Abstract
Silencing of ribosomal RNA genes (rDNA) requires binding of the chromatin remodelling complex NoRC to RNA that is complementary to the rDNA promoter. NoRC-associated RNA (pRNA) folds into a conserved stem–loop structure that is required for nucleolar localization and rDNA silencing. Mutations that disrupt the stem–loop structure impair binding of TIP5, the large subunit of NoRC, to pRNA and abolish targeting of NoRC to nucleoli. Binding to pRNA results in a conformational change of TIP5, as shown by enhanced sensitivity of TIP5 towards trypsin digestion. Our results indicate an RNA-dependent mechanism that targets NoRC to chromatin and facilitates the interaction with co-repressors that promote heterochromatin formation and rDNA silencing.
Keywords: RNA polymerase I, noncoding RNA, intergenic spacer, RNA structure, protein conformation
Introduction
In eukaryotes, most DNA does not have any protein coding ability; many of these sequences, however, are transcribed, generating an extensive noncoding (nc) RNA transcriptome that correlates with the increasing complexity of regulatory networks in higher eukaryotes. Thus, ncRNAs are important as biological regulators, rather than simply being the by-products of background transcription. Recently, transcripts originating from a spacer promoter located about 2 kb upstream from the pre-ribosomal RNA transcription start site have been shown to be important in ribosomal RNA gene (rDNA) silencing (Mayer et al, 2006). The data suggested that short-lived intergenic transcripts are processed into 150–250 nucleotide (nt) RNAs that overlap the rDNA promoter. This ‘promoter RNA' (pRNA) is stabilized by binding to TIP5 (TIF interacting protein 5), the large subunit of the chromatin remodelling complex NoRC. NoRC is associated with silent rDNA and mediates heterochromatin formation and transcriptional silencing of a fraction of rDNA repeats (Santoro et al, 2002; Santoro & Grummt, 2005; Zhou & Grummt, 2005). Antisense-mediated depletion of pRNA leads to displacement of NoRC from nucleoli, decreased rDNA methylation and activation of RNA polymerase I (Pol I) transcription, indicating the requirement of pRNA in rDNA silencing (Mayer et al, 2006).
Here, we have studied the molecular principles that control NoRC–pRNA recognition. We show that TIP5 recognizes a phylogenetically conserved hairpin structure that is contained within the minimal RNA binding domain. Mutations that disrupt the hairpin alleviate the interaction with TIP5, whereas compensatory mutations that maintain the hairpin structure restore binding to TIP5. Furthermore, RNase footprinting and protease sensitivity experiments show that TIP5 binds to pRNA in an induced fit mechanism, resulting in structural changes that might facilitate NoRC function. Depletion of pRNA causes translocation of NoRC from nucleoli to the nucleoplasm, whereas ‘refeeding' with ectopic pRNA restores nucleolar localization. This targeting process requires both the hairpin structure of pRNA and sequences upstream from the hairpin. The results show an important role of RNA in targeting a chromatin remodelling complex to a gene promoter to trigger heterochromatin formation and transcriptional silencing.
Results
NoRC recognizes a conserved hairpin within pRNA
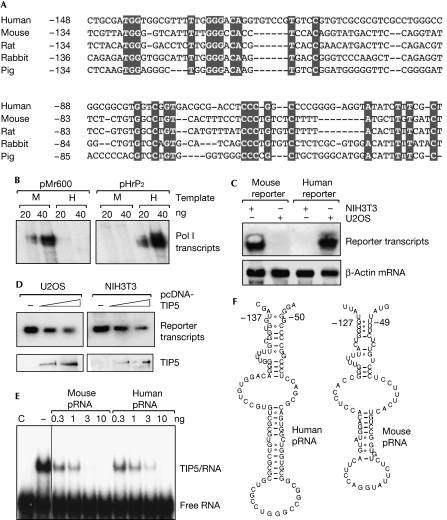
Mammalian rDNA promoters share little sequence homology (Fig 1A), which accounts for the species specificity of rDNA transcription that requires factors from the same or closely related organisms. In cell-free transcription assays, transcription of the murine template requires factors present in nuclear extracts from mouse cells, whereas transcription of human rDNA templates requires human factors (Fig 1B). Similarly, in transient transfection assays, the murine Pol I reporter was efficiently transcribed in mouse but not in human cells, and vice versa (Fig 1C). By contrast, NoRC-dependent repression of Pol I transcription was not species specific. Overexpression of murine TIP5 repressed Pol I transcription in both human and mouse cells (Fig 1D), which is an unexpected result, given that NoRC function requires binding of TIP5 to pRNA—that is, RNA complementary to the rDNA promoter (Mayer et al, 2006).
