Abstract
Cytoplasmic localization and localized translation of messenger RNAs contribute to asymmetrical protein distribution. Recognition of localized mRNAs by RNA-binding proteins can occur in the cytoplasm or, alternatively, co- or post-transcriptionally in the nucleus. In budding yeast, mRNAs destined for localization are bound by the She2 protein before their nuclear export. Here, we show that a specific transcript, known as ASH1 mRNA, and She2 localize specifically to the nucleolus when their nuclear export is blocked. Nucleolar She2 localization is enhanced in a She2 mutant that cannot bind to RNA. A fusion protein of the amino terminus of She3 and She2 (She3N-She2) fails to enter the nucleus, but does not impair ASH1 mRNA localization. Instead, these cells fail to distribute Ash1 protein asymmetrically, which is caused by a defective translational control of ASH1 mRNA. Our results indicate that the nucleolar transit of RNA-binding proteins such as She2 is necessary for the correct assembly of translationally silenced localizing messenger ribonucleoproteins.
Keywords: mRNA localization, nucleolus, Saccharomyces cerevisiae, SHE2, translational control
Introduction
Cytoplasmic localization and the localized translation of messenger RNAs contribute to asymmetrical protein distribution in cell differentiation or neuronal function (St Johnston, 2005; Palacios, 2007). Recognition of localized mRNAs by RNA-binding proteins can occur in the cytoplasm or, in the case of nucleocytoplasmic shuttling proteins, co- or post-transcriptionally in the nucleus (Oleynikov & Singer, 2003; Kress et al, 2004; Giorgi & Moore, 2007). Furthermore, localized viral RNAs not only require nucleocytoplasmic shuttling of their corresponding binding proteins, but also their passage through nuclear subcompartments such as the nucleolus (Hiscox, 2007; Kim et al, 2007a).
In the budding yeast Saccharomyces cerevisiae, more than 30 mRNAs are localized to the bud tip, and among these ASH1 mRNA encodes a transcriptional repressor that controls mating-type switching (Shepard et al, 2003; Cosma, 2004; Aronov et al, 2007). These mRNAs are recognized by the RNA-binding protein She2, which can shuttle between the cytoplasm and the nucleus (Böhl et al, 2000; Kruse et al, 2002; Niessing et al, 2004). Once exported from the nucleus, the She2–RNA complex associates with a motor protein complex containing the class V myosin Myo4 and She3 (Böhl et al, 2000; Long et al, 2000), and is subsequently transported along actin filaments to the bud tip. During transport, the mRNA is translationally silenced by the action of two additional RNA-binding proteins, Khd1 (Irie et al, 2002; Paquin et al, 2007) and Puf6 (Gu et al, 2004).
Here, we show that nuclear shuttling of She2 is required for correct translational control of the mRNA, but not for mRNA localization. Furthermore, She2 and its target mRNA ASH1 accumulate in the yeast nucleolus when their nuclear export is blocked, indicating that nucleolar passage is required for the correct assembly of a localizing ribonucleoprotein (RNP) complex.
Results And Discussion
She2 accumulates in nucleoli during mRNA export block
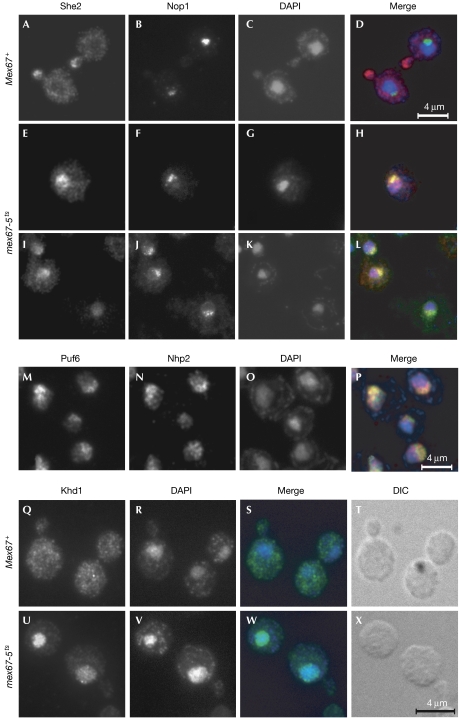
Previous experiments have shown that blocking mRNA export in a temperature-sensitive mex67-5 yeast mutant results in nuclear accumulation of Myc-tagged She2 protein (Kruse et al, 2002). When repeating these experiments with an antibody raised against recombinant She2, we observed the typical bud-tip localization of She2 in wild-type cells (Mex67+; Fig 1A). Strikingly, in Mex67-5 cells at 37°C, untagged She2 was confined to a subregion of the nucleus. Most of the She2 protein was found in a region opposite the 4,6-diamidino-2-phenylindole (DAPI)-stained chromatin-rich material (Fig 1E,G,I,K). Co-staining with an antibody against the nucleolar marker protein Nop1 (Fig 1B,F,J; Aris & Blobel, 1988) showed that She2 accumulated at sites overlapping with the characteristic crescent-shaped nucleoli (Fig 1H,L). These data indicate that She2 accumulates in nucleoli when mRNA-mediated export is blocked.
