The Keystone Symposium on Forkhead Transcription Factor Networks in Development, Signalling and Disease took place between 13 and 17 January 2008, at the Zermatt Resort and Spa in Midway, Utah, USA, and was organized by K. Zaret, K. Kaestner and S. Mango.
Introduction
Subjects ranging from chromatin remodelling to ageing, from human disease to avian song, and from metabolism and the cell cycle to structural biology and differentiation were all covered at this keystone symposium, and illustrate the diversity of forkhead transcription factors and their regulatory functions. Forkhead transcription factors, or ‘FOX' proteins, are unified as a family of more than 40 conserved proteins, each of which contains a DNA-binding domain of approximately 110 amino acids dubbed the ‘forkhead box'. The members of the family are subdivided alphabetically and numerically into subfamilies based on evolutionary conservation (Kaestner et al, 2000). Forkhead factors have essential roles in regulatory networks that are involved in development, signalling, metabolism and human disease. Here, we summarize the new information presented at the meeting, to emphasize the breadth of biological areas in which forkhead factors have crucial regulatory roles. We apologize to the many colleagues whose exciting work we could not include owing to space limitations.
Epigenetics and forkheads
The evolutionarily conserved forkhead box has been structurally characterized by both X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, and resembles a butterfly or ‘winged helix' that is reminiscent of the globular chromatin-binding domain of linker histones H1 and H5 (Clark et al, 1993; Weigelt et al, 2002). However, unlike linker histones, which compact the chromatin structure, binding by forkhead factors relieves chromatin compaction at target enhancer and/or promoter sites, allowing the entry of other transcriptional activators to induce target gene expression (Cirillo et al, 2002). Relatively little is known about the mechanisms that define how forkhead factors choose among highly similar binding sites in the genome, and, more importantly, the developmental and biological consequences of this choice.
The keynote address from M. Brown (Boston, MA, USA) emphasized the essential role that chromatin modifications have in specifying forkhead factor binding and steroid receptor-regulated gene expression. Genome-wide profiles of FOXA1 binding in MCF7 breast cancer cells have shown that the distribution of histone H3 lysine 4 di-methylation (H3K4me2) defines differential cell-specific FOXA1 recruitment sites in chromatin. Intriguingly, although H3K4me2 is required for the maintenance of FOXA1 binding, FOXA1 is not required for either the genesis or the maintenance of H3K4me2 (Lupien et al, 2008), which raises the question of how this marker is specified. Overlapping forkhead and oestrogen receptor binding motifs occur at the enhancers of more than 50% of all oestrogen receptor-responsive genes. Knockdown of FOXA1 in MCF7 cells leads to the loss of DNase hypersensitivity and oestrogen receptor binding at these enhancers, which generally lie at a considerable distance from core promoters (Carroll et al, 2005). The ability of FOXA1 to read a histone-modification signature and alter chromatin structure—allowing the subsequent binding of lineage-specific transcription factors—emphasizes how the expression and function of forkhead factors must be carefully regulated. P. Koeffler (Los Angeles, CA, USA) supported this concept with data on a growth-inhibitory role for FOXA1 in breast cancer, in which dysregulated levels of this transcription factor might act as a potential prognostic marker.
New work described by K. Zaret (Philadelphia, PA, USA) suggests that, once bound to chromatin, FOXA factors themselves might act as true epigenetic markers that remain stable throughout mitosis. Data from fluorescence recovery after photobleaching (FRAP) experiments show that FOXA1–green fluorescent protein (GFP) fusion proteins exhibit highly stable chromatin binding in vivo, approaching a level that rivals linker histones. Stable binding by FOXA1 during mitosis points to the tantalizing hypothesis that FOXA binding is an epigenetic marker of active chromatin, when other trans-acting factors are evicted. Stable chromatin binding during mitosis has been observed for both FOXC1 (M. Walter, personal communication) and FOXI1 (Yan et al, 2006), suggesting that this might be a general way in which forkhead factors potentiate sustained developmental regulatory control.
Stable mitotic binding might prove to be deleterious for forkhead factors that dictate inducible gene expression, as exemplified by the FOXO factors. These regulate target genes in glucose metabolism, stress resistance, cell-cycle arrest and DNA-damage response (Barthel et al, 2005; Birkenkamp & Coffer, 2003). Accordingly, dysfunction of these proteins has been proposed to have a role in ageing and disease. Protein kinase B (PKB)/AKT-mediated phosphorylation of FOXO factors inhibits FOXO-mediated transactivation by promoting cytoplasmic sequestration (Biggs et al, 1999). In addition, data presented by L. Cirillo (Milwaukee, WI, USA) suggest that acetylation regulates the stability of FOXO factors that are bound to chromatin. Cyclic acetylation and deacetylation of nuclear FOXO factors might regulate their ‘dwell time' on chromatin, potentially shifting the nuclear/cytoplasmic balance and their availability for inducible gene activation/repression.