Figure 1.
Transcription of ribosomal RNA genes but not NoRC-dependent silencing is species specific. (A) Alignment of mammalian rDNA promoter sequences. Conserved bases are highlighted, and numbers refer to the position of nucleotides with respect to the polymerase I (Pol I) transcription start site. Sequence alignment was performed using the CLUSTALW 1.74 software. (B) Pol I transcription is species specific. In vitro transcription assays contained either a linearized mouse (pMr600) or human (pHrP2) rDNA template and 20 or 40 μg nuclear extract (NE) from human (HeLa; H) or mouse (FM3A; M) cells. (C) In vivo reporter assays. NIH3T3 cells (left) and U2OS cells (right) were transfected with 2.5 μg of a human (pHrP2-BH) or murine (pMr1930-BH) Pol I reporter plasmid. Transcripts from the reporter plasmids and β-actin mRNA were analysed on Northern blots. (D) NoRC represses transcription in both mouse and human cells. U2OS (left panel) and NIH3T3 cells (right panel) were co-transfected with pcDNA-HA/Flag-TIP5 (4 and 8 μg) and either the human reporter pHrP2-BH or the mouse reporter pMr1930-BH, and RNA was analysed on Northern blots. The Western blot (bottom) shows the expression of HA/Flag-tagged TIP5 in U2OS and NIH3T3 cells. (E) Murine TIP51−598 binds to human and mouse pRNA with similar affinity. In vitro transcripts corresponding to selected regions of murine rDNA were used in electrophoretic mobility shift assays to compete for binding of TIP51−598 to radiolabelled run-off transcripts from pBluescript (MCS-RNA). (F) Computer-predicted secondary structure of human (left) and mouse (right) pRNA. The numbers refer to the position of nucleotides with regard to the Pol I transcription start site. Secondary structure prediction of RNA folds was performed using the ‘RNA Alifold' program of the Vienna RNA structure prediction package. HA, haemagglutinin; pRNA, promoter RNA; rDNA, ribosomal RNA genes.
To examine whether TIP5 interacts with both human and mouse pRNA, we performed electrophoretic mobility shift assays (EMSAs) using unspecific radiolabelled T7 run-off transcripts (MCS-RNA). The competition assays in Fig 1E show that pRNA binds with high affinity to TIP5, displacing unspecific MCS-RNA from TIP5, regardless of whether human or mouse pRNA was used as a competitor. Thus, TIP5 binds to pRNA from both species with similar affinity, which is consistent with NoRC silencing rDNA in both species.
In search for structural motifs that are shared by pRNA from various species, a computational approach showed that RNAs corresponding to rDNA promoters of several mammals, including human, mouse, rat and pig, can fold into a common secondary structure. The most striking feature of this hypothetical structure is a hairpin that contains rDNA sequences from nucleotides −127 to −49 in mouse and from −137 to −50 in human pRNA (Fig 1F).
TIP5 recognizes the stem–loop structure of pRNA
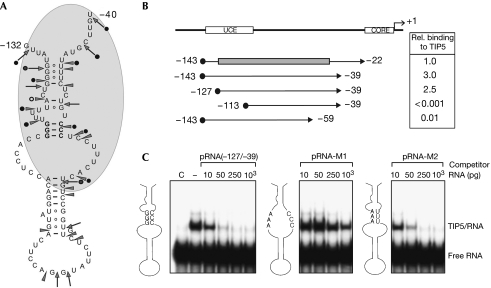
RNase structure mapping and footprinting experiments showed that TIP5 binding protects sequences near the base of the hairpin (supplementary Fig S1 online). As summarized in Fig 2A, the results support the algorithm-predicted hairpin in pRNA, showing that TIP5 specifically recognizes the basal part of the hairpin, thus stabilizing the RNA structure.