Figure 1.
She2 but not Khd1 accumulates in nucleoli when messenger RNA export is inhibited. (A–L) She2 localization. Representative cells of (A–D) wild-type (Mex67+) and (E–L) mex67-5ts strains that were grown for 30 min at 37°C and co-stained for She2 and the nucleolar marker protein Nop1 are shown. Nuclei were stained with DAPI. Wild-type cells show normal localization of She2 at the bud tip (A–D), whereas in mex67-5 cells the signals of She2 and Nop1 largely overlap (H,L, yellow signal). (M–P) Puf6 localization. Puf6-Myc9 in a wild-type strain is detected in a crescent–shaped region and colocalizes with nucleolar Nhp2. (Q–X) Khd1 localization. (Q–T) Staining of Khd1-HA6 shows cytoplasmic distribution of Khd1 in wild-type cells. In mex67-5 cells, Khd1-HA6 accumulates in nuclei when shifted to 37°C. Note that, in contrast to She2, nuclear Khd1-HA6 largely overlaps with the DAPI-stained region (W). DAPI, 4,6-diamidino-2-phenylindole; DIC, differential interference contrast.
To investigate whether other proteins that bind to localized mRNAs shuttle between the nucleolus and the cytoplasm, we investigated the localization of Puf6 and Khd1 that were epitope-tagged at the carboxyl terminus. Even in a wild-type background, Puf6-Myc9 was heavily enriched in nucleoli (Fig 1M), which is consistent with its second proposed function in ribosome biogenesis that occurs in the nucleolus (Nissan et al, 2002). By contrast, Khd1-HA6 was cytoplasmic under normal growth conditions (Mex67+; Fig 1Q). The blocking of mRNA export (mex67-5) led to the accumulation of Khd1-HA6 in nuclei (Fig 1U) but not in nucleoli, indicating that this protein shuttles between the nucleoplasm and the cytoplasm. A similar behaviour has been reported for Npl3, a shuttling mRNA export factor (Thomsen et al, 2003). In conclusion, nucleolar shuttling of She2 is specific, indicating that localization of She2 to the nucleolus could be of functional significance.
She2 binds to double-stranded RNA that contains stem loops in its target mRNAs (Niessing et al, 2004). Thus, it seems that nuclear-trapped She2 is attracted to sites with high concentrations of structured RNAs such as pre-ribosomal RNAs in the nucleolus. To investigate this possibility, we generated a mutant of She2 (N36S and R63K) that was unable to bind to and localize ASH1 mRNA (Gonsalvez et al, 2003). Recombinant wild-type and She2-N36S-R63K mutant proteins were used in filter-binding assays to assess binding to a specific RNA target (E3 localization element of ASH1 mRNA) or to an unspecific transactivating response region (TAR) RNA stem loop from human immunodeficiency virus (HIV; Niessing et al, 2004). Although wild-type She2 bound to ASH1 E3 with an equilibrium dissociation constant (KD) of 0.14 μM, binding to TAR was 6.5-fold lower (Fig 2A). By contrast, the She2-N36S-R63K mutant protein did not show significant binding to ASH1 E3 mRNA or to TAR RNA. Expression of She2-N36S-R63K from its endogenous promoter in she2Δ yeast cells showed that the mutant protein is enriched in nucleoli (Fig 2B), even when mRNA export is not blocked. This finding indicates that She2 enters the nucleolus but becomes trapped in this subcompartment if it fails to bind to RNA. A similar finding has been reported for the mammalian Staufen 2 protein that is involved in cytoplasmic mRNA localization in neurons. Staufen 2 mutants that are unable to bind to RNA also accumulate in nucleoli (Macchi et al, 2004), which suggests a shuttling of both proteins between the cytoplasm and the nucleolus.
