The dramatic difference in half-life between the Δ43 and the Δ45 constructs suggested an important element in this region. The Δ45 deletion mutant lacks the RL of the FHRL sequence as well as all of the amino acids C-terminal of it (Figure 1). The sequence FHRL within the gp180/CPD cytoplasmic domain resembles the tyrosine-based retrieval sequence YxxL seen in numerous transmembrane proteins (Sandoval and Bakke, 1994; Marks et al., 1997). To examine whether the FHRL sequence is important for retrieval, a point mutation was constructed to convert the phenylalanine to an alanine. At steady state this AHRL mutant displayed juxtanuclear staining that substantially overlaps the distribution of syntaxin 6 (Figure 5). When cells expressing the AHRL mutant were continuously labeled with antiserum to gp180 for 1 h, this antiserum was largely internalized to a juxtanuclear compartment that partially overlaps with the syntaxin 6-containing compartment (Figure 5). In addition, the internalized AHRL mutant shows a diffuse punctate pattern that does not overlap with syntaxin 6 (Figure 5).
To examine the timecourse of the internalization of the AHRL mutant, cells were exposed to antiserum at 4°C, and then the antiserum was removed and the cells were incubated at 37°C for various times. In these experiments, cell surface staining is seen after 5 min of internalization, indicating an impairment in internalization compared with full-length gp180 (Figure 6); however at 30 min, a punctate staining pattern is seen throughout the cell. After 2 h of incubation, more of the internalized antiserum is concentrated in a juxtanuclear region. These results show that the FHRL sequence is required for efficient internalization from the cell surface to the TGN.
Pulse–chase analysis with [35S]Met was performed to examine whether the FHRL motif played a role in the stability of the protein. The AHRL point mutant was stable for the first 2 h of the chase; however, longer chase periods showed it to have a half-life of approximately 5 h (not shown), which is shorter than that for full-length gp180, which has a half-life of >7 h. This result suggests that the FxxL motif is an important determinant of the routing or retention of gp180 and that mutation of this motif leads to increased degradation.
Colocalization of Δ24 Mutant with a Lysosomal Marker
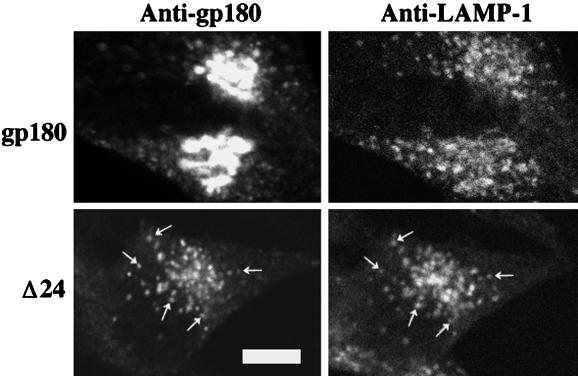
To examine whether the deletion mutants are targeted to lysosomes, cells expressing either the Δ24 mutant or full-length gp180 were treated with lysosomal protease inhibitors and CHX. Although the 2-h CHX treatment without lysosomal protease inhibitors eliminated staining for the Δ24 mutant (Figure 3), in the presence of lysosomal protease inhibitors a punctate pattern of staining is observed (Figure 7). The distribution of the Δ24 mutant in the CHX-treated cells shows substantial overlap with the distribution of LAMP-1 (Figure 7), a lysosomal marker (Fukuda, 1991). In contrast, full-length gp180 shows only partial overlap with the LAMP-1 distribution (Figure 7), consistent with the predominant localization of gp180 to the TGN.
Figure 7.
Distribution of full-length gp180 and the Δ24 deletion mutant in cells treated with lysosomal protease inhibitors and CHX. AtT-20 cells were treated with lysosomal protease inhibitors for 2 h, and then with a combination of lysosomal protease inhibitors and CHX for another 2 h as described in MATERIALS AND METHODS. Cells were fixed and costained with rabbit polyclonal antiserum to gp180 (left panels) and a mouse monoclonal antibody LAMP-1 (right panels). Arrows indicate representative colocalization of the Δ24 mutant and LAMP-1. Bar, 5 μm.