Figure 2.
TIP5 recognizes the secondary structure of promoter RNA. (A) Schematic depicting the secondary structure of mouse pRNA as shown by RNase footprinting. Cleavages by RNase T1 are indicated by arrows, and RNase V1 cleavages by arrowheads. Cleavages that are decreased as a consequence of human TIP5 binding are indicated by black circles. The area of pRNA that is shielded from RNase cleavage is shaded in grey. The three G-C base pairs mutated in pRNA-M1 are indicated in bold. (B) Mapping of the minimal TIP5 interacting RNA domain. Black circles indicate T7 promoter-derived sequences common to all run-off transcripts used. The grey box marks the part of pRNA that is involved in hairpin formation. The numbers on the right are taken from Table 1, representing the relative binding affinities of murine TIP51−598. (C) Mutations that disrupt the secondary structure abrogate TIP5 binding to pRNA. Mouse pRNA and mutants pRNA-M1 and pRNA-M2 comprising the minimal sequence required for TIP5 binding (from −127 to −39) were used in EMSA competition assays containing murine TIP51−598 and labelled MCS-RNA. The schemes indicate nucleotide exchanges in pRNA-M1 and -M2 and the predicted structure of the individual pRNAs. pRNA, promoter RNA.
To define the minimal RNA sequence that interacts with TIP5, we synthesized pRNAs comprising different lengths of promoter sequences and assayed the ability of these transcripts to compete for TIP5 binding to unspecific MCS-RNA. This analysis showed that TIP5 bound with high affinity to pRNAs containing rDNA sequences −127 to −39 (Fig 2B). Truncation at the 5′ or 3′ end lowered the binding affinity at least 100-fold, indicating that sequences from −127 to −39 are crucial for binding to TIP5 (Table 1). Strikingly, this sequence contains the predicted hairpin structure (from −127 to −49), suggesting that NoRC recognizes this specific RNA structure.
Table 1.
Binding of murine TIP51−598 to promoter-proximal murine and human pRNA transcripts
| pRNA transcripts | Relative binding |
|---|---|
| Murine | |
| rDNA −168/+7 | 50 |
| rDNA −205/−1 | 350 |
| rDNA −143/−22 | 500 |
| rDNA −143/−39 | 500 |
| rDNA −127/−39 | 400 |
| rDNA −143/−59 | 5 |
| rDNA −113/−39 | <0.1 |
| rDNA −127/−39 (M1) | 0.4 |
| rDNA −127/−39 (M2) | 330 |
| Human | |
| rDNA −143/−22 | 125 |
| Radiolabelled MCS-RNA was incubated with purified TIP51−598 and increasing amounts of the indicated competitor RNAs, and RNP complexes were analysed by electrophoretic mobility shift assay. Numbers of pRNA transcripts denote positions relative to the pre-rRNA transcription start site. The amount of total RNA required to reduce binding of TIP5 to 50% was set to 1.0. rDNA, ribosomal RNA genes. | |
To examine whether the hairpin fold is required for NoRC binding to pRNA, we disrupted the basal part of the predicted stem structure by replacing three guanosine residues (nucleotides −113, −114 and −115) with adenosine residues, and measured the relative binding affinity of TIP5 to wild-type and mutant pRNA in EMSA competition assays. Mutations that prevent base pairing between nucleotides −113 to −115 and −60 to −58, and therefore destroy the stem–loop (pRNA-M1), abrogate binding to TIP5, whereas introduction of compensatory C to U mutations (pRNA-M2), allowing hairpin formation, restored high-affinity binding (Fig 2C). These results are consistent with TIP5 recognizing an evolutionarily conserved RNA conformation, suggesting that the secondary structure rather than the primary pRNA sequence is selected.