Figure 2.
An RNA-binding mutant of She2 accumulates in the nucleolus. (A) Filter-binding analysis of the ASH1 E3 localization element (left) or unrelated human immunodeficiency virus type I (HIV-I) TAR RNA (right) to She2 (black) and mutant She2-N36S-R63K (red). Binding constants (KD) of wild-type She2 to ASH1 E3 RNA and HIV-I TAR RNA are shown. Mutant She2-N36S-R63K showed no significant binding to either RNA. (B) Representative cells of strain RJY2838 expressing the She2-N36S-R63K mutant were stained for She2 and Nop1. She2-N36S-R63K mutant (left panel) accumulates in a region that overlaps with Nop1 signals (right panel; merge), indicating nucleolar accumulation of She2-N36S-R63K.
ASH1 mRNA can accumulate in nucleoli
If nucleolar accumulation of She2 during inhibition of mRNA export reflects a trapped intermediate during the assembly or maturation of localized messenger RNPs (mRNPs), we should also detect a She2 target mRNA in the nucleolus. This assumption is supported by the finding that nucleolar accumulation of a reporter mRNA can be achieved by the insertion of the ASH1 E3 localization element (Brodsky & Silver, 2000). Therefore, we determined the localization of endogenous ASH1 by fluorescent in situ hybridization in wild-type (Mex67+) and mex67-5 mutant cells. In wild-type cells, ASH1 is seen at the bud tip and in an intense fluorescent intranuclear spot that might reflect the genomic site of ASH1 transcription; this spot never overlapped with the nucleolar marker ITS2 (internal transcribed spacer 2; Fromont-Racine et al, 2003; supplementary Fig 1 online). Blocking of mRNA export in mex67-5 cells resulted in the loss of ASH1 at the bud tip without affecting the nuclear spot. The persistent spot might reflect the retention of ASH1 transcripts at the transcription site, as observed for other mRNAs when mRNA export is blocked (Jensen et al, 2001). As this RNA retention requires the function of the nuclear exosome, we disrupted the gene that encodes the nuclear exosome-specific subunit Rrp6 (ribosomal RNA processing 6; Hilleren et al, 2001). This resulted in colocalization of ASH1 mRNA with ITS2 in the nucleolus (supplementary Fig 1C online), which is in contrast to bulk poly A+ RNA that has been detected at the nuclear periphery in Δrrp6 mex67-5 cells at 37°C (Thomsen et al, 2003). Nucleolar localization of ASH1 requires She2, as deletion of SHE2 in Δrrp6 mex67-5 cells blocks nucleolar accumulation of this mRNA (supplementary Fig 1D online). These results indicate that not only She2 but also ASH1 mRNA can be trapped in the nucleolus, which might reflect an intermediate step necessary for later cytoplasmic transport.
She2 can be retained in the cytoplasm
To investigate why She2 might traverse the nucleolus, we tethered She2 to a cytoplasmic protein that blocks its entry into the nucleus. Out of several fusion partners tested, only the fusion of She2 C-terminally to the N-terminal domain of She3 did not interfere with She2 localization, suggesting that cytoplasmic transport of this fusion is not affected (Fig 3; data not shown). The N-terminal She3 domain binds strongly to the actin-associated Myo4 myosin motor protein (Böhl et al, 2000). By means of the interaction of Myo4 with the cytoskeleton, the resulting protein complex should be effectively retained in the cytoplasm. The She3N–She2 fusion protein (She3N–S2), if expressed in she2Δ cells, can be efficiently immunoprecipitated by She2 antibodies (Fig 3B, arrow in the right panel). Co-immunoprecipitation and reverse transcription–PCR (RT–PCR) analysis showed that She3N–S2 binds to ASH1 mRNA with an efficiency similar to that of She2 (Fig 3C). Furthermore, She3N–S2 localizes to the bud in wild-type cells at 25 and 37°C (Fig 3D, upper panels), indicating that it behaves in a similar manner to She2. However, in contrast to She2, She3N–S2 is retained in the cytoplasm when mRNA export is blocked (Fig 3D, lower panels). Thus, She3N–S2 is a useful tool for investigating the significance of nuclear or nucleolar shuttling of She2.
Figure 3.
