Abstract
The contention that quantitative profiles of biomolecules contain information about the physiological state of the organism has motivated a variety of high-throughput molecular profiling experiments. However, unbiased discovery and validation of biomolecular signatures from these experiments remains a challenge. Here we show that the Arabidopsis thaliana (Arabidopsis) leaf ionome, or elemental composition, contains such signatures, and we establish statistical models that connect these multivariable signatures to defined physiological responses, such as iron (Fe) and phosphorus (P) homeostasis. Iron is essential for plant growth and development, but potentially toxic at elevated levels. Because of this, shoot Fe concentrations are tightly regulated and show little variation over a range of Fe concentrations in the environment, making them a poor probe of a plant's Fe status. By evaluating the shoot ionome in plants grown under different Fe nutritional conditions, we have established a multivariable ionomic signature for the Fe response status of Arabidopsis. This signature has been validated against known Fe-response proteins and allows the high-throughput detection of the Fe status of plants with a false negative/positive rate of 18%/16%. A “metascreen” of previously collected ionomic data from 880 Arabidopsis mutants and natural accessions for this Fe response signature successfully identified the known Fe mutants frd1 and frd3. A similar approach has also been taken to identify and use a shoot ionomic signature associated with P homeostasis. This study establishes that multivariable ionomic signatures of physiological states associated with mineral nutrient homeostasis do exist in Arabidopsis and are in principle robust enough to detect specific physiological responses to environmental or genetic perturbations.
Keywords: Arabidopsis, biomarker, ionomics, multivariable signature
There has been an explosion of interest in the use of biomolecular signatures (e.g., RNA, protein, and metabolites) as a way to predict both the early onset of disease and possibly the susceptibility of certain individuals to a particular disease (e.g., refs. 1–3). There is also strong interest in using such molecular signatures as a data reduction tool to screen large multivariable datasets, for identification of the molecular networks that link a particular physiology to the genes that control it (e.g., ref. 4). To be practical, such molecular signatures should be tightly linked to the particular physiology of interest, and there is a growing consensus that signatures composed of multiple components are likely to be most useful (5). Although in recent years there has been an explosion of high-throughput profiling experiments, unbiased discovery and validation of molecular signatures is frequently hampered by limited availability of biological samples, difficulties in handling large datasets, and heterogeneous sources of variation.
The shoot ionome of a plant represents its mineral nutrient and trace element content (6), and is controlled by multiple physiological processes starting in the rhizosphere, and ending with evapotranspiration and phloem recycling. Alterations in any of the processes that transport inorganic ions from the soil solution to the shoot could potentially affect the shoot ionome. Because of this, the shoot ionome is likely to be very sensitive to the physiological state of the plant, with different ionomic signatures being reflective of different physiological states. Because the shoot is a much more accessible tissue for profiling than roots, such shoot ionomic signatures would be useful as markers for the particular physiological condition with which they are associated. Using a high-throughput elemental profiling and data handling pipeline to rapidly analyze the shoot elemental composition of thousands of Arabidopsis plants, we have identified and used multivariable ionomic signatures that are diagnostic for a plants response to reduced Fe or P nutrition.
Iron is an essential nutrient required for many biochemical processes in plants, including photosynthesis and respiration, where it participates in electron transport processes. Plants respond to Fe deficiency by increasing both Fe bioavailability and Fe transport (7). In dicotyledonous plants, including Arabidopsis, this is achieved through Strategy 1 processes in the roots that include increases in H+ efflux, ferric-chelate reductase (FRO) activity, and the accumulation of the Fe2+ transporter IRT1 facilitating uptake of Fe. Here we show that these mechanisms are capable of maintaining stable concentrations of shoot Fe in plants across a 10-fold range of available Fe concentrations in the growth media. Because of this homeostasis, shoot Fe concentrations are not a good indicator of the Fe response state of a plant, making it difficult to detect the Fe status of plants by simply measuring shoot Fe concentrations. However, by evaluating the Arabidopsis shoot ionome in plants grown under different Fe nutritional conditions, we have established a multivariable shoot ionomic signature, consisting of manganese (Mn), cobalt (Co), zinc (Zn), molybdenum (Mo), and cadmium (Cd), that is indicative of a plants Fe nutritional status. This signature has been validated against known Fe response genes (IRT1 and FRO2). Using a logistic regression model (LRM), trained on this multivariable ionomic signature, we can successfully classify the Fe response state of individual plants. This model can detect alterations in the Fe response status of a plant before the onset of changes in shoot Fe concentrations that are driven by a breakdown in the Fe homeostatic mechanisms. It is this failure of the Fe homoeostatic mechanisms that generally leads to chlorosis and reduced plant growth, the classic symptoms of Fe deficiency. We establish the utility of this LRM model by performing an in silico“metascreen” of ionomic data previously collected on 880 Arabidopsis mutants and natural accessions, with the successful identification of previously known Fe mutants.