Binding to pRNA induces a structural change of TIP5
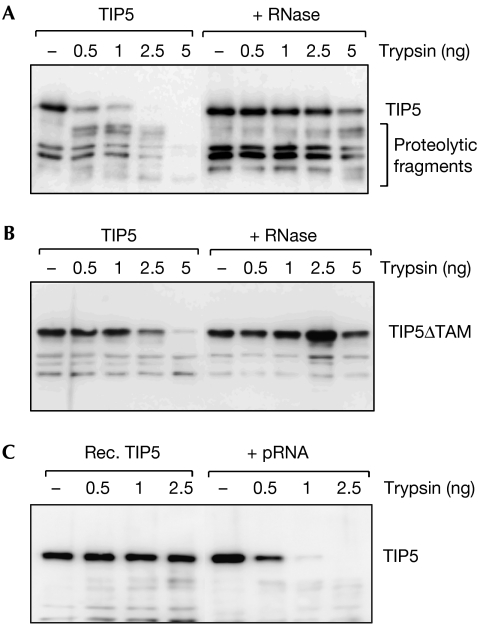
To examine whether binding to RNA induces a conformational change in TIP5, we preincubated immunopurified TIP5 in the absence or presence of RNase A before digestion with limited amounts of trypsin. The immunoblot in Fig 3A shows that TIP51−731 was efficiently cleaved by low concentrations of trypsin. However, after treatment with RNase A, TIP51−731 was about 20-fold more resistant towards proteolysis, indicating that in the absence of RNA the protein has adopted a more compact structure. In support of this, TIP5ΔTAM, a TIP5 mutant that lacks the TAM domain and is deficient in RNA binding (Mayer et al, 2006), was also more resistant towards trypsin digestion (Fig 3B).
Figure 3.
Interaction with promoter RNA changes the conformation of TIP5. (A) RNase treatment confers a more compact structure to TIP5. Immunopurified HA/Flag-murine TIP51−731 (50 ng) was incubated with increasing amounts of trypsin in the absence or presence of RNase A, and cleavage products were analysed on immunoblots. (B) Same as in (A) except that murine TIP51−731 lacking the RNA-binding domain (TIP5ΔTAM) was used. (C) Interaction with pRNA renders TIP5 hypersensitive towards proteolysis. Recombinant (Rec.) murine TIP51−598 was preincubated with 200 ng yeast transfer RNA or with mouse pRNA (−127/−39), and subjected to partial proteolysis and immunoblotting. HA, haemagglutinin; pRNA, promoter RNA.
Finally, we compared the protease sensitivity of TIP5 in the absence or presence of exogenous pRNA. TIP5 was first treated with RNase A, incubated with a control RNA (yeast transfer RNA, lanes 1–4) or mouse pRNA (lanes 5–8), and then subjected to partial proteolysis (Fig 3C). Binding to pRNA, but not tRNA, induced the trypsin-hypersensitive ‘open' protein structure that is characteristic of cellular TIP5. These results indicate that on pRNA binding, TIP5 undergoes a conformational change in the region encompassing the TAM domain, and this structural change might facilitate the interaction with other proteins required for NoRC function.
pRNA regulates the nucleolar localization of NoRC
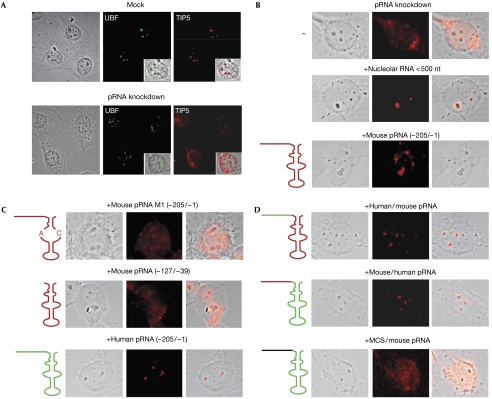
Depletion of pRNA by antisense locked nucleic acid (LNA)-DNA gapmers leads to translocation of NoRC from nucleoli to the nucleoplasm (Mayer et al, 2006). To show the direct role of pRNA in targeting NoRC to nucleoli, we sought to restore the nucleolar localization of NoRC in pRNA-depleted NIH3T3 cells by adding an excess of exogenous pRNAs to permeabilized cells. As shown in Fig 4A, knockdown of pRNA (supplementary Fig S2 online) resulted in translocation of NoRC into the nucleoplasm without affecting the nucleolar localization of upstream binding factor (UBF). Notably, ‘feeding' of pRNA-depleted cells with size-fractionated nucleolar RNA (<500 nt), but not with tRNA, restored the nucleolar localization of NoRC (Fig 4B). Similarly, in vitro transcripts comprising mouse or human pRNA sequences from −205 to −1 rescued NoRC localization (Fig 4B,C). However, mutant pRNA in which the predicted stem structure has been disrupted (pRNA-M1) did not restore NoRC localization. Thus, exogenous pRNA is able to trigger relocalization of NoRC into nucleoli, provided that its secondary structure is kept intact.