RNA transport mediated by a She3N–She2 fusion protein results in inefficient sorting of Ash1. (A) Schematic diagram of the She3N–She2 (She3N–S2)fusion protein. Numbers indicate amino acids used of the corresponding protein part. (B) Immunoprecipitation of She2 and She3N–S2 using She2 antibody. She2 (white arrowhead) or the She3N–S2 fusion protein (black arrow) were detected at similar levels in total extracts (T) and immunopellets (P) of a wild-type strain or strain RJY2414 (She3N–S2). The asterisk indicates the heavy chain of the antibody that co-eluted with the antigen. S, supernatant. (C) Immunoprecipitates were subjected to reverse transcription–PCR and analysed for bound ASH1 RNA. SDS E, elution with 10% SDS; G E, elution with 100 mM glycine (pH 2.5), −RT, control lacking reverse transcriptase. ASH1 co-precipitated with She2 and She3N–S2, but not in the absence of She2 (Δshe2). (D) Cytoplasmic retention of She3N–S2 after inhibition of mRNA export. In strain RJY2422 (Mex67 She3N–S2), She3N–S2 localized to the bud tip (upper panel). Nucleolar accumulation of She2 (middle panels) but not She3N–S2 (lower panels) was observed in mutant mex67-5 cells. (E) Cytoplasmic retention of She2 did not interfere with mRNA localization. In situ hybridization in strains expressing She2 (wild type) or She3N–S2 indicated normal ASH1 localization (left panels). ASH1 mRNA localization efficiency was quantified from 150 binucleate cells (right panel). The black columns show category percentage for wild type and grey columns for She3N–S2. DAPI, 4,6-diamidino-2-phenylindole; DIC, differential interference contrast.
Retention of She2 affects ASH1 translation
If nucleolar shuttling of She2 is required for correct mRNA localization, she2Δ cells expressing the She3N–S2 fusion protein should show defects in ASH1 mRNA localization. However, SHE3N–S2 cells localized ASH1 almost as efficiently as wild-type cells (70% versus 76% of anaphase cells with ASH1 at the bud tip; Fig 3E). In contrast to mRNA localization, asymmetrical distribution of Ash1 protein was strongly affected in these cells. In wild-type post-anaphase (binucleate) cells, 36% carried Ash1-Myc9 only in the daughter cell nucleus (Fig 4, ‘asymmetrical') and 33% had a much stronger signal in the daughter cell nucleus than the mother cell nucleus (‘partly asymmetrical'). In SHE3N–S2 cells, the number of cells with an asymmetrical Ash1 distribution was reduced to 20%, whereas the number of cells with partly asymmetrical or symmetrical Ash1 distribution increased concomitantly (Fig 4B, grey columns). An additional fusion of a tandem SV40 nuclear localization signal (NLS) N-terminally to the She3N–S2 protein reversed the loss of asymmetrical Ash1-Myc9 distribution (Fig 4B, white columns). The observed differences are statistically significant, as determined by a Pearson's χ2 test (P-values of 0.02 in the case of SHE3N–S2 versus wild type, and 0.002 in the case of SHE3N–S2 versus NLS-SHE3N–S2). Our findings support a model in which nuclear shuttling of She2 is required for asymmetrical Ash1 distribution and argues against the possibility that an N-terminal fusion of She3 to She2 simply interferes with the association of a putative cytoplasmic cofactor.
Figure 4.
Loss of asymmetrical Ash1 distribution in She3N–S2 cells. (A) Cellular distribution of nuclear Myc-tagged Ash1 was categorized as asymmetrical, partly asymmetrical or symmetrical. (B) Ash1-Myc9 distribution in a wild-type strain (She2; black columns), an She3N–S2 strain (She3N–She2; grey columns) and a strain expressing an She3–She2 fusion protein containing a tandem nuclear localization signal at the N terminus (2 × NLS-She3N–She2; white columns) was quantified by counting 351 binucleate cells. DAPI, 4,6-diamidino-2-phenylindole; NLS, nuclear localization signal.
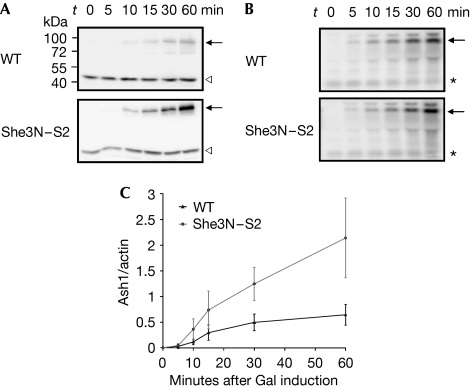
Loss of Ash1 asymmetry despite correct mRNA localization might be caused by impaired translational repression during cytoplasmic ASH1 transport, resulting in Ash1 protein in the mother cell. To test for premature translation in SHE3N–S2 cells, we compared the rates of Ash1-Myc9 protein synthesis with rates in wild-type cells. Ash1-Myc9 protein levels were significantly enhanced in SHE3N–S2 cell extracts even at early time points of induction (Fig 5A,C), whereas no difference in ASH1-Myc9 mRNA synthesis was detected (Fig 5B). A similar premature translation of Ash1-Myc9 was observed in cells deleted for the nucleolar proteins Loc1 and Puf6, which have also been implicated in translational control of ASH1 mRNA (supplementary Fig 2 online; Gu et al, 2004; Komili et al, 2007). We conclude from our data that She2 must enter the nucleus and most probably the nucleolus for the correct assembly of a translational repressor complex on ASH1 mRNA.
Figure 5.
Accelerated Ash1 synthesis in cells expressing She3–She2 fusion protein. After induction of Ash1-Myc9 from a GAL1 promoter in wild-type (WT) or She3N–S2 cells, samples were taken at the indicated time points and Ash1/actin protein ratios were determined by western blot analysis. (A) She3N–S2 cells show an accelerated Ash1 synthesis (arrow) compared with wild-type cells; the white arrowheads indicate actin signal. (B) Increase in ASH1 mRNA levels was similar in wild-type and She3N–S2 cells. ASH1 levels (black arrows) were determined by northern blot analysis and normalized against intensities of an unspecific crossreacting RNA (asterisks). (C) Plot of average Ash1-Myc9/actin ratios from three independent experiments.
The nucleolus is primarily implicated in ribosomal RNA processing and subunit assembly, but other functions such as ribonucleoprotein particle biogenesis have recently been described (Boisvert et al, 2007). So far, a functional significance of the nucleolar transit of shuttling RNA-binding proteins has been reported only for viral RNPs or non-mRNA-containing RNPs such as the signal recognition particle (SRP; Grosshans et al, 2001; Hiscox, 2007). Long-distance movement of specific plant viral RNPs requires passage through the nucleolus for their correct maturation (Hiscox, 2007). SRP assembly requires import of SRP core proteins into the nucleolus where they assemble with the 7S RNA component (Grosshans et al, 2001). Similar to ASH1 and She2, the 7S RNA component can be trapped only in the nucleolus of mutant cells, and not in the nucleolus of wild-type cells. This suggests that, in both cases, the assembly and exit from the nucleolus occur too rapidly to allow detection under steady-state conditions.
If nucleolar transit is important, how could prevention of nuclear entry of She2 lead to improper translational control? Blocking nucleolar import of the open reading frame 3 (ORF3) protein of a plant umbravirus prevents the correct formation of viral RNPs (Kim et al, 2007a). ORF3-mediated nucleolar transit of the viral RNA is required for the integration of an authentic nucleolar protein, fibrillarin, into the RNP complex and subsequent systemic transport (Kim et al, 2007b). Interestingly, similar to fibrillarin, yeast Puf6 is a nucleolar protein and also a component of the ASH1 mRNP that binds to She2 in vivo and is required for translational control (Gu et al, 2004; Deng et al, 2008). We propose that the nucleolar transit of She2 might be required to correctly assemble Puf6 into the ASH1 mRNP for full translational repression of ASH1 during transport.
The examples provided by the shuttling RNA-binding proteins Staufen 2 (Macchi et al, 2004) and She2 suggest that the nucleolus has a role in the assembly of cytoplasmic mRNP localization complexes. Similar to the nucleoplasm (Giorgi & Moore, 2007), the nucleolus might be a transit station to nurture localizing RNP complexes and to prime them for translational regulation during cytoplasmic transport.
