The European Science Foundation (ESF)–European Molecular Biology Organization (EMBO) Symposium on Antiviral Applications of RNA Interference took place between 5 and 10 April 2008, in Sant Feliu de Guixols, Spain, and was organized by J. Kurreck and B. Berkhout.
Introduction
Small RNAs are important regulators of gene expression that operate through a range of RNA-silencing mechanisms. Of these, RNA interference (RNAi)—gene silencing through small-interfering RNAs (siRNAs) derived from exogenous double-stranded RNA (dsRNA)—and the microRNA (miRNA) pathway—gene silencing guided by small RNAs encoded in the genome—are the best known. The discovery that dsRNA induces RNAi (Fire et al, 1998) and the appreciation of its antiviral activity revolutionized our view of viruses in many ways. The European Science Foundation (ESF)–European Molecular Biology Organization (EMBO) Symposium on Antiviral Applications of RNA Interference brought together about 50 scientists working at the interface between virology and RNAi. Here, I review the main issues that were discussed at the meeting.
RNAi as an antiviral response in plants and insects
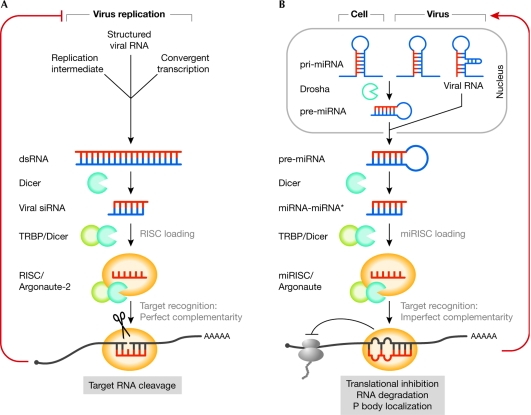
RNAi provides an antiviral defence mechanism in plants, nematodes and insects. The cleavage of viral dsRNA by the ribonuclease Dicer generates viral siRNAs (v-siRNAs), which are incorporated into the RNA-induced silencing complex (RISC) and direct RISC activity onto messenger RNAs (mRNAs) in a sequence-specific manner. Recognition of a complementary sequence then triggers the endonucleic cleavage (slicer activity) or translational inhibition of the viral target RNAs by an Argonaute family member (Fig 1).
Figure 1.
RNA silencing in virus infections. (A) RNA interference (RNAi) as an antiviral defence mechanism. Viruses might produce double-stranded RNAs (dsRNAs) that are processed by the RNAi machinery and thereby programme the RNA-induced silencing complex (RISC) to degrade viral RNAs. (B) Cellular or viral microRNAs (miRNAs), or viral RNAs with structural similarity to pre-miRNAs, regulate viral and host gene expression, and contribute to viral pathogenesis. miRISC, microRNA-induced silencing complex; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA transcript; siRNA, small-interfering RNA; TRBP, TAR RNA-binding protein.
In plants, RNAi acts as a systemic antiviral defence against cytoplasmic RNA and nuclear DNA viruses (Ding & Voinnet, 2007). Arabidopsis thaliana encodes four Dicer-like genes (DCLs) that exhibit both specialization and redundancy. RNA viruses are mainly targeted by DCL4 and by the redundant activity of DCL2. All four DCLs are involved in targeting nuclear DNA viruses (geminiviruses and the pararetrovirus cauliflower mosaic virus (CaMV; Ding & Voinnet, 2007). Substrates for Dicer include dsRNA replication intermediates and local base-paired RNA structures of cytoplasmic RNA viruses (Hamilton & Baulcombe, 1999; Molnar et al, 2005), or overlapping transcripts of geminiviruses (Chellappan et al, 2004). T. Hohn (Basel, Switzerland) reported that v-siRNAs map across the genome of CaMV, with up to 1,000-fold enrichment in hotspots in the structured translational leader sequence.
As a counter-defence, many, if not all, plant viruses encode viral suppressors of RNA silencing (VSRs). The discovery that viruses might even encode multiple VSRs underlines the selection pressure exerted by RNA silencing (Ding & Voinnet, 2007). J. Garcia (Madrid, Spain) discussed the example of cucumber vein yellowing virus—a potyvirus that lost the typical silencing suppressor, helper component-proteinase (HcPro), through a recombination event, but evolved VSR activity in another protein, P1b, which arose through gene duplication (Valli et al, 2008).