Furthermore, we establish that the approach of using the shoot ionome as a multivariable signature of a physiological state can also be used to detect the P response state of Arabidopsis. After validation of the P status of Arabidopsis using the known P response gene PAP23, encoding a purple acid phosphatase (8), an LRM using shoot concentration of boron (B), P, Co, copper (Cu), Zn, and arsenic (As) was developed. This validated model was able to detect with >90% accuracy the P nutritional status of Arabidopsis.
Here we demonstrate that multivariable shoot ionomic signatures associated with mineral ionome homeostasis do exist in Arabidopsis and are robust enough to be used for the development of statistical tools for the unsupervised detection of specific physiological responses to environmental or genetic perturbations.
Results
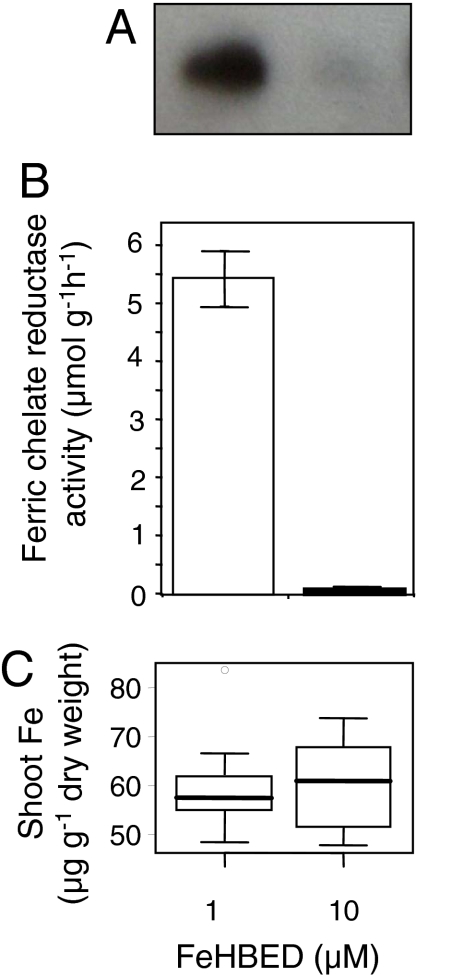
Ionomic Variation Observed Under Changing Fe Nutrition.
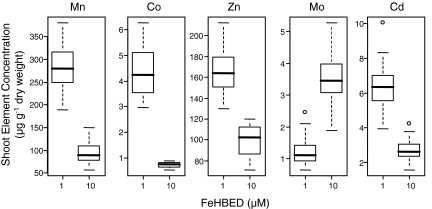
To begin modeling the effect of the Fe nutritional status of the plant on the shoot ionome, we chose FeHBED fertilization regimes that provide sufficient and deficient levels of Fe. HBED [N,N′-di-(2-hydroxybenzoyl)-ethylenediamine-N,N′-diacetic acid] was chosen to provide soluble Fe to the plants for its high selectivity for Fe(III) (9); addition of HBED is unlikely to alter the bioavailability of other elements. Under the Fe-sufficient conditions chosen (10 μM FeHBED), both ferric chelate reductase activity and IRT1 accumulation were low, indicting the Fe-sufficient status of these plants (Fig. 1 A and B), whereas the Fe-deficient conditions (1 μM FeHBED) initiated a strong Fe-deficiency response, with an ≈50-fold induction of ferric chelate reductase activity and increased accumulation of IRT1. Significantly, Fe accumulation in shoot tissues of plants grown under either Fe-sufficient or Fe-deficient conditions (10 or 1 μM FeHBED) was found to remain constant (Fig. 1C). This clearly illustrates that the Strategy 1 responses, tracked by using IRT1 expression and ferric-chelate reductase activity, are sufficient to maintain shoot Fe concentrations even when Fe in the growth medium changes 10-fold. However, even though shoot Fe concentrations are found not to vary under Fe-deficient or -sufficient conditions, we observe significant relationships between the shoot concentration of Mn, Co, Zn, Mo, and Cd (Fig. 2), the Fe concentration in the fertilizer solution, and the response of IRT1 and ferric-chelate reductase (Fig. 1 A and B). No other elements showed significant relationships with the Fe in the fertilizer solution in this and two other experiments where FeHBED was altered between 1 and 10 μM. As the Fe in the fertilizer solution is reduced, and plants respond by increasing IRT1 expression and ferric-chelate reductase activity, we observe increases in the shoot concentrations of Mn, Co, Zn, and Cd, all of which can be transported by IRT1 (10), and decreases in the concentration of Mo in shoot tissue (Fig. 2).