Methods
Fluorescence microscopy. Indirect immunofluorescence microscopy and in situ hybridization against ASH1 mRNA were performed as described previously (Long et al, 1997; Böhl et al, 2000). For double staining of She2 and Nop1, yeast spheroplasts were sequentially incubated with She2 antibody, rabbit IgG antibody coupled to AlexaFluor®594, Nop1 antibody (MCA-38F3, Encor Biotechnology, Gainesville, FL, USA) and mouse IgG antibody coupled to AlexaFluor488. For double staining of Puf6-Myc9 and Nhp2 (Henras et al, 2001), primary antibodies were replaced by Myc (9E10) and Nhp2 antibodies. Dual in situ hybridizations were performed by using TexasRed- and AlexaFluor488-labelled antisense oligonucleotide probes against ASH1 (Long et al, 1997) and ITS2 (5′-T*AGGCCAGCAATTTCAAGT*AACTCCAAAGAGTATCACTC*-3′), respectively. The asterisks indicate the nucleotides modified for the attachment of AlexaFluor488 fluorochromes. Images were acquired by using Openlab software (Improvision, Tübingen, Germany), processed and mounted with Adobe Photoshop 10.
Immunoprecipitation. Co-immunoprecipitation of She2 and subsequent detection of ASH1 mRNA by RT–PCR was performed as described previously (Böhl et al, 2000). After removing an aliquot that acted as the input control (total), cell lysates were incubated for 90 min with purified She2 antibodies that had been coupled to magnetic protein G beads. These She2 antibody beads had been blocked at 4°C with 0.5% BSA, 200 μg/ml insulin and 0.1 mg heparin. She2 and associated components were sequentially eluted with glycine (pH 2.5) and 1% SDS. One-third of the eluate was used to detect She2 and the rest was extracted with phenol–chloroform, treated with DNase and subjected to RT–PCR analysis. ASH1 RNA was detected with oligonucleotides 5′-TACATGGATAACTGAATCTC-3′ and 5′-CAGGATGACCAATCTATTGC-3′.
In vivo ASH1 translation assay. Yeast strains were grown in 2% raffinose-containing medium overnight at 30°C, diluted and grown to an OD600=0.8–1.0 before ASH1 expression was induced by the addition of 4% galactose (w/v). Cells (20 OD600 units) from each culture were collected at regular intervals from 0 to 60 min and the cell pellets were frozen in liquid nitrogen. Total protein was extracted from 10 OD600 units of cells and protein concentration was determined. One OD280 unit of lysate was used for western blot analysis of She2 and actin. Signals were detected by chemiluminescence in a Fuji LAS-3000 mini and normalized against each other. To assess ASH1 mRNA levels, RNA was extracted from 10 OD600 unit of cells and ASH1 mRNA signal was detected by northern blotting and quantified by using a probe against the ASH1 ORF. Normalization of the ASH1 signal was performed against an unspecific crossreacting band recognized by the ASH1 probe.
A description of plasmid and yeast strain construction, two figures and three tables are given in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank F. Fuchs, S. Lange and A. Tresch for technical advice. This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB646-TP A5 and FOR855), the Helmholtz Association, Boehringer Ingelheim Fonds and the Elite Network of Bavaria.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aris JP, Blobel G (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol 107: 17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE (2007) mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol 27: 3441–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhl F, Kruse C, Frank A, Ferring D, Jansen R-P (2000) She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 19: 5514–5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA (2000) Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP (2004) Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep 5: 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W (2008) Translation of ASH1 mRNA is repressed by Puf6p–Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ (2007) The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol 18: 186–193 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Lehmann KA, Ho DK, Stanitsa ES, Williamson JR, Long RM (2003) RNA–protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA 9: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Deinert K, Hurt E, Simos G (2001) Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J Cell Biol 153: 745–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH (2004) A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 18: 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M (2001) Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 29: 2733–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Hiscox JA (2007) RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K, Herskowitz I (2002) The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J 21: 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M (2001) A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell 7: 887–898 [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JW, Taliansky M (2007a) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Macfarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JW, Taliansky M (2007b) Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad Sci USA 104: 11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL (2004) Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol 165: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, Ellenberg J, Jansen RP (2002) Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol 159: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P (2000) She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Brownawell AM, Grunewald B, DesGroseillers L, Macara IG, Kiebler MA (2004) The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem 279: 31440–31444 [DOI] [PubMed] [Google Scholar]
- Niessing D, Huttelmaier S, Zenklusen D, Singer RH, Burley SK (2004) She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell 119: 491–502 [DOI] [PubMed] [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH (2003) Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM (2007) How does an mRNA find its way? Intracellular localisation of transcripts. Semin Cell Dev Biol 18: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P (2007) Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell 26: 795–809 [DOI] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD (2003) Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA 100: 11429–11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D (2005) Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 6: 363–375 [DOI] [PubMed] [Google Scholar]
- Thomsen R, Libri D, Boulay J, Rosbash M, Jensen TH (2003) Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information