DISCUSSION
A major finding of the present study is the identification of motifs involved in the retention and trafficking of CPD/gp180. The [35S]Met pulse–chase analysis (Figure 4) revealed that two distinct regions of the cytoplasmic tail of gp180 play a large role in the half-life of the protein. One domain present in the C-terminal 12 residues appeared to be important for preventing the degradation of gp180, either by retention in the TGN or by retrieval from the endosomal–lysosomal pathway. Point mutations of the CKII sites within this region resembled the Δ12 deletion mutant, suggesting that these CKII sites contribute to the trafficking and retention of gp180. The other domain, present in the Δ43 construct but absent from the Δ45 construct, has a positive effect on the turnover of gp180 because deletion of this domain (i.e., the Δ45 construct) resulted in a longer half-life. A point mutation of the YxxL-like sequence (FxxL) in this region showed that the aromatic residue of this sequence contributes to the routing and retention of gp180.
The finding that the gp180/CPD cytoplasmic domain is essential for TGN localization and trafficking is consistent with previous studies on other TGN proteins (Luzio et al., 1990; Tausk et al., 1992; Bos et al., 1993; Wong and Hong, 1993; Bosshart et al., 1994; Molloy et al., 1994; Sandoval and Bakke, 1994; Schafer et al., 1995; Marks et al., 1997). Although some sequence elements within the cytoplasmic domains are similar between gp180/CPD and other TGN proteins, the activity of individual elements to promote TGN localization and retrieval differ. For TGN38, the retrieval sequence YQRL was found to be important for TGN localization and retrieval (Bos et al., 1993; Humphrey et al., 1993; Wong and Hong, 1993). For furin, the sequence YKGL can internalize the protein from the cell surface into endosomes but is not sufficient for retrieval back to the TGN (Schafer et al., 1995). Instead, furin has an acidic cluster containing CKII sites that can independently mediate both TGN retrieval and retention (Jones et al., 1995; Schafer et al., 1995; Voorhees et al., 1995). The internalization activity of both sequence elements suggests their cooperation in the retrieval into endosomes, but the subsequent delivery to the TGN requires an additional sorting step mediated by the acidic cluster of furin. In the present study, we have shown that the sequence (FHRL) of CPD/gp180 is important for the efficient internalization and retrieval to the TGN as demonstrated by the impaired retrieval of the AHRL point mutant (Figure 6). This mutant also demonstrates that the CPD/gp180 acidic region does not efficiently and independently mediate retrieval to the TGN, as does the acidic region of furin.
The CKII sites in CPD/gp180 are similar to sites found in PAM and furin (SESEEE in PAM, SDSEED in furin, and TDTEEE in CPD/gp180). All three of these sequences have two phosphorylatable residues separated by a single amino acid within a larger acidic-rich region and are located near the C terminus. Thus, it is tempting to speculate that this sequence similarity might represent functional similarity. Although deletion of the CKII sites and surrounding acidic cluster of PAM did not alter the trafficking of this protein (Milgram et al., 1996), deletion of this region of furin affects uptake of the protein to the TGN (Jones et al., 1995; Voorhees et al., 1995). Recently, a protein designated PACS-1 has been shown to interact with the cytosolic tail of furin when the CKII sites are phosphorylated (Wan et al., 1998). Furin with alanine substitutions in the CKII sites was detected in the tips of the AtT-20 cell processes (Dittie et al., 1997). In some experiments, we found a small amount of staining of the CPD/gp180 ADA mutant in the tips of the cells. Although it is possible that this represents a small amount of sorting into mature secretory vesicles, there are examples of nonregulated pathway proteins that have been detected in the tips of AtT-20 cell processes (Matsuuchi and Kelly, 1991; Song and Fricker, 1997). Further studies are needed to examine whether the CPD/gp180 with mutated CKII sites enters mature secretory vesicles.