Figure 4.
Promoter RNA is required for nucleolar localization of NoRC. (A) Exogenous pRNA restores the nucleolar localization of NoRC in pRNA-depleted cells. NIH3T3 cells were transfected with 50 nM of control (mock) or antisense LNA-DNA (pRNA knockdown), and TIP5 and UBF were visualized by immunostaining. The insets show an overlay of phase-contrast images and immunostaining. (B) Permeabilized NIH3T3 cells were incubated with transfer RNA, nucleolar RNA (<500 nt) or in vitro-synthesized mouse pRNA (−205/−1), and NoRC localization was visualized with TIP5 antibodies (red). Phase-contrast images, an immunofluorescence of TIP5 and an overlay with the respective phase-contrast image are shown (left to right). (C) Same as in (B) but permeabilized cells were incubated with mouse pRNA-M1 (−205/−1), mouse pRNA (−127/−39) and human pRNA (−205/−1) as indicated. The schematics show characteristics of the pRNA variants used for refeeding. Mouse rDNA sequences are drawn in red, and human rDNA sequences in green. (D) Same as in (B) but permeabilized cells were incubated with chimaeric human/mouse or mouse/human pRNAs (−205/−1) as indicated. In the bottom panels, pRNA sequences from −205 to −128 were replaced by unrelated sequences from the multiple cloning site (MCS/mouse pRNA). Mouse rDNA sequences are drawn in red, human rDNA sequences in green and unspecific MCS sequences in black. pRNA, promoter RNA; rDNA, rRNA genes.
Next, we examined whether the pRNA hairpin loop—that is, the minimal sequence required for NoRC binding to pRNA—is sufficient for targeting NoRC to nucleoli or whether other sequences are crucial for NoRC localization. For this, we used the ‘refeeding' assay to compare the ability of ‘long' pRNA (from −205 to −1) and ‘short' pRNA (from −127 to −39) to mediate nucleolar relocation of NoRC after knockdown of endogenous pRNA. Although ‘short' pRNA bound to NoRC with higher affinity than the ‘long' pRNA (Table 1), it was not able to restore the nucleolar localization of NoRC (Fig 4C, upper panel), indicating that sequences adjacent to the stem–loop are necessary for pRNA-mediated nucleolar localization of NoRC. To examine whether these adjacent regions are species specific, we generated chimaeric ‘human–mouse' or ‘mouse–human' pRNAs in which rDNA sequences from −205 to −126 from one species were fused to the hairpin fold (−127 to −39) from the other species, and assayed these RNAs for their ability to restore NoRC localization. In accordance with the cross-species relevance of pRNA in NoRC function, both pRNA chimaera facilitated NoRC translocation to nucleoli (Fig 4C). However, if the adjacent region was replaced by an unrelated RNA sequence, the hybrid RNA was much less able to relocate NoRC (Fig 4C). This shows that TIP5 binding to the hairpin structure of pRNA is necessary but not sufficient for targeting NoRC to nucleoli, and NoRC recruitment to nucleolar chromatin requires 5′ adjacent sequences of pRNA that cover important regulatory elements such as the upstream terminator T0 and the upstream control element.