Developmental redundancy resolved into adult specificity
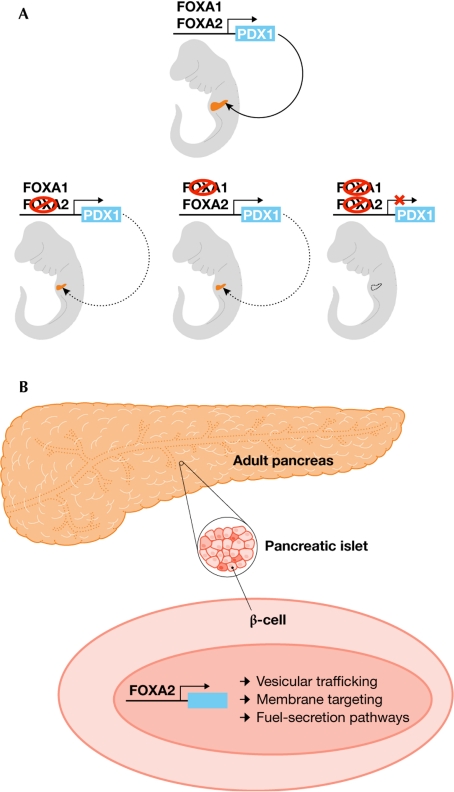
The forkhead proteins within a subfamily often show functional redundancy in controlling processes in the embryo, but have distinct functions in the adult organism—regulating metabolism, stress response, cell-cycle control or neuronal plasticity, for example. Data presented by K. Kaestner (Philadelphia, PA, USA) suggest that, although FOXA1 and FOXA2 show redundant and overlapping functions during early pancreatic development in the embryo, FOXA2 exhibits specific biological functions in the adult endocrine pancreas (Fig 1). Both FOXA1 and FOXA2 bind to the enhancer region of the gene encoding the pancreatic transcription factor PDX1, which is essential for pancreas development. Elimination of PDX1 expression requires the knockout of FOXA1 and FOXA2, and the resulting mice have no pancreas (Fig 1A). However, the situation changes in the adult endocrine pancreas, where function is solely dependent on FOXA2 (Fig 1B). Expression profiling identified many new targets of FOXA2 in islet cells, including genes involved in vesicular trafficking, membrane targeting and fuel-secretion pathways, which comprise a transcription network regulated by FOXA2 in mature β-cells (Gao et al, 2007).
Figure 1.
Embryonic compensation versus adult specificity. (A) Forkhead box A1 (FOXA1) and FOXA2 show overlapping and compensatory activation of the pancreatic transcription factor PDX1 expression during embryonic development of the pancreas. Loss of PDX1 expression and the development of mice lacking a pancreas require the ablation of both FOXA1 and FOXA2. (B) By contrast, FOXA2 alone exhibits specific biological functions in the adult endocrine pancreas and regulates a transcription network of genes in mature β-cells.
As in the case of β-cells of the endocrine pancreas, FOXA2 works together with FOXA1 during the embryonic specification of dopaminergic neurons. However, data presented by S.-L. Ang (London, UK) raise the intriguing possibility that the timing and duration of midbrain dopaminergic (mDA) neuron differentiation are regulated by the concentration of FOXA1/2 in these cells (Ferri et al, 2007). Loss-of-function studies in mice bearing different numbers of FoxA1/2 alleles suggest that FOXA1 and FOXA2 cooperate to regulate the differentiation of mDA neurons in a dosage-dependent manner, whereby FOXA1/2 proteins are required at progressively higher levels as the mDA cells mature.
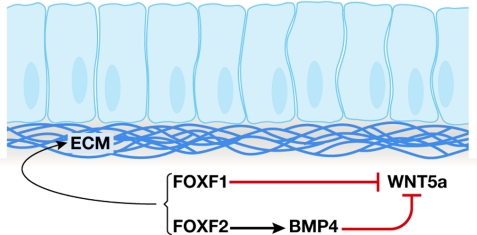
Similarly, the dosage of FOXF1 and FOXF2 factors has an important role in both development and proliferation in the intestinal epithelium (Fig 2). P. Carlsson (Goteborg, Sweden) showed that both of the FoxF genes are important for the development of the gut, where they promote the production of the extracellular matrix (ECM), and inhibit epithelial proliferation by reducing Wingless (WNT) and increasing bone morphogenetic protein (BMP) signalling from the surrounding mesenchyme. Tissue layers of the intestine disintegrate in FoxF2−/− mice (Ormestad et al, 2006). The importance of FOXF proteins as signalling hubs in the gut is illustrated by the influence of FOXF gene dosage. FOXF1 and FOXF2 block production of WNT5a by two distinct mechanisms, and inactivation of either one has dire consequences for the regulatory control of proliferation in intestinal epithelia.
Figure 2.
Forkhead factors working towards a common goal. Both forkhead box F1 (FOXF1) and FOXF2 are required for normal embryonic development and integrity of the intestine. FOXF1 and FOXF2 act by distinct and non-overlapping mechanisms, and together maintain intestinal epithelial integrity by promoting extracellular matrix (ECM) production through inhibition of Wingless 5a (WNT5a). In the absence of FOXF2, specific layers of the gut disintegrate. BMP4, bone morphogenetic protein 4.
Forkhead factors: a crucial balance
Human genetic disorders have been attributed to mutations in genes encoding forkhead transcription factors, further highlighting how the expression and function of these factors must be tightly controlled. In exciting work that established a strong correlation between elevated FOXA2 expression in dopaminergic neurons and Parkinson disease, R. McKay (Bethesda, MD, USA) showed that the correct dosage of FOXA2 is essential for the maintenance of adult dopaminergic neurons. Intriguingly, aged FoxA2-heterozygous mice spontaneously develop deficits in motor skills, which are accompanied by an asymmetric loss of motor neurons from the substantia nigra, similar to that seen in Parkinson disease (Kittappa et al, 2007). Families with a high incidence of Parkinson disease carry single-nucleotide polymorphisms (SNPs) mapping to the FOXA2 gene. The potential treatment of dopamine neuron disease might therefore lie in controlling FOXA2 expression through stem cell-based and/or pharmacological approaches.
S. Fisher (Oxford, UK) and colleagues previously discovered a heterozygous mutation of FOXP2 in a family in which members suffered from a severe speech and language disorder that involved problems with learning and produced complex orofacial movements. At the meeting, Fisher and colleagues showed that heterozygous mice carrying the equivalent mutation had abnormal synaptic plasticity in the neural circuits that mediate motor-skill learning (Groszer et al, 2008). C. Scharff (Berlin, Germany) underscored the role of FOXP2 in learned vocalizations, complete with digitized reproductions of bird song. Juvenile male zebra finches learn highly structured and stereotyped songs from adult conspecifics at a time when they also show increased FOXP2 expression in Area X, which is a basal ganglia structure necessary for song acquisition (Haesler et al, 2004). RNA interference (RNAi)-depletion of FOXP2 in Area X of juvenile finches led to incomplete and inaccurate imitations of the tutor finch. These data are consistent with a conserved role of FOXP2 in the neural circuits mediating motor learning, which might have been recruited for vocal learning in songbirds and humans.
M. Walter (Edmonton, Canada) continued the theme of the crucial dosage of forkheads. Walter previously linked mutations in the human FOXC1 gene with Axenfeld–Rieger syndrome, which is an autosomal-dominant malformation of the eye associated with glaucoma (Mears et al, 1998). Notably, many FOXC1 mutations observed in Axenfeld–Rieger syndrome prevent post-translational modifications or protein–protein interactions that normally keep FOXC1 silent. Genes that are involved in stress responses are direct targets of FOXC1 in ocular cells, including FOXO1a, which regulates the cell cycle, apoptosis and survival under oxidative stress (Birkenkamp & Coffer, 2003). Multiple lines of evidence indicate that the pathology associated with glaucoma might result, in part, from oxidative damage caused by FOXC1 dysfunction, and an imbalance between the generation and removal of reactive oxygen species.
Forkheads regulate cell-cycle progression
Forkhead factors function—in combination with other transcription regulatory proteins—to coordinate developmental and physiological processes. This is perhaps especially apparent for forkhead factors that regulate the cell cycle. In yeast, the G2/M phase-specific expression of 30 genes comprising the Cyclin B (Clb2) cluster is controlled by the yeast forkhead factors Fkh1 and Fkh2 (Wittenberg & Reed, 2005). J. Mellor (Oxford, UK) presented an intriguing mechanism by which the forkhead factors Fkh1 and Fkh2 regulate CLB2 transcription. Fkh2 activates transcription of the CLB2 gene, which encodes a B-type cyclin that is required during G2/M (Sherriff et al, 2007). During G1, Fkh2 represses CLB2 by promoting anti-sense transcription from the 3' end. Fkh1, along with the stress-response factor Centrosome binding factor 1 (Cbf1), induces sense transcription and represses anti-sense transcription induced by Fkh2. This sense/anti-sense tug-of-war is not restricted to CLB2, as Fkh1 and Fkh2 similarly regulate HMS2.
In mammalian cells, the forkhead factor FOXM1 controls progression into S phase by upregulating expression of the CDK2-activating phosphatase CDC25A, and downregulating protein levels of the CDK inhibitors p21CIP1 and p27KIP (Wang et al, 2005). H.-J. Park (Chicago, IL, USA)—a student of the late R. Costa, who largely defined the forkhead family—presented elegant work on the regulated stability of the FOXM1 protein. FOXM1 degradation is stimulated by its amino-terminal interaction with the anaphase-promoting complex (APC)/cyclosome (C) complex and its adaptor cadherin 1, type 1, E-cadherin (CDH1). Waves of ubiquitin-mediated proteolysis and the active degradation of FOXM1 in late mitosis and early G1 prevent entry into S phase. The precision in FOXM1 regulation is underscored by its role in hepatocellular carcinoma (Kalinichenko et al, 2004), supporting its importance in cell-cycle control.
Forkheads in metabolism and lifespan
Reduced food intake, which is known as dietary restriction, is associated with increased lifespan in worms, flies and mammals (van der Horst & Burgering, 2007); however, the genetic components that regulate the effect of dietary restriction on lifespan have remained elusive. A. Dillin (La Jolla, CA, USA) described an adult-specific function for the forkhead factor PHA-4 (FoxA) in the regulation of dietary restriction-mediated longevity in Caenorhabditis elegans. The role of pha-4 in lifespan determination is specific for dietary restriction, as it does not affect the extended longevity caused by other genetic pathways that regulate aging such as insulin/insulin-like growth factor 1 (IGF-1) signalling (IIS), abnormal dauer formation 16 (daf-16)/FoxO and reduced mitochondrial activity (Panowski et al, 2007). This is despite the fact that DAF-16 and PHA-4 bind overlapping sites in vitro, and Suppressor of mitogen-activated protein kinase 1 (SMK-1) might regulate both DAF-16 and PHA-4 to modulate the IIS and dietary restriction pathways, respectively. Interestingly, S. Mango (Salt Lake City, UT, USA) described that pha-4-mediated lifespan extension in worms is antagonized by target of rapamycin (TOR) signalling. The pha-4 gene is not required for lifespan extension through IIS; rather, inhibition of TOR-dependent protein translation and subsequent extension of lifespan requires pha-4, and TOR is intimately linked to dietary restriction-mediated longevity.
Summary
The broad strokes emphasized by this conference reveal that forkhead transcription factors have remarkable and crucial roles in numerous pathways that control normal development and homeostasis. The molecular mechanisms exploited by forkhead factors continue to be revealed and clearly hold great promise for a better understanding of many processes, including ageing and disease. As noted by several speakers, the last (and only) previous meeting to focus on forkhead transcription factors was held more than 10 years ago. In light of the tremendous growth in this field, the high level of enthusiasm expressed by the attendees and continuing revelations about the regulatory importance of forkhead factors in biology, we suggest a decreased interval between future meetings.

Lisa Cirillo

Michelle Barton
References
- Barthel A, Schmoll D, Unterman TG (2005) FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 16: 183–189 [DOI] [PubMed] [Google Scholar]
- Biggs WH III, Meisenhelder J, Hunter T, Cavenee WK, Arden KC (1999) Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA 96: 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp KU, Coffer PJ (2003) Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans 31: 292–297 [DOI] [PubMed] [Google Scholar]
- Carroll JS et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Cirillo L, Lin FR, Cuesta I, Jarnik M, Zaret K (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK (1993) Co-crystal structure of the HNF3/fork head DNA recognition motif resembles histone H5. Nature 364: 412–420 [DOI] [PubMed] [Google Scholar]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL (2007) Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 134: 2761–2769 [DOI] [PubMed] [Google Scholar]
- Gao N, White P, Doliba N, Golson ML, Matschinsky FM, Kaestner KH (2007) Foxa2 controls vesicle docking and insulin secretion in mature β cells. Cell Metab 6: 267–279 [DOI] [PubMed] [Google Scholar]
- Groszer M et al. (2008) Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr Biol 18: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C (2004) FoxP2 expression in avian vocal learners and non-learners. J Neurosci 24: 3164–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14: 142–146 [PubMed] [Google Scholar]
- Kalinichenko VV et al. (2004) Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev 18: 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD (2007) The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol 5: e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ et al. (1998) Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld–Rieger anomaly. Am J Hum Genet 63: 1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P (2006) Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133: 833–843 [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A (2007) PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447: 550–555 [DOI] [PubMed] [Google Scholar]
- Sherriff JA, Kent NA, Mellor J (2007) The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol Cell Biol 27: 2848–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM (2007) Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol 8: 440–450 [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2–Cks1) ubiquitin ligase. Mol Cell Biol 25: 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt J, Climent I, Dahlman-Wright K, Wikstrom M (2002) Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4). Biochemistry 40: 5861–5869 [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI (2005) Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24: 2746–2755 [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM (2006) The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol 26: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]