In addition to the YxxL-like motif and the CKII sites within the acidic cluster, it is likely that other motifs within the tail of CPD/gp180 are involved in the routing of this protein. Additional domains within the C-terminal tail are predicted from the extremely high conservation of the entire tail among human, rat, and duck. Inspection of the sequence reveals additional motifs that have been previously reported to function in the trafficking of other proteins, such as the di-leucine sequence. CPD/gp180 also contains a di-methionine that may be related to the di-leucine sequence. It is also possible that additional sequences will be found that affect the interaction of transmembrane proteins with cytosolic proteins.
The finding that gp180 is transiently expressed on the cell surface is consistent with previous studies that suggested but did not clearly establish its expression on the cell surface (Kuroki et al., 1995; Varlamov and Fricker, 1998). Our finding further supports the potential role for gp180 as a receptor for duck hepatitis B virus. The internalization of gp180 from the cell surface of AtT-20 cells and also of primary duck hepatocytes (our unpublished observations) suggests that viral entry could proceed through an endocytic pathway. In support of this possibility, it has been shown that infection with duck hepatitis B virus requires endocytosis (Kock et al., 1996); however, the expression of gp180 alone in hepatoma cell lines is not sufficient for productive virus infection even though the virus is internalized (Breiner et al., 1998), indicating that other cellular factors are required to reconstruct the viral entry pathway as has been shown for the human immunodeficiency virus (Feng et al., 1996).
CPD/gp180 functions in the processing of proteins in the secretory pathway. Based on the broad pH optimum of CPD/gp180 between pH 5 and 7 (Song and Fricker, 1995; Eng et al., 1998), the enzyme will be catalytically active in multiple compartments within both the secretory and endocytic pathways as well as when exposed on the cell surface. A recent analysis of gp180 revealed that the first two carboxypeptidase-like domains are enzymatically active, but not the third carboxypeptidase-like domain (Eng et al., 1998). Because the third domain is highly conserved between distant species (82% amino acid identity between rat CPD and duck gp180), it is likely that this region has a biological function. Analysis of the predicted active site amino acids of CPD/gp180 suggests that the third domain will bind but not cleave peptides (Eng et al., 1998). Recently, we have shown that the third domain binds the duck hepatitis B virus (Eng et al., 1998). Taken together with the results of the present study, the possibility that CPD/gp180 functions in the transport of molecules in endocytic and exocytic pathways should be considered.
ACKNOWLEDGMENTS
Confocal microscopy was performed in the Analytical Imaging Facility of the Albert Einstein College of Medicine. Antibodies to syntaxin 6 were generously provided by Dr. Richard Scheller and Jason Bock (Stanford University). This work was supported primarily by National Institutes of Health grant DK-51271 and also by Research Scientist Development Award DA-00194. The DNA sequencing facility of the Albert Einstein College of Medicine is supported in part by Cancer Center grant CA-13330.
REFERENCES
- Bock JB, Klumperman J, Davanger S, Scheller RH. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos K, Wraight C, Stanley KK. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters PJ, Bonifacino JS. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1166. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiner KM, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B virus. J Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LG, Dibner MD, Battey JF. Basic Methods in Molecular Biology. Amsterdam: Elsevier; 1986. [Google Scholar]
- Dittie AS, Thomas L, Thomas G, Tooze SA. Interaction of furin in immature secretory granules from neuroendocrine cells with the AP-1 adaptor complex is modulated by casein kinase II phosphorylation. EMBO J. 1997;16:4859–4870. doi: 10.1093/emboj/16.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng FJ, Novikova EG, Kuroki K, Ganem D, Fricker LD. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J Biol Chem. 1998;273:8382–8388. doi: 10.1074/jbc.273.14.8382. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G-protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fricker LD. Carboxypeptidase E. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- Fricker LD. Peptide processing exopeptidases: amino- and carboxypeptidases involved with peptide biosynthesis. In: Fricker LD, editor. Peptide Biosynthesis and Processing. Boca Raton: CRC Press; 1991. pp. 199–230. [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- Fukuda M. Lysosomal membrane glycoproteins. J Biol Chem. 1991;266:21327–21330. [PubMed] [Google Scholar]
- Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–694. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1993;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BG, Thomas L, Molloy SS, Thulin CD, Fry MD, Walsh KA, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock J, Borst EM, Schlicht HJ. Uptake of duck hepatitis B virus into hepatocytes occurs by endocytosis but does not require passage of the virus through an acidic intracellular compartment. J Virol. 1996;70:5827–5831. doi: 10.1128/jvi.70.9.5827-5831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K, Cheung R, Marion PL, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Brake B, Banting G, Howell KE, Braghetta P, Stanley KK. Identification, sequencing and expression of an integral membrane protein of the trans- Golgi network (TGN38) Biochem J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Ohno H, Kirchhausen T, Bonifacino JS. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- Matsuuchi L, Kelly RB. Constitutive and basal secretion from the endocrine cell line, AtT-20. J Cell Biol. 1991;112:843–852. doi: 10.1083/jcb.112.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgram SL, Mains RE, Eipper BA. Identification of routing determinants in the cytoplasmic domain of a secretory granule-associated integral membrane protein. J Biol Chem. 1996;271:17526–17535. doi: 10.1074/jbc.271.29.17526. [DOI] [PubMed] [Google Scholar]
- Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–23. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggert JK, Fricker LD, Varlamov O, Nishina PM, Rouille Y, Steiner DF, Carroll RJ, Paigen BJ, Leiter EH. Hyperproinsulinemia in obese fat/fat mice associated with a point mutation in the carboxypeptidase E gene and reduced carboxypeptidase E activity in the pancreatic islets. Nat Genet. 1995;10:135–142. doi: 10.1038/ng0695-135. [DOI] [PubMed] [Google Scholar]
- Reaves B, Banting G. Perturbation of the morphology of the trans-Golgi network following brefeldin A treatment: redistribution of a TGN-specific integral membrane protein, TGN38. J Cell Biol. 1992;116:85–94. doi: 10.1083/jcb.116.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval IV, Bakke O. Targeting of membrane proteins to endosomes and lysosomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settle SHJ, Green MM, Burtis KC. The silver gene of Drosophila melanogaster encodes multiple carboxypeptidases similar to mammalian prohormone-processing enzymes. Proc Natl Acad Sci USA. 1995;92:9470–9474. doi: 10.1073/pnas.92.21.9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Fricker LD. Purification and characterization of carboxypeptidase D, a novel carboxypeptidase E-like enzyme, from bovine pituitary. J Biol Chem. 1995;270:25007–25013. doi: 10.1074/jbc.270.42.25007. [DOI] [PubMed] [Google Scholar]
- Song L, Fricker LD. Tissue distribution and characterization of soluble and membrane-bound forms of metallocarboxypeptidase D. J Biol Chem. 1996;271:28884–28889. doi: 10.1074/jbc.271.46.28884. [DOI] [PubMed] [Google Scholar]
- Song L, Fricker LD. The pro region is not required for the expression or intracellular routing of carboxypeptidase E. Biochem J. 1997;323:265–271. doi: 10.1042/bj3230265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Rehli M, Krause SW, Skidgel RA. Sequence of human carboxypeptidase D reveals it to be a member of the regulatory carboxypeptidase family with three tandem active site domains. Biochem J. 1997;327:81–87. doi: 10.1042/bj3270081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausk FA, Milgram SL, Mains RE, Eipper BA. Expression of a peptide processing enzyme in cultured cells: truncation mutants reveal a routing domain. Mol Endocrinol. 1992;6:2185–2196. doi: 10.1210/mend.6.12.1491698. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Fricker LD. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the Golgi and recycling from the cell surface. J Cell Sci. 1998;111:877–885. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Wong SH, Hong W. The SXYQRL sequence in the cytoplasmic domain of TGN38 plays a major role in trans-Golgi network localization. J Biol Chem. 1993;268:22853–22862. [PubMed] [Google Scholar]
- Xin X, Varlamov O, Day R, Dong W, Bridgett MM, Leiter EH, Fricker LD. Cloning and sequence analysis of cDNA encoding rat carboxypeptidase D. DNA Cell Biol. 1997;16:897–909. doi: 10.1089/dna.1997.16.897. [DOI] [PubMed] [Google Scholar]