Discussion
Here, we have used RNA binding assays in conjunction with partial proteolysis and RNA replenishment experiments to decipher the structure–function relationship of NoRC binding to pRNA. We have shown that mammalian rDNA-specific pRNAs fold into a conserved secondary structure, and that this structure is required for high-affinity binding of TIP5. Mutations that affect the maintenance of the hairpin stem impair TIP5 binding, whereas introduction of compensatory base changes restores the interaction with TIP5. The results indicate that TIP5 recognizes a specific stem–loop structure within pRNA that is conserved across mammalian species. The interaction of TIP5 with this specific RNA fold might provide the selection pressure necessary for the conservation of RNA conformation, and might allow NoRC to tolerate mutational changes without loss of function. Such plasticity of the interacting partners would allow for evolution of a broad spectrum of RNA binding specificities despite the selective pressure that conserves the primary sequence of the RNA-binding site. Of note, the fact that unrelated sequences can fold into similar secondary structures raises the exciting possibility that other RNA binding partners might interact with TIP5 and guide NoRC to non-ribosomal target genes outside of the nucleolus to induce heterochromatin formation and transcriptional silencing.
The structure and function of proteins are affected and regulated by interacting partners, including RNAs (Williamson, 2000). In accordance with this, we found that association with RNA induces a conformational change of TIP5. If bound to RNA, TIP5 adapts an ‘open', trypsin-hypersensitive structure, whereas both RNase A treatment and deletion of the RNA-binding domain confer a more compact structure that is less sensitive towards proteolysis. This implies an induced fit mechanism—a common feature of RNA–protein complexes (Puglisi et al, 1995)—using the binding energy to drive conformational changes to ensure that the RNA–protein complex exerts its function only after the appropriate conformational adaptation has occurred. Such a structural change might facilitate or modulate the interaction of TIP5 with other proteins required for NoRC function such as histone deacetylases (HDACs) and DNA methyltransferases (DNMTs; Santoro & Grummt, 2005; Yuan et al, 2007). As TIP5 is apparently more extended if bound to RNA, this ‘open' configuration would allow for additional interaction surfaces to become available.
Regarding the role of pRNA in targeting NoRC to rDNA, we have previously shown that heterochromatin formation and nucleolar localization of NoRC require the interaction of TIP5 with pRNA (Mayer et al, 2006). After depletion of pRNA, NoRC was no longer nucleolar, being distributed throughout the nucleoplasm. Here, we have used a ‘refeeding' or ‘RNA replenishment' assay, involving a brief incubation of permeabilized, pRNA-depleted cells with size-fractionated nucleolar RNA or with in vitro transcripts. These assays showed that exogenous pRNA was able to re-establish the nucleolar localization of NoRC in pRNA-depleted cells. Notably, rescue of NoRC localization required pRNA containing sequences from −205 to −1—that is, the part of rDNA that comprises essential regulatory elements, the core promoter, the upstream control element and the upstream terminator T0. Consistent with the prevalent use of RNA to target chromatin-associated factors to subcellular structures, our results show that pRNA is necessary and sufficient to target NoRC to nucleoli. As complex rearrangements in the chromatin structure are unlikely to occur within the short time needed for NoRC relocalization, pRNA seems to have a role in targeting NoRC to nucleolar chromatin. This targeting process requires the stem–loop structure that is required for TIP5 binding (nucleotides −127/−39), as well as further upstream sequences (up to nucleotide −205). Notably, both mouse and human pRNA sequences are able to target NoRC to nucleoli, whereas replacement of the upstream part of pRNA by unrelated sequences impairs nucleolar localization of NoRC, underscoring the role of the hairpin in TIP5 binding and upstream pRNA sequences in targeting NoRC to nucleoli. Given that silencing of a fraction of rDNA repeats has an important role in genome stability (Kobayashi et al, 2004; Peng & Karpen, 2007), NoRC-associated pRNA might control the number of silent rDNA repeats, thereby contributing to rDNA stability, nucleolar structure and longevity.