RNAi also mediates antiviral defence in Drosophila melanogaster. Flies that are defective in RNAi—Dicer-2 (Dcr-2), R2D2 and Argonaute-2 (Ago-2) null mutants—are hypersensitive to the (+) stranded RNA viruses, flock house virus (FHV), cricket paralysis virus (CrPV) and Drosophila C virus (DCV; Galiana-Arnoux et al, 2006; van Rij et al, 2006; Wang et al, 2006). Ago-2 and R2D2 mutants are also hypersensitive to the dsRNA virus Drosophila X virus but, paradoxically, Dcr-2 mutants are not (Zambon et al, 2006). J.-L. Imler (Strasbourg, France) reported that the (−) stranded RNA virus, vesicular stomatitis virus (VSV), does not produce detectable levels of dsRNA during replication, yet is sensitive to natural antiviral RNAi in Drosophila. v-siRNAs have been detected in FHV and CrPV infections of Drosophila (Galiana-Arnoux et al, 2006; Wang et al, 2006), and in alphavirus infections of several other insect species (Sanchez-Vargas et al, 2004). Furthermore, VSRs have been identified in three viruses that infect different insect species: FHV (B2), CrPV and DCV (1A) (Li et al, 2004; van Rij et al, 2006; Wang et al, 2006).
In addition to its function as an antiviral effector mechanism, RNAi seems to regulate other antiviral responses. Imler reported that the putative antiviral factor vago is upregulated in DCV and sindbis virus (SINV) infection in a Dicer-2-dependent manner. A separate RNAi-independent Janus kinase (Jak)–signal transducer and activator of transcription (Stat) pathway is responsible for virus-specific induction of virus induced RNA-1 (vir-1) expression.
RNAi in Drosophila is thought to be a cell-autonomous phenomenon. However, M-C. Saleh (Paris, France) reported that the systemic spread of viral dsRNA is essential to the antiviral immune response. Viral dsRNA, released either through cell lysis or through a specialized shedding mechanism, is taken up by uninfected cells to generate virus-specific adaptive immunity that prevents virus spread.
Does RNAi have natural antiviral activity in mammals?
Whether RNAi has natural antiviral activity in mammals is a hotly debated issue. Virus-derived siRNAs could not be detected by small-RNA cloning in tissue-culture models for hepatitis C virus (HCV) and yellow fever virus (Pfeffer et al, 2005). Encephalomyocarditis virus, lymphocytic choriomeningitis virus, coxsackie virus B3 and influenza A virus (all of which are RNA viruses), as well as vaccinia virus (a DNA virus that is known to produce dsRNA), replicated to similar, or even lower, viral titres in Dicer−/− macrophages (Otsuka et al, 2007). R. van Rij (Nijmegen, The Netherlands) reported that SINV replicated with similar kinetics in Dicer−/− and wild-type murine fibroblasts, yet this virus is under natural RNAi control in Drosophila and other insects (Sanchez-Vargas et al, 2004).
The evolution of VSR activity suggests a strong selection pressure by RNAi. The interferon antagonists, nonstructural protein 1 (NS1) of influenza A virus, E3L of vaccinia virus, viral protein 35 (VP35) of Ebola virus and the non-structural protein NSs of La Crosse virus (LACV), are putative RNAi suppressors (Haasnoot et al, 2007; Li et al, 2004; Lichner et al, 2003; Soldan et al, 2005). The physiological significance of these observations, however, is not clear. For example, overexpression of the RNase III dsRNA-binding domain from the bacterium Escherichia coli suppressed RNAi in plants (Lichner et al, 2003), even though E. coli is not able to perform RNAi.
A defect in VSR-mutant virus replication in wild-type insects and plants—and rescue of this defect in an RNAi-defective background—provides strong genetic support for the importance of RNAi in antiviral defence (Deleris et al, 2006; Galiana-Arnoux et al, 2006; Wang et al, 2006). However, replication defects of influenza virus lacking NS1 and LACV lacking NSs are absent in dsRNA-dependent protein kinase R (PKR)−/− and type I interferon receptor −/− mice, respectively (Bergmann et al, 2000; Blakqori et al, 2007). This indicates that the suppression of interferon responses, rather than RNAi suppression, is the main function of these viral proteins.
Even more controversy surrounds the role of RNA silencing in human immunodeficiency virus (HIV)-1 infection. Small-RNA cloning could not detect v-siRNAs from HIV-1-infected cells (Landgraf et al, 2007; Lin & Cullen, 2007; Pfeffer et al, 2005). Although a single specific v-siRNA corresponding to the structured Rev responsive element was detected in HIV-1 infected cells (Bennasser et al, 2005), this observation could not be reproduced by another laboratory (Lin & Cullen, 2007), possibly owing to the use of a different cell line. F. Kashanchi (Washington, DC, USA) reported work that identified small RNAs corresponding to the TAR element—an imperfect, roughly 50 nucleotide (nt) hairpin structure—by an RNase-protection assay (Klase et al, 2007; Ouellet et al, 2008). Personal discussions with Kashanchi and Z. Bentwich (Rehovot, Israel) highlighted the recent confirmation of this result using small-RNA cloning. K-T. Jeang (Bethesda, MD, USA) reported the identification of several small viral RNAs from HIV-1-infected cells by an oligonucleotide-tiling array and deep sequencing.
The TAR RNA-binding protein (TRBP) favours HIV-1 replication through several RNAi-independent mechanisms, but is also a partner of Dicer in RISC. A. Gatignol (Montreal, Canada) reported that knockdown of TRBP and Dicer results in a decrease in virus production (Christensen et al, 2007). This finding seems to conflict with an observed increase in viral replication on Dicer and Drosha knockdown, as noted by M. Benkirane (Montpellier, France) and discussed below, which might be due to differences in experimental set-up.
Do retroviruses inhibit RNA-silencing pathways? The RNA-binding proteins HIV-1 Tat and primate foamy virus (PFV)-1 Tas were identified as inhibitors of RNA silencing (Bennasser et al, 2005; Haasnoot et al, 2007; Lecellier et al, 2005); however, this observation could not be confirmed by others (Lin & Cullen, 2007). Gatignol presented data that suggest that HIV-1 infection inhibits RNAi initiated by endogenous miRNAs targeting a perfect complementary site in a reporter construct.
Recently, it has been shown that endogenous siRNAs in Drosophila and mouse map to transposable elements, including retrotransposons (reviewed by Birchler & Kavi, 2008). These results suggest that repression of transposons is a natural function of RNAi in animals, as was previously observed in Caenorhabditis elegans and plants.
Viral miRNAs
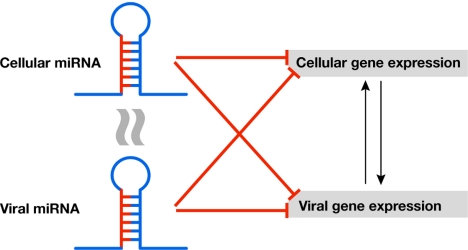
With the benefit of hindsight, it might be of little surprise that viruses exploit a mechanism for gene regulation as versatile as the miRNA pathway. An RNA hairpin as small as 70 nt is sufficient to instruct the RISC. Furthermore, miRNA genes do not encode proteins and are therefore not immunogenic. Viral miRNAs have been identified in nuclear DNA viruses, such as polyoma viruses SV40 and SA12, and in many members of the herpesvirus family. As with their cellular counterparts, the functions of most viral miRNAs await elucidation, yet several themes are beginning to emerge (Fig 2).
Figure 2.
Cellular and viral microRNAs and their targets. See text for details. The curved lines indicate orthology of a viral miRNA to a cellular miRNA. miRNA, microRNA.
Regulation of cellular gene expression. Cytomegalovirus miR-UL122, for example, downregulates the major histocompatibility complex class I-related chain B, which is the ligand of the natural killer cell-activating receptor NKG2D, resulting in reduced killing by natural killer cells and thereby providing a mechanism for immune evasion (Stern-Ginossar et al, 2007).
Regulation of viral gene expression. The SV40 miRNA is fully complementary to the early viral transcript and, through RNAi, regulates expression of the T antigens, rendering infected cells less susceptible to lysis by cytotoxic T cells (Sullivan et al, 2005). B. Cullen (Durham, NC, USA) reported that viral miRNAs within or adjacent to the herpes simplex virus-1 non-coding latency-associated transcript (LAT) gene contribute to viral latency by targeting infected cell protein 0 (ICP0) and ICP4, which are important for the reactivation of latent herpesvirus infection. One of these miRNAs, miR-H2-3, is atypical in that its target site is located in the coding region of ICP0. Furthermore, although perfectly complementary to its target, it acts by translational inhibition rather than by RNA cleavage through RNAi. The context of the target sequence seems to be responsible for this activity.
Regulation of host gene expression by orthology to a cellular miRNA. The Kaposi's sarcoma associated herpesvirus (KSHV) miR-K12-11 phenocopies the cellular miR-155 through a shared seed region, thereby exploiting a pre-existing cellular regulatory pathway in B cells that might contribute to KSHV-associated B-cell tumours (Gottwein et al, 2007).
Adenoviruses express high levels of virus-associated (VA) RNA I and II, which are important for inhibition of the PKR response. G. Akusjarvi (Uppsala, Sweden) highlighted the fact that these RNAs also function as substrates for Dicer, generating small RNAs (mivaRNAs) that are incorporated into a functional RISC (Andersson et al, 2005; Aparicio et al, 2006; Xu et al, 2007). P. Fortes (Pamplona, Spain) identified several targets for mivaRNAs using a combination of microarray analysis and bioinformatics; however, how these targets contribute to viral pathogenesis remains to be established.
Cellular miRNAs
Host miRNAs might indirectly influence viral replication by regulating the expression of an essential host factor (Fig 2). The cellular miRNAs miR-17-5p and miR-20a target the histone acetyltransferase P300/CBP-associated factor (PCAF), which is a cofactor for the HIV-1 transactivator Tat, thereby inhibiting viral replication. In turn, HIV-1 infection specifically inhibits the expression of these miRNAs (Triboulet et al, 2007). Benkirane reported that knockdown of Dicer and Drosha, and of the P-body components RCK/p54 and GW182, increases viral replication in HIV-1-infected lymphocytes. Furthermore, a physical association of viral RNA with RCK/p54, Dcp1a and Argonaute 2 points to a direct interaction between HIV-1 and P bodies.
Do cellular miRNAs directly target viral genomes? HIV-1 infection results in major changes in the cellular miRNA profile (Triboulet et al, 2007; Yeung et al, 2005). J. Haasnoot (Amsterdam, The Netherlands) reported that many miRNA target sites are predicted to cluster in the HIV-1 genome. The relevance of these target sites in the viral life-cycle is under investigation. Cellular miRNAs might have a role in the post-transcriptional latency of HIV-1 in resting CD4+ T cells (Huang et al, 2007). Furthermore, miR-32 mediates the translational inhibition of PFV-1 (Lecellier et al, 2005). Similarly, miR-24 and miR-93 inhibit the replication of VSV by targeting the viral L and P genes (Otsuka et al, 2007). Finally, it has been shown that part of the antiviral activity of interferon-β is mediated by upregulation of miRNAs with direct targets in the HCV genome (Pedersen et al, 2007).
It is unclear, however, why functional miRNA target sites are retained in viruses (especially in fast-replicating viruses that establish high viraemia, such as HIV-1, HCV and VSV), as viruses readily escape from miRNA suppression through single mismatches. Indeed, viruses in which the miRNA target sites were mutated were viable in vitro (PFV-1; Lecellier et al, 2005), and replicated to higher titres with increased pathogenicity in mice (VSV; Otsuka et al, 2007).
HCV provides another notable example of the interplay between virus and cellular miRNAs, as this virus depends on a direct interaction with the liver-specific miR-122 for replication through an unknown mechanism (Jopling et al, 2005).
RNAi as an antiviral therapy
Treatment options for many important viral pathogens are lacking, but are urgently needed. RNAi-based antiviral therapy holds promise for the treatment of many viral diseases. Such nucleic acid-based therapy can be extremely versatile, as it can be adapted to target different virus strains, subtypes or species by merely changing the nucleotide sequence of the therapeutic siRNA. Numerous in vitro and animal studies have provided proof-of-concept: major viral pathogens, including HIV-1, respiratory viruses such as influenza and respiratory syncytial virus (RSV), and hepatitis viruses, have been shown to be susceptible to siRNA treatment (van Rij & Andino, 2006).
Major hurdles to therapeutic applications of RNAi include the delivery of the siRNA into the target organ and across cellular membranes into the relevant cell types, the bioavailability and stability of siRNAs, and the prevention of off-target effects and toxicity. These issues are being addressed through chemical alterations of the therapeutic RNA by backbone modifications, 2′ sugar modifications and the use of specific delivery methods—such as cholesterol conjugation, and formulation into liposomes and in complex with cationic polymers—as discussed by J. Kjems (Aarhus, Denmark), J. Kaufmann (Berlin, Germany) and S. Moschos (London, UK). M. Kay (Stanford, CA, USA) reported that toxicity in mice arising from saturation of endogenous RNAi/miRNA pathways by adeno-associated virus-mediated delivery of short-hairpin RNA (shRNA) could be reduced by optimizing the shRNA dose and sequence, and, under experimental conditions, by the overexpression of Exportin 5 and Argonaute 2.
Another hurdle is the fact that viruses readily escape from RNAi suppression by single mutations, deletions of the target sites and mutations outside the target sites. O. ter Brake (Amsterdam, The Netherlands) reminded us that the simultaneous use of multiple siRNAs successfully delays or even prevents viral escape (Gitlin et al, 2005; Schubert et al, 2005; ter Brake et al, 2006). Alternatively, viral untranslated regions (UTRs) are often highly conserved and are therefore attractive targets for antiviral therapy that might not allow viral escape; however, structural elements in UTRs might limit the accessibility to RISC, as noted by J. Kurreck (Stuttgart, Germany) and J. Kjems (Aarhus, Denmark). E. Pauls (Badalona, Spain) and S. Pinkert (Berlin, Germany) noted that targeting host factors known to be involved in viral replication, such as entry receptors or other cofactors, might be an alternative approach to limit viral escape. Genome-wide RNAi is a powerful approach to map the global networks of genes that affect virus replication and to identify cellular targets for antiviral therapy, as discussed by Jeang, C. Coyne (Pittsburgh, PA, USA) and A. Karlas (Berlin, Germany).
G. Hartmann (Bonn, Germany) is evaluating the antiviral potential of triphosphorylated siRNAs, which combine RNAi activity with the immunostimulatory properties of Toll-like receptor 7 (TLR7) and retinoic acid-inducible gene I (RIG-I) activation in an in vivo model for hepatitis B virus.
Transduction of haematopoietic stem cells (HSCs) with HIV-1-specific shRNAs generates a pool of CD4+ T cells that are resistant to HIV-1. Ter Brake reported the successful transduction of human fetal liver CD34+ cells with a lentiviral vector containing anti-HIV shRNAs, and the subsequent transplantation into recombination-activating gene (RAG)−/− mice. Mature transduced CD4+ T cells are tested ex vivo for their HIV-1 susceptibility. A clinical trial has recently started to evaluate the safety of autologous HSC transplantation after transduction with a lentivirus expressing an anti-HIV-1 shRNA in combination with a ribozyme targeting the coreceptor, chemokine (C-C motif) receptor 5 (CCR5), and a TAR decoy RNA in acquired immune deficiency syndrome (AIDS)-related lymphoma patients (J. Rossi, personal communication).
Conclusion
Despite the enormous progress made in recent years, important questions remain. Do the miRNA and RNAi pathways provide antiviral immunity in mammals? What is the full regulatory potential of viral miRNAs? Will RNAi live up to its promise as an antiviral therapy? A clinical trial evaluating the efficacy of siRNA treatment of RSV infection in humans (DeVincenzo et al, 2008) will shed some light on whether the current optimism about antiviral applications of RNAi is justified.
Finally, there was a reminder that more surprises lie ahead, when O. Voinnet (Strasbourg, France) reported an RNAi-based synergism between a bacterium and a virus. The Pseudomonas syringae bacterium injects, through its type III secretion system, suppressors of the RNAi and miRNA pathways, and thereby interferes with the miRNA-regulated defence responses of the plant host. This might be a quirk of the RNA-silencing system of the plant, in which, unlike in animals, the antiviral RNAi and miRNA pathways converge at the slicer Argonaute 1. Yet, it might also indicate that RNA-silencing pathways have a broader function in basal defence than previously anticipated, and act as central regulators of immune responses.

Ronald P. van Rij
Acknowledgments
I thank Jens Kurreck and Ben Berkhout for organizing a stimulating meeting. I thank the meeting participants for giving permission to cite their work and for helpful suggestions, and apologize to those colleagues whose work could not be discussed or referenced due to space limitations. I also thank Carla Saleh, Marco Vignuzzi and Ben Berkhout for critical reading of the manuscript. The illustration on the meeting poster was produced by Ebbe Sloth Andersen. R.P.v.R. is funded by a fellowship from the Nijmegen Centre for Molecular Life Sciences.
References
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G (2005) Suppression of RNA interference by adenovirus virus-associated RNA. J Virol 79: 9556–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P (2006) Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 80: 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT (2005) Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22: 607–619 [DOI] [PubMed] [Google Scholar]
- Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T (2000) Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol 74: 6203–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Kavi HH (2008) Slicing and Dicing for small RNAs. Science 320: 1023–1024 [DOI] [PubMed] [Google Scholar]
- Blakqori G et al. (2007) La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol 81: 4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Pita J, Fauquet CM (2004) Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virol 78: 7465–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen HS et al. (2007) Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J Virol 81: 5121–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- DeVincenzo J et al. (2008) Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antiviral Res 77: 225–231 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL (2006) Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat Immunol 7: 590–597 [DOI] [PubMed] [Google Scholar]
- Gitlin L, Stone JK, Andino R (2005) Poliovirus escape from RNAi: siRNA-target recognition and implications for therapeutic approaches. J Virol 79: 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E et al. (2007) A viral microRNA functions as an orthologue of cellular miR-155. Nature 450: 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B (2007) The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Huang J et al. (2007) Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Jopling C, MinKyung Y, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of Hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F (2007) HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 8: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560 [DOI] [PubMed] [Google Scholar]
- Li WX et al. (2004) Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 101: 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichner Z, Silhavy D, Burgyan J (2003) Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J Gen Virol 84: 975–980 [DOI] [PubMed] [Google Scholar]
- Lin J, Cullen BR (2007) Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 81: 12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J (2005) Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol 79: 7812–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M et al. (2007) Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27: 123–134 [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P (2008) Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 36: 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449: 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S et al. (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE (2004) RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res 102: 65–74 [DOI] [PubMed] [Google Scholar]
- Schubert S, Grunert HP, Zeichhardt H, Werk D, Erdmann VA, Kurreck J (2005) Maintaining inhibition: siRNA double expression vectors against coxsackieviral RNAs. J Mol Biol 346: 457–465 [DOI] [PubMed] [Google Scholar]
- Soldan SS, Plassmeyer ML, Matukonis MK, Gonzalez-Scarano F (2005) La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J Virol 79: 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N et al. (2007) Host immune system gene targeting by a viral miRNA. Science 317: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D (2005) SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686 [DOI] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M, Berkhout B (2006) Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 14: 883–892 [DOI] [PubMed] [Google Scholar]
- Triboulet R et al. (2007) Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Valli A, Dujovny G, Garcia JA (2008) Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from cucumber vein yellowing ipomovirus. J Virol 82: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Andino R (2006) The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol 24: 186–193 [DOI] [PubMed] [Google Scholar]
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20: 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW (2006) RNA interference directs innate immunity against viruses in adult Drosophila. Science 312: 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Segerman B, Zhou X, Akusjarvi G (2007) Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J Virol 81: 10540–10549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT (2005) Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology 2: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Vakharia VN, Wu LP (2006) RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol 8: 880–889 [DOI] [PubMed] [Google Scholar]