Fig. 1.
Biochemical assessment of the Fe response status and Fe accumulation of Arabidopsis (Col-0): immunoblot showing IRT1 protein accumulation in roots (A), ferric chelate reducatse activity in roots (B), and Fe accumulation in leaves (C) of Arabidopsis Col-0 plants grown under Fe-deficient (1 μM FeHBED) and Fe-sufficient (10 μM FeHBED) conditions. Data for the ferric chelate reductase activity represents the mean ± SD of three batches of roots from approximately three plants each. The immunoblot represents IRT1 protein in a pooled sample of roots from ≈18 plants with 10 μg of total protein loaded in each lane, and the experiment replicated twice with a representative blot shown. Box plots represent the interquartile range (IQR), the bisecting line represents the median, the whiskers represent 1.5 times the IQR, the dots represent outlier points, and the data are from 21 individual plants. All plants were grown for 5 weeks in soil.
Fig. 2.
Box plot of the Mn, Co, Zn, Mo, and Cd concentrations of Arabidopsis (Col-0) shoots when grown with varying concentrations of FeHBED. For each concentration, the box represents the interquartile range (IQR), the bisecting line represents the median, the whiskers represent 1.5 times the IQR, and the dots represent outlier points. All five elements are significantly different (P < 0.001) between treatments. Plants (n = 21) were grown for 5 weeks in soil.
Classification of the Fe Response Status Based on the Shoot Ionome.
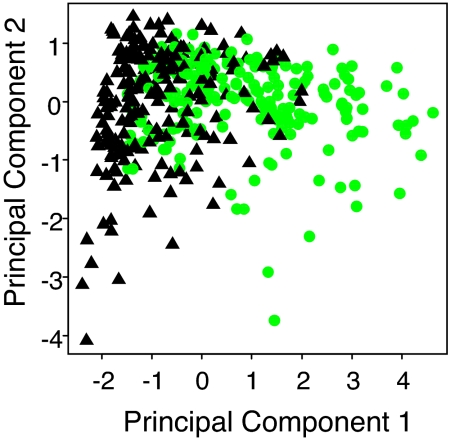
Given that shoot concentrations of Mn, Co, Zn, Mo, and Cd vary with the Fe nutritional status of Arabidopsis, we wanted to establish whether the concentration of these elements in shoot tissue could be used to discriminate plants responding to sufficient and deficient Fe. Principal component analysis (PCA) is a nonsupervised dimension reduction method. It attempts to select a small number of orthogonal coordinates (expressed as linear combinations of inputs) to maximize the overall explained variation in the data, regardless of class labels (11, 12). PCA with the shoot concentrations of Mn, Co, Zn, Mo, and Cd from a random selection of 200 plants grown under Fe-sufficiency and 200 plants from Fe-deficiency conditions revealed systematic, nonrandom structure within the data (Fig. 3). We observed that the principal components 1 and 2 provided reasonable separation between Fe-sufficient and -deficient plants (Fig. 3).
Fig. 3.
PCA based on the shoot concentrations of Mn, Co, Zn, Mo, and Cd of Arabidopsis (Col-0) plants grown with low (1 μM FeHBED) or high (10 μM FeHBED) Fe fertilization. The analysis was performed on data from n = 200 plants for each group. Green circles and black triangles represent data from plants grown under low and high Fe conditions, respectively.
Given that the PCA established that shoot concentrations of Mn, Co, Zn, Mo, and Cd contain information that can discriminate plants based on their Fe status, we developed a statistical model that could be used to classify plants as either responding to Fe deficiency or Fe sufficiency, based on their shoot Mn, Co, Zn, Mo, and Cd concentrations. To develop a statistical classification model, we randomly selected a training set of 200 plants responding to Fe sufficiency and 200 to Fe deficiency, from the larger set of 1,769 Fe-sufficient and 407 Fe-deficient plants. Thus, the training set can be considered representative of a larger population of plants grown under conditions of either Fe sufficiency or Fe deficiency. Using this training set, we derived a classification model for plants responding to Fe sufficiency versus Fe deficiency based on logistic regression (13). A logistic regression takes as input the values of multiple predictor variables, such as the shoot ionome, and outputs a probability of showing a response to Fe deficiency, in this example. Classification is then performed by selecting a probability cutoff. Plants with probabilities above this cutoff are classified as responding to Fe deficiency. To determine an optimal set of predictor variables, we considered eight logistic regressions that take as input combinations of shoot concentrations of Mn, Co, Zn, Mo, and Cd and their interactions [supporting information (SI) Table S1; also available as SI are SI Text, Figs. S1–S4, Table S2, Dataset S1, and the script and input files discussed below, which are available at www.ionomicshub.org/publications] and selected the model with the best predictive ability. We measured the predictive ability of each model using the area under the receiver–operator curve, which plots sensitivity (i.e., the fraction of plants responding to Fe deficiency classified as such) versus one minus specificity (i.e., the fraction of plants responding to Fe sufficiency that are classified as such) for each model under various probability cutoffs. The model maximizing the area under the curve has the best predictive ability.
Because each fitted logistic regression model is optimized with respect to the training dataset, the training set-based estimates produce an inadequate estimate of performance of the model on future data. To obtain a more realistic estimate of performance of each of the eight logistic regressions, we separated the training set into 11 randomly selected subsets, estimated the predictive error of the model on each subset using bootstrap, and reported the estimates of the area under the receiver–operator curve and the associated standard error (Table S1). The bootstrap is an established statistical technique frequently used for logistic regression and involves repeated resampling of the training set and model fitting for improved estimation and prediction (14). Our final selected model is the simplest logistic regression (i.e., the regression with the smallest number of predictor variables) that had the area under the receiver–operator curve, which was within the 99% confidence interval (only a 2% difference in area) of the area of the best-performing model. Models built by using any four of the five elements were 4–20% worse than the five-element model.
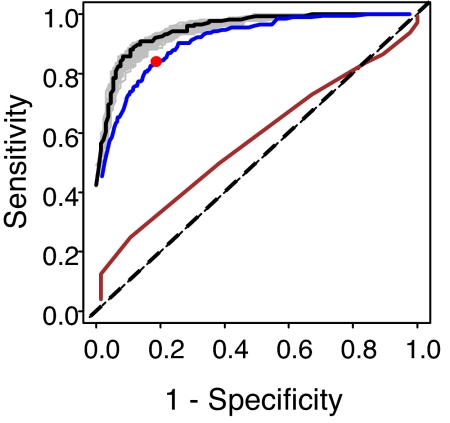
The next step of the analysis was to determine an optimal probability cutoff for classification of the Fe response status of plants in future experiments. To this end, we used bootstrap to repeatedly fit the selected logistic regression to the resampled training set of 200 plants responding to Fe sufficiency and 200 responding to Fe deficiency (Fig. 4, gray lines), determined the probability cutoff that maximized sensitivity and specificity at each bootstrap iteration, and calculated the optimal cutoff as the median value of optimal cutoffs over all bootstrap iterations (Fig. 4, red dot). Median values were used because they are less sensitive to outliers.
Fig. 4.
Receiver–operator curves of the various logistic regression models. Black line, simple model built and tested on training set; gray lines, simple models built on the bootstrapped set and tested on training set; blue line, simple model built on training set and tested on validation set; brown line, model built solely by using shoot Fe concentrations and tested on the full training set; red dot, cutoff level chosen for further studies; dotted line, unity, a model with no predictive ability.
Finally, the performance of our selected model combined with the selected probability cutoff was characterized by using a validation dataset, namely 1,569 plants grown under Fe-sufficient and 207 under Fe-deficient conditions not selected for the training set (Fig. 4, blue line). Because the validation set was not used at any stage of model fitting and selection, it provides an unbiased estimate of the performance of logistic regression model in future experiments. The logistic regression model produced a sensitivity of 82% (false negative rate of 18%) and a specificity of 84% (false positive rate of 16%) when applied to the validation set, which results in a >80% accuracy in assigning the Fe response status of a given plant, based on its shoot ionome. A logistic regression model built by using only the shoot Fe concentrations has no predictive power (Fig. 4, compare brown line with dashed line).
To test whether the logistic regression model was specific for a response to Fe nutritional status, we grew Col-0 wild-type plants in growth media fertilized with nutrient solution containing either reduced or elevated concentrations, compared with the normal nutrient solution, of B, Ca, Mg, Mn, Cu, Zn, P, and nitrate, all under sufficient Fe (Table 1). The model did not identify significant numbers of plants (i.e., greater than the error rate predicted from the modeling) grown in any of these conditions as responding to Fe-deficient growth conditions.
Table 1.
Prediction of Fe status of Arabidopsis Col-0 plants grown in altered nutrient environments
| Element (Hoagland's) | Level | Concentration | Plants predicted to be Fe-deficient, % |
|---|---|---|---|
| P (250 μM) | Low | 62.5 μM | 7* |
| High | nd | nd | |
| Mg (0.5 mM) | Low | 0.0 | |
| High | 4 mM | 0 | |
| Mn (2.25 μM) | Low | 0.0 | 0 |
| High | 18.3 μM | 11* | |
| B (11.5 μM) | Low | 0.0 | 0 |
| High | 90 μM | 11* | |
| Ca (1 mM) | Low | 0.0 | 0 |
| High | 8 mM | 0 | |
| NO3 (2.5 mM) | Low | 0.0 | 0 |
| High | 24 mM | 0 | |
| Zn (0.2 μM) | Low | 0.0 | 0 |
| High | 1.6 μM | 11* | |
| Cu (75 nM) | Low | 0.0 | 0 |
| High | 0.6 μM | 0 |
Each element in the nutrient solution was either increased or decreased/removed from the standard concentration in our normal Hoagland's nutrient solution (shown below element name). The Fe model was used to predict the Fe response status of the plants. All of the Arabidopsis (Col-0) plants grown at the same time with the normal nutrient solution had <25% of the plants predicted to be in the low-Fe state. nd, not determined.
*Values do not exceed the miscall rate predicted from the modeling and are therefore not significant. Nine plants were grown in each treatment except for P where n = 108.
Identification of Plants Responding to Fe Deficiency Under Fe-Sufficient Growth Conditions.
The trained and validated logistic regression model was tested to establish its utility for the identification of Arabidopsis lines showing altered Fe homeostasis. We used the logistic regression model to perform a metascreen of previous collected shoot ionomic data of 880 homozygous T-DNA insertional mutants, fast neutron and EMS mutants, and natural accessions, a dataset of over 70,000 plants, for plants with altered Fe responses. We used the model to predict the Fe status of all plants from lines run in experiments where ≥75% of the Col-0 wild-type plants were classified as Fe-sufficient, and the test line had more than three replicate plants. Lines where >75% of the plants were identified as responding to Fe deficiency by the logistic regression model were classified as Fe-deficient (Dataset S1). This approach identified the known Fe mutants frd3-1 and frd1-1 (15–18), which have mutations in FRD3 encoding an Fe-citrate transporter involved in translocation of Fe from roots to shoots (16), and FRO2 (19), a gene encoding a ferric chelate reductase known to be required for normal Fe homeostasis.
Response to Phosphate Deficiency.
To determine whether this approach would be applicable for other environmental conditions, we developed a multivariable, shoot ionomic model of the response of Arabidopsis to varying phosphate levels in the growth media (SI Text). Plants were watered with fertilizer solution containing either 62.5 μM phosphate (deficient) or 250 μM phosphate (sufficient). The occurrence of a P response in plants grown in these conditions was confirmed by using expression of a purple acid phosphatase (PAP23) know to be elevated in roots under P deficiency (8, 20) (Fig. S1). Unlike Fe as described above (Fig. 1), plants grown in P-deficient conditions accumulated less shoot P than those grown in sufficient conditions (Fig. S2), so shoot P could be used as a marker for P-deficient growth conditions. However, a six-element logistic regression model based on the shoot concentrations of B, P, Co, Cu, Zn, and As performed significantly better than P alone, with a 4% false negative rate and a 6% false positive rate when detecting whether plants were responding to P deficiency. This P response model was also shown to be insensitive to alterations in the concentrations of B, Ca, Mg, Mn, Fe, Cu, Zn, and nitrate in the nutrient solution (Table S2), confirming the specificity of the model for detecting plants responding to P-deficient growth conditions.
Discussion
Iron concentrations in Arabidopsis shoots are tightly regulated and do not vary even when the concentration of available Fe in the growth media is changed 10-fold (Fig. 1). Such homeostasis involves the Strategy I response that includes enhanced activity of a root-specific ferric-chelate reductase FRO2, increased Fe uptake via the Fe-transporter IRT1, and increased acidification of the rhizosphere (for review, see ref. 7). Not only do these responses lead to increased Fe uptake, but a side effect of these changes is an increase in the uptake and accumulation of Mn, Co, Zn, and Cd (10, 18, 21) via elevated IRT1 expression (10). Furthermore, the reduced rhizospheric and apoplastic pH caused by this Strategy 1 response is also likely to reduce the availability of molybdate in the rhizosphere, because the solubility of molybdate is reduced as the pH is reduced (22). Such decreased solubility of molybdate would be expected to cause reduced Mo bioavailability and plant uptake (22). In support of this model, we recently observed that hyperacidification of the apoplast/rhizosphere, by overexpression of the vacuolar pyrophosphatase (AVP1-1) in Arabidopsis (23), causes an 81–85% reduction in shoot Mo accumulation (experimental trays 369, 407, and 1,066; data available at www.purdue.edu/dp/ionomics). Interestingly, Mn, Co, Zn, and Cd were not significantly altered in this line, suggesting that rhizosphere pH changes in the AVP1-1 overexpression line affect Mo only.
Similar to Fe homeostasis, plants also respond to changes in P availability in the environment to maintain adequate supplies of inorganic phosphate for essential biochemical processes. These involve biochemical responses both to conserve internal inorganic phosphate and to enhance the bioavailability and uptake of external P (for review, see ref. 24). Side-effects of these responses to P deficiency are the alterations in shoot B, P, Co, Cu, Zn, and As concentrations we observe. Under P deficiency, the expression of several high-affinity phosphate transporters, which are thought to be required for As uptake, are increased (25–28). Also, the suggestion that under P deficiency Arabidopsis scavenges metals such as Zn to limit the formation of complexes with P (8) may explain the increased shoot Zn observed under P deficiency. However, the molecular basis for interactions between P nutrition and the accumulation of B, Co, and Cu is less clear.
It is these multivariable shoot ionomics signatures, created as a side-effect of specific biochemical responses to Fe- and P-deficient growth conditions, that the trained logistic regression model detects. Using the receiver–operator curves of the logistical regression model is ideal for evaluating the predictive value of these multivariable molecular signatures, because these curves illustrate the strength of the model, as well as the tradeoffs between false positive and false negative values (29). Not only are these multivariable shoot ionomic signatures identified here sensitive to the Fe or P response status of Arabidopsis, they also appear to be specific to these responses. Elevation or reduction in the concentration of B, Ca, Mg, Mn, Cu, Zn, and nitrate in the Arabidopsis growth media, under Fe- and P-sufficient conditions, did not mimic either the Fe or P response ionomic signatures, or cause the trained logistic regression model to falsely detect an Fe- or P-deficiency response. Furthermore, P deficiency was not detected by the Fe model nor Fe deficiency by the P model (Table 1 and Table S2).
The ability to use the trained logistic regression model to detect the Fe or P response status of plants using the shoot ionome opens up the possibility of performing unsupervised in silico metascreens on previously collected shoot ionomic data sets to identify plants with altered Fe or P homeostasis. Here we performed such a metascreen to detect plants with altered Fe homeostasis, using part of the large ionomic data set curated at the Purdue Ionomics Information Management System (ref. 30; www.ionomicshub.org). This approach identified the known Fe mutants frd3-1 and frd1-1, clearly verifying that this approach works. Such success suggests that this type of model is a useful data-reduction tool for the rapid screening of ionomic data from many thousands of lines for the identification of mutants and natural variants that have altered Fe homeostasis.
Given that the shoot ionome represents the summation of multiple biochemical and physiological processes, including rhizosphere chemistry, apoplastic and symplastic transport, vascular transport, and transpiration, any process that affects these steps will possibly alter the shoot ionome and will potentially be open to analysis by measurement of the particular ionomic signature that is associated with the process under study.
In conclusion, here we establish the concept that ionomic signatures can be identified in multivariable datasets that can be used to detect specific physiological states. Furthermore, these signatures can be identified in tissues that are not directly related to the primary site of the physiological response, alleviating the problem of sampling inaccessible tissues. We apply this method to allow detection of both the Fe and P response status of Arabidopsis, and use it to perform an unsupervised in silico metascreen of a large shoot ionomic dataset, for the identification of Arabidopsis with altered Fe homeostasis.
Materials and Methods
Plant Growth and Ionomic Analysis.
Seeds for Arabidopsis accessions used in this study were obtained from the Arabidopsis Biological Resource Center (ABRC). For most experiments, lines were planted in two rows of a 20-row (10.5 × 21 inches) tray. The planting pattern was varied across trays to reduce positional effects. For the nutrient conditional screen plants were grown in groups of six rectangular pots (8.5 × 13 cm) for each nutrient to be tested (B, Ca, Mg, Mn, Cu, Zn, nitrate, and phosphate). For all experiments, seeds were sown onto moist soil and stratified at 4°C for 48–72 h. After stratification, plants were grown in a climate-controlled room at 19–24°C with 10 h of photosynthetically active light at 80–100 μE for 36–40 days. The soil was Sunshine Mix #2 (Sun Gro Horticulture) amended with various elements including Cd, Co, Li, Na, Ni, As, and Se at subtoxic concentrations (31). Plants were bottom watered with 0.25× Hoagland's solution (1.5 mM KNO3, 1 mM Ca(NO3)2·4H2O, 0.5 mM MgSO4·7H2O, 0.25 mM NH4H2PO4, 11.5 μM H3BO3, 2.25 μM MnCl2·4H2O, 0.2 μM ZnSO4·7H2O, 0.075 μM CuSO4·5H2O, 0.016 μM Na2MoO4·2H2O) supplemented with varying amounts of Fe as FeHBED (Strem Chemical). However, for the nutrient conditional screen, plants were watered with 0.25× Hoagland's solution in which selected nutrients were provided at 0× and 2× that found in standard Hoagland's solution. In the magnesium experiment, 0.5 mM Na2SO4 was added so that sulfate was not varied and magnesium provided as MgCl2·H2O. For the nitrogen experiment, KNO3 and Ca(NO3)2·4H2O were replaced with 1 mM CaCl2·2H2O and 1.5 mM KCl, and nitrogen was added as NaNO3. For the calcium experiment, 1.5 mM KCl and 1 mM NaNO3 replaced KNO3 and Ca(NO3)2·4H2O; hence, calcium was provided as CaCl2·2H2O. All plants were watered twice per week, and pots were rotated horizontally to help reduce gradient effects for light, temperature, and humidity.
Plants were sampled for ICP-MS analysis by removing two to three leaves (0.001–0.005 g fresh weight) and washing with 18 MΩ water before being placed in Pyrex digestion tubes. Sampled plant material was dried for 24 h at 92°C and weighed before open-air digestion in Pyrex tubes using 0.7 ml of concentrated HNO3 (OmniTrace grade; EM Science) at 112°C for 5 h. Each sample was diluted to 6.0 ml with 18 MΩ water and analyzed for Li, B, Na, Mg, P, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, and Cd on an ELAN DRC-e ICP-MS (PerkinElmer/Sciex), using an Apex High-Sensitivity Inlet System (Elemental Scientific) with a self-aspirating PFA nebulizer drawing 150 μl/min. Methane was used as a reaction gas at a flow rate of 0.55 ml/min1 while measuring Fe. NIST traceable calibration standards (ULTRA Scientific) were used for the calibration. All experiments were managed by using the Purdue Ionomics Information Management System (PiiMS) (30), and all ionomic data are publicly available for viewing, download, and reanalysis at www.purdue.edu/dp/ionomics.
Immunoblot Analysis of IRT1.
Immunoblotting was performed as described in ref. 32.
Assay of Ferric Chelate Reductase Activity.
Ferric chelate reductase activity in roots of Fe-sufficient (10 μM FeHBED) or Fe-deficient (1 μM FeHBED) plants was quantified as described in ref. 18.
Multivariate Analysis and Classification.
We retrieved data for the analysis from the PiiMS database on April 4, 2007, and imported the data into the R statistical package (v2.5.1; cran.us.r-project.org). The R-script and input files used are available from the authors upon request. The PiiMS query retrieved ionomic data for 7,862 Col-0 plants. From this dataset, 1,769/407 Col-0 plants were from experiments that were fertilized with either 10 μM FeHBED (High) or 1 μM FeHBED (Low), respectively. These plants are referred to as the “Known Set” because their Fe-status is known. The Known Set was standardized for each element by subtracting the median of the set and dividing by the median absolute deviation. A random sampling of 200 Col-0 High and 200 Col-0 Low Fe plants was taken as the training set. PCA was performed by using the five elemental concentrations and the princomp function from the R stats package with all of the default settings. Logistic regression models were calculated by using the lrm and validate.lrm functions in the Design (v2.0–12) package (cran.us.r-project.org). Models with all five elements, all combinations of four elements, and all five with interaction terms were evaluated. The models were bootstrapped with B = 40, and the area under the receiver–operator curve was calculated from the bootstrap set. This process was repeated for 11 different randomly chosen test sets, and the results were analyzed to pick the optimal model (see Results and Table S1). The chosen model was bootstrapped again to choose the appropriate cutoffs (Fig. 4). To predict the Fe response status of the lines in the database, the full dataset was queried on May 12, 2007 and all lines annotated with the genetic structure “segregating” and generation “2” removed as they constituted F2 mapping lines or screening populations. Data were normalized, processed, and fit by the model as described above.
Supplementary Material
Acknowledgments.
We thank Marina Tikhonova and Elena Yakubova for plant cultivation and harvesting. This work was supported by National Science Foundation Plant Genome Research Program Grant DBI-0077378 (to M.L.G., D. Eide, J. Harper, D.E.S., J. Schroeder, and J. Ward), National Science Foundation Arabidopsis 2010 Program Grant IOS-0419695 (to M.L.G., J. Harper, D.E.S., J. Schroeder, and J. Ward), and Indiana 21st Century Research and Technology Fund Grant 912010479 (to D.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804175105/DCSupplemental.
References
- 1.Bitarte N, Bandres E, Zarate R, Ramirez N, Garcia-Foncillas J. Moving forward in colorectal cancer research, what proteomics has to tell. World J Gastroenterol. 2007;13:5813–5821. doi: 10.3748/wjg.v13.i44.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–952. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- 3.Ward M. Biomarkers for Alzheimer's disease. Exp Rev Mol Diagn. 2007;7:635–646. doi: 10.1586/14737159.7.5.635. [DOI] [PubMed] [Google Scholar]
- 4.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 6.Salt DE, Baxter I, Lahner B. Ionomics and the study of the plant ionome. Annu Rev Plant Biol. 2008;59:709–733. doi: 10.1146/annurev.arplant.59.032607.092942. [DOI] [PubMed] [Google Scholar]
- 7.Kim SA, Guerinot ML. Mining iron: Iron uptake and transport in plants. FEBS Lett. 2007;581:2273–2280. doi: 10.1016/j.febslet.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Misson J, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA. 2005;102:11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leplatte. F, Murase I, Martell AE. New multidentate ligands: Chelating tendencies of N,N′-di(2-hydroxybenzyl ethylenediamine)-N,N′-diacetic acid. J Am Chem Soc. 1967;89:837. [Google Scholar]
- 10.Vert G, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson K. On lines and planes of closest fit to systems of points in space. Philos Mag. 1901;2:559–572. [Google Scholar]
- 12.Johnson R, Wichern D. Applied Mutlivariate Statistical Analysis. Old Tappan, NJ: Pearson Prentice Hall; 2007. [Google Scholar]
- 13.Hosmer D, Lemeshow S. Applied Logistic Regression. New York: Wiley; 1989. [Google Scholar]
- 14.Harrell F. Regression Modeling Strategies. Berlin: Springer; 2001. [Google Scholar]
- 15.Delhaize E. A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1996;111:849–855. doi: 10.1104/pp.111.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers EE, Guerinot ML. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell. 2002;14:1787–1799. doi: 10.1105/tpc.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi Y, Guerinot ML. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996;10:835–844. doi: 10.1046/j.1365-313x.1996.10050835.x. [DOI] [PubMed] [Google Scholar]
- 19.Robinson NJ, Proctor CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 20.Ward JT, Lahner B, Yakubova E, Salt DE, Ragothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampey RA, et al. An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics. 2006;174:1841–1857. doi: 10.1534/genetics.106.061044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marschner H. Mineral Nutrition of Higher Plants. Boston: Academic; 1995. [Google Scholar]
- 23.Li J, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 24.Ticconi CA, Abel S. Short on phosphate: Plant surveillance and countermeasures. Trends Plants Sci. 2004;9:548–555. doi: 10.1016/j.tplants.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Shin H, Shin HS, Dewbre GR, Harrison MJ. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- 26.Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DA, Chen A, Schroeder JI. ars1, an Arabidopsis mutant exhibiting increased tolerance to arsenate and increased phosphate uptake. Plant J. 2003;35:637–646. doi: 10.1046/j.1365-313x.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- 28.Clark G, Dunlop J, Phung H. Phosphate adsorption by Arabidopsis thaliana: Interactions between phosphorus status and inhibition by arsenate. Aus J Plant Physiol. 2003;27:959–965. [Google Scholar]
- 29.Baker SG. The central role of receiver operating characteristic (ROC) curves in evaluating tests for the early detection of cancer. J Natl Cancer Inst. 2003;95:511–515. doi: 10.1093/jnci/95.7.511. [DOI] [PubMed] [Google Scholar]
- 30.Baxter I, et al. Purdue Ionomics Information Management System. An integrated functional genomics platform. Plant Physiol. 2007;143:600–611. doi: 10.1104/pp.106.092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahner B, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- 32.Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.