Methods
Plasmids. pMr1930-BH and pHrP2-BH, pMr600 and pHrP2 have been described previously (Budde & Grummt, 1999; Mayer et al, 2005). pcDNA-haemagglutinin (HA)/Flag-TIP5 encodes full-length murine TIP5 and pcDNA-HA/Flag-TIP51−731 encodes amino acids 1–731 of murine TIP5 fused to an amino-terminal HA/Flag tag.
Transfections and RNA analysis. A total of 3 × 105 NIH3T3 cells or 8 × 105 HEK293T cells were transfected with 1 μg of Pol I reporter plasmids and various amounts of pcDNA-HA/Flag-TIP5. Transcripts from reporter plasmids were monitored on northern blots. To normalize for RNA loading, blots were probed for β-actin messenger RNA. Alternatively, transcripts were analysed by real-time quantitative PCR with primers specific for the rDNA reporter and glyceraldehyde-3-phosphate dehydrogenase mRNA.
Expression and purification of recombinant proteins. His-tagged murine TIP51−598 and human TIP5510−611 were expressed in Escherichia coli BL21(DE3) codon+ and purified on Ni-NTA agarose. For expression in mammalian cells, HEK293T cells were transfected with pcDNA-HA/Flag-TIP5, and TIP5 was purified with HA or Flag antibodies.
In vitro transcription assays. A 25 μl assay sample contained 50 ng of template DNA (pMr600/EcoRI or pHrP2/EcoRI), 30 μg of nuclear extract proteins from exponentially growing FM3A or HeLa cells, 12 mM Hepes–KOH (pH 8.0), 0.1 mM EDTA, 5 mM MgCl2, 80 mM KCl, 10 mM creatine phosphate, 12% (v/v) glycerol, 0.66 mM each of ATP, GTP and CTP, 0.01 mM UTP and 0.5 mCi [α-32P]UTP (5,000 Ci/mmol). After incubation for 60 min at 30°C, RNA was extracted and analysed on 4.5% polyacrylamide gels.
Electrophoretic mobility shift assays. Radiolabelled MCS-RNA was synthesized by T7 RNA polymerase using pBluescript-KS/EcoRI as a template. A 1–2 fmol portion of RNA was incubated for 5 min on ice with 1–3 pmol of recombinant TIP5 in 20 μl binding buffer (20 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 100 mM KCl and 0.2 mM EDTA). After the addition of competitor RNA, incubation was continued for 15 min. RNA–protein complexes were analysed on 6% native polyacrylamide gels and visualized by autoradiography.
Refeeding assay and immunofluorescence. To examine the ability of pRNA to restore NoRC localization after knockdown of endogenous pRNA, NIH3T3 cells were transfected with antisense LNA-DNA gapmers (5′-CAGGtatgacttccaGGTA-3′; LNA bases, uppercase; DNA bases, lowercase) using TransIT-TKO Transfection Reagent. After 48 h, cells were permeabilized for 3 min with 0.04% Triton X-100 in 20 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 0.5 mM EDTA and 25% glycerol and incubated for 30 min with 100 ng/ml of RNA in the presence of 25 U/ml RNasin (Promega, Madison, WI, USA). Cells were fixed with methanol (7 min) and acetone (1 min) at −20°C, blocked with 1% BSA and immunostained with TIP5 and UBF antibodies.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank K. Schmitz for helpful suggestions and critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB/Transregio 5, SP, Epigenetics'), the European Union-Network ‘Epigenome' and the Fonds der Chemischen Industrie.
Footnotes
The authors declare that they have no conflict of interest.
References
- Budde A, Grummt I (1999) p53 represses ribosomal gene transcription. Oncogene 18: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) Sir2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Mayer C, Bierhoff H, Grummt I (2005) The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev 19: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Chen L, Blanchard S, Frankel AD (1995) Solution structure of a bovine immunodeficiency virus Tat–TAR peptide–RNA complex. Science 270: 1200–1203 [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2005) Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR (2000) Induced fit in RNA–protein recognition. Nat Struct Biol 7: 834–837 [DOI] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y (2007) Activation of RNA polymerase I transcription by Cockayne Syndrome group B (CSB) protein and histone methyltransferase G9a. Mol Cell 27: 585–595 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Grummt I (2005) The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol 15: 1434–1438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures