Abstract
The imprinted insulin-like growth factor 2 (IGF2) gene is expressed predominantly from the paternal allele. Loss of imprinting (LOI) associated with hypomethylation at the promoter proximal sequence (DMR0) of the IGF2 gene was proposed as a predisposing constitutive risk biomarker for colorectal cancer. We used pyrosequencing to assess whether IGF2 DMR0 methylation is either present constitutively prior to cancer or whether it is acquired tissue-specifically after the onset of cancer. DNA samples from tumour tissues and matched non-tumour tissues from 22 breast and 42 colorectal cancer patients as well as peripheral blood samples obtained from colorectal cancer patients [SEARCH (n=case 192, controls 96)], breast cancer patients [ABC (n=case 364, controls 96)] and the European Prospective Investigation of Cancer [EPIC-Norfolk (n=breast 228, colorectal 225, controls 895)] were analysed. The EPIC samples were collected 2–5 years prior to diagnosis of breast or colorectal cancer. IGF2 DMR0 methylation levels in tumours were lower than matched non-tumour tissue. Hypomethylation of DMR0 was detected in breast (33%) and colorectal (80%) tumour tissues with a higher frequency than LOI indicating that methylation levels are a better indicator of cancer than LOI. In the EPIC population, the prevalence of IGF2 DMR0 hypomethylation was 9.5% and this correlated with increased age not cancer risk. Thus, IGF2 DMR0 hypomethylation occurs as an acquired tissue-specific somatic event rather than a constitutive innate epimutation. These results indicate that IGF2 DMR0 hypomethylation has diagnostic potential for colon cancer rather than value as a surrogate biomarker for constitutive LOI.
INTRODUCTION
The insulin-like growth factor 2 (IGF2) gene is expressed predominantly from the paternal allele and is located within a cluster of imprinted genes on chromosome 11p15 (1). Loss of IGF2 imprinting (LOI) manifested by biallelic expression of IGF2 has been observed in 30–70% of patients with adult onset cancers such as breast (2), hepatocellular carcinoma (3), ovarian tumours (4,5), bladder cancer (6) and colorectal cancer (7–15). Additionally, IGF2 expression is up-regulated in many cancers including uterine leiomyomas (16), colon cancer (17), adrenal cortical tumours (18) and Wilms tumours (19). Biallelic IGF2 expression is also part of the aetiology of Beckwith Wiedemann Syndrome (BWS), which is characterized by foetal overgrowth and a propensity to develop embryonic and childhood cancers such as rhabdomyosarcoma and Wilms tumours (20–22).
Imprinted genes are associated with CpG rich regions that have allele-specific DNA methylation known as differentially methylated regions (DMRs). At the IGF2-H19 locus, these DMRs have been well characterized in both humans and mice (23–27). Reciprocal imprinting of IGF2 and H19 is regulated by a conserved DMR located 2–5 kb upstream of H19 that acts as a CTCF mediated insulator that controls access of the IGF2 and H19 promoters to downstream enhancers (28–31). In mice, the H19 DMR acts as an imprinting control region that acquires methylation in the male germ cells and subsequently influences the methylation of the Igf2 DMRs in cis after fertilization (25,32). The H19 DMR and Igf2 DMRs have been shown in mice to physically interact in an allele-specific manner to form higher order chromatin loops that maintain or preclude Igf2 promoter–enhancer interactions (33,34). In humans, the IGF2 gene has a DMR corresponding to the mouse Igf2 DMR0 which is methylated on the silent maternal allele and is placenta specific (24,35). Unlike mice, the human DMR0 is not placenta specific (26) and is methylated on the paternal allele in all tissues (36). The DMR0 has been shown to contain promoter activity (26), but its function in the higher order chromatin looping structure has not yet been elucidated. Biallelic IGF2 expression follows gain of methylation on the maternal allele at the H19 and IGF2 DMRs in a subset of BWS patients (20,36). Although it has widely been assumed that cancers with LOI will have the same methylation epigenotypes as BWS, we have previously shown that the methylation patterns associated with constitutive LOI at the IGF2 and H19 genes in BWS patients are not identical to the tumour-associated LOI in Wilms patients (36). Other studies have shown that the H19 insulator is not invariably hypermethylated together with LOI in colorectal cancer (15). Loss of methylation at the IGF2 DMR0 has been reported to be associated with IGF2 LOI in cancer studies including Wilms tumour (36,37), esophageal cancer (38) and colorectal cancer (15).
Constitutive IGF2 LOI predisposes to Wilms tumour and possibly colorectal cancer (14). LOI associated with IGF2 DMR0 hypomethylation was reported in peripheral blood at a significantly higher frequency in patients with a history of colorectal cancer than patients without (14,15). Since IGF2 DMR0 is normally methylated on the expressed paternal allele, it is unlikely that its demethylation directly leads to reactivation of the maternal allele. Moreover, IGF2 DMR0 hypomethylation has now been reported in osteosarcomas (39), bladder- (6) and ovarian cancer (5) without an association with LOI. Thus, IGF2 DMR0 hypomethylation in cancer is likely to be indicative of changes in IGF2 transcription from the active allele independent of the imprinting status. Therefore, IGF2 DMR0 methylation levels may be useful indicators of cancer and constitutive IGF2 DMR0 methylation levels could be predictive of cancer risk. In this study, we measured the prevalence of IGF2 DMR0 hypomethylation in tumour tissue samples from colorectal cancer and breast cancer patients and compared this to IGF2 imprinting status in these tissues. We also investigated whether IGF2 hypomethylation is present in peripheral blood prior to the onset of breast or colorectal cancer by screening a series of samples selected from the European Prospective Investigation of Cancer (EPIC-Norfolk) (40) cohort.
Our results indicate that IGF2 DMR0 hypomethylation is highly prevalent in cancer and detected more frequently than LOI. Hypomethylation is not present constitutively in peripheral blood prior to cancer suggesting that it is an acquired somatic epimutation and therefore a useful indicator of breast and colorectal cancer.
RESULTS
Validation of methylation pyrosequencing assay
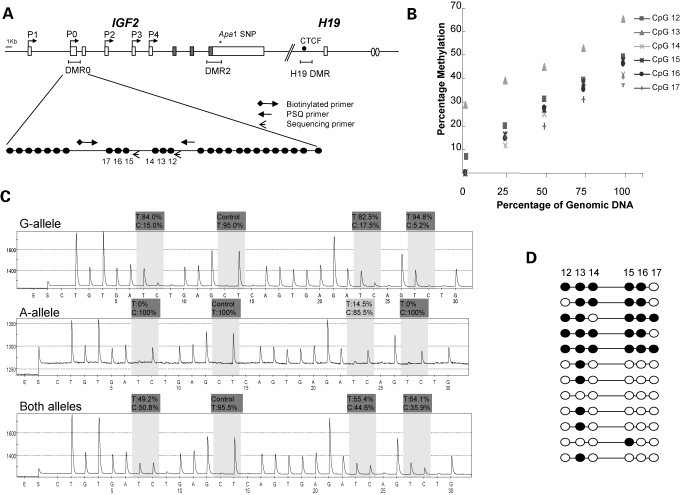
Pyrosequencing is a real-time sequencing methodology that has been successfully used to quantify bisulphite modified cytosine to thymine changes (41,42). We designed and validated a pyrosequencing assay for IGF2 methylation within the DMR0 region (Fig. 1A) which included six CpGs, three of which were previously reported to be hypomethylated (CpG15–17) in peripheral blood samples from colorectal patients with LOI at IGF2 (15). The pyrosequencing assay is linear (Fig. 1B) and IGF2 methylation levels between 7 and 35% consistent with hypomethylation were obtained when genomic DNA from pooled lymphocyte samples was diluted 2- and 4-fold with unmethylated DNA. Methylation levels between 42 and 50% consistent with normal monoallelic methylation were obtained when genomic DNA was undiluted with unmethylated DNA. Based on this result, we predict that if DMR0 methylation levels of 35% and less were obtained in a heterogeneous cell population such as a tumour sample, this would be indicative of 30% or more cells having no DMR0 methylation on the paternal IGF2 allele.
Figure 1.
Pyrosequencing assay to detect methylation levels at the IGF2 DMR0. (A) The human IGF2 gene showing positions of DMRs relative to exons and active promoters on the paternal allele. IGF2 promoters access enhancers downstream of H19 (ovals) when the CTCF binding site within the H19 DMR is methylated. IGF2 and H19 genes are 100 kb apart. The DMR0 region within the IGF2 has been reported to be hypomethylated in colorectal cancer. The expanded region shows the 26 CpGs within this region and the CpGs that we assayed for methylation by pyrosequencing. Our assay encompassed a 255 bp region, which includes six CpGs (CpG12–17 NCBI36 2125904–2126160). CpG15–17 were previously reported to be hypomethylated in colorectal patients with LOI at IGF2 (15). Arrows indicate position and direction of biotinylated, pyrosequencing (PSQ) and sequencing primers. Circles indicate individual CpGs on expanded DMR0. (B) Linearity of pyrosequencing assay: unmethylated template was obtained by PCR amplification of target DMR0 sequence prior to bisulphite conversion. After bisulphite conversion, the unmethylated template was serially diluted with PCR product obtained from bisulphite converted genomic DNA. At 100% genomic DNA, we have close to 50% methylation which is as expected for normal imprinting. (C) Pyrosequencing profiles for allele-specific methylation compared to total methylation. Total methylation levels reliably reflected the average of the two alleles. (D) Methylation diagrams for 12 cloned PCR products from bisulphite-treated DNA showing that alleles separate into methylated and unmethylated alleles and that methylation at CpG13 is not allele-specific. Filled and open circles denote methylated and unmethylated CpGs.
Our assay reliably measures methylation levels in a population of PCR products representative of both parental alleles. This was verified by comparing methylation levels of individual alleles to total methylation levels in a panel of 10 peripheral blood DNA samples from healthy individuals that were informative for a G/A polymorphism (Fig. 1C). IGF2 DMR0 methylation levels were 0–20% at unmethylated alleles and between 80 and 100% at methylated alleles. The average methylation between G and A-alleles in each sample was between 35 and 55% consistent with the total methylation levels obtained with our standard pyrosequencing assay. Conventional bisulphite sequencing of cloned PCR products showed that the methylation levels of CpGs analysed were concordant and that amplicons separated into methylated and unmethylated alleles, with the exception of CpG13 (Fig. 1D). Thus, our assay measuring total methylation levels in a population of PCR products could reliably distinguish between normal differentially imprinted methylation (45–50%) and hypomethylation (≤35%).
Baseline methylation levels for IGF2 DMR0
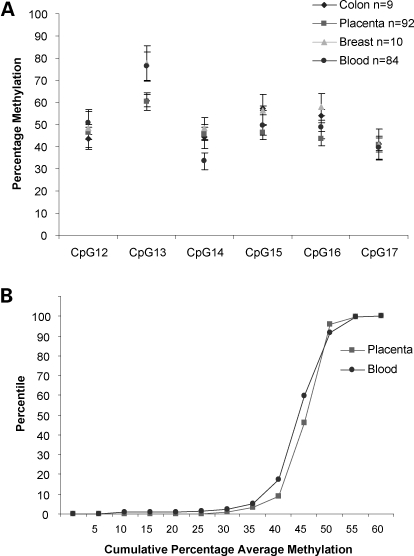
Baseline IGF2 methylation levels were established for the DMR0 region in 233 non-cancerous peripheral blood, 189 term placentae, 10 normal colon and 10 normal breast tissue samples. Figure 2A shows that methylation at individual CpGs in this region were interrelated with CpGs 14 and 17 having the lowest methylation, while CpG13 was higher in all tissues. Allele-specific methylation assays described above indicated that CpG13 was not monoallelically methylated in blood. This CpG13 lies within a homopolymeric sequence tract and was refractory to pyrosequencing. We therefore excluded CpG13 when calculating average methylation levels for the IGF2 DMR0 in subsequent population studies. Baseline methylation levels (methylation average of CpG12, 14–17) for peripheral blood, colon, term placentae and breast tissue samples were 45, 48, 45 and 50%, respectively.
Figure 2.
Distribution of average CpG methylation across IGF2 DMR0 and population frequency of methylation in a cohort of peripheral blood and placentae. (A) Graph showing average methylation levels of each CpG across the IGF2 DMR0 in peripheral blood, normal colon, normal breast and term placentae samples. (B) Cumulative frequency distribution of average methylation levels for CpG12, 14–17 in the IGF2 DMR0 in placentae and peripheral blood. At the 90% percentile average methylation levels for the DMR0 was 48%; at the 10% percentile, the average methylation level was less than 35%.
Our sample sizes for peripheral blood and placentae were large enough to establish that average methylation at the IGF2 DMR0 for these sample sets followed a unimodal negatively skewed frequency distribution with median methylation levels 43.8% (IQR 41.2, 46.9) in blood and 45.2% (IQR 42.8, 46.7) in placentae. The skewness was −1.126 and −2.445 for blood and placentae samples, respectively (D′ Agostino–Pearson normality test, P < 0.0001) and this was due to a dominant left tail demonstrating individuals with methylation levels less than 35% (Fig. 2B). It is also noteworthy that methylation levels did not exceed 60% and that less than 1% of individuals had methylation levels above 55% indicating that hypermethylation of IGF2 DMR0 is rare in peripheral blood and placenta samples of normal individuals. It has been suggested that 10% of normal individuals are constitutively hypomethylated for the IGF2 DMR0 (14). In our assay, 7.5% of peripheral blood and 4% of placental samples had less than 35% methylation. We therefore set 35% methylation as a cut-off for of hypomethylation.
We examined IGF2 imprinted status in three placental RNA samples that were informative for the Apa1 polymorphism and which were hypomethylated at IGF2 DMR0, these were all mono allelic indicating that in placenta constitutive hypomethylation of DMR0 does not correlate with LOI. RNA was available from 70 of the peripheral blood samples, and 30 of these were informative for the Apa1 polymorphism, IGF2 expression levels were very low and we did not detect LOI in any of the samples including 1 sample that had hypomethylation of IGF2 DMR0. The results confirm that in normal tissue, IGF2 hypomethylation is not associated with LOI.
IGF2 methylation levels in colorectal cancer
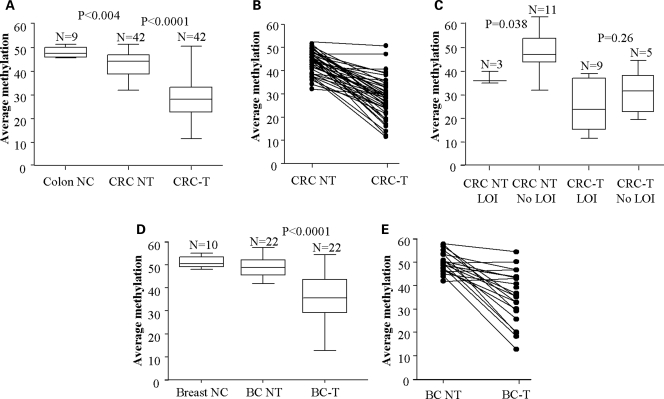
We examined tumour tissue and matching normal control tissue from 42 colorectal cancer patients. Median methylation at the IGF2 DMR0 in tumours were 28.6% (IQR 22.0; 32.3) compared to adjacent normal tissue 45.3% (IQR 38.9; 47.1). Methylation levels ≤35% were found in 34 (80%) of tumours compared to 4 (10%) adjacent normal colon tissue samples (Fig. 3A). IGF2 DMR0 methylation was reduced in tumours compared to paired normal tissue in 38 (90%) patients (Fig. 3B). Methylation levels for individual CpGs within the DMR0 were interrelated similar to controls and are as shown in Supplementary Material, Figure S1.
Figure 3.
IGF2 DMR0 methylation levels in colorectal and breast cancers compared to normal tissue from same patients and non-cancer patients: prevalence of hypomethylation in breast and colorectal cancer tumours. (A) DMR0 methylation levels in DNA from colon tissue from patients with no cancer (Colon NC), and colorectal cancer patients' tumour tissue (CRC-T) and matched control biopsies of non-tumour tissues (CRC-NT). Tumours have significantly lower methylation than non-tumour tissue from same patients and this is lower than in colons from patients without cancer. (B) Line plots showing individual data comparisons between colorectal tumour (CRC-T) and matched control biopsies of non-tumour tissues (CRC NT). (C) LOI relative to IGF2 DMR0 methylation in colorectal cancer tissue: colorectal cancer patients with LOI have lower methylation in colon tissue than patients without LOI. Colorectal tumours (CRC-T) have lower methylation than non-tumour tissues (CRC NT) regardless of LOI. (D) DMR0 methylation levels in DNA from breast tissue from individuals with no cancer (Breast NC) and breast cancer patients' tumours (BC-T) and matched control biopsies of non-tumour tissue (BC-NT). Breast cancer tumour tissues have significantly lower methylation levels than non-tumour tissue. Non-tumour tissue from breast cancer patients has a wider variation of methylation levels than tissue from non-cancer patients, but median levels are not significantly different. (E) Line plots showing individual data comparisons between breast tumour (BC-T) and matched control biopsies of non-tumour tissues (BC NT). (Box and whisker plots represent Median and IQR with max and min values. P-values calculated by Wilcoxon Rank sum test.)
IGF2 methylation levels relative to genomic imprinting status in colorectal cancer
Previous studies have shown that IGF2 DMR0 hypomethylation is associated with LOI in colorectal cancer (15) and Wilms tumour (36,37). Fourteen colorectal cancer patients were heterozygous for the Apa1 polymorphism in exon 9 of IGF2 enabling us to examine the RNA for LOI. Nine (64%) of our informative cases were biallelic for IGF2 expression in their tumours with median methylation levels of 23.9% (range 11.6–39.0) compared to five tumours which showed normal imprinting with median methylation level 32.9% (range 19.4–47.1 Wilcoxon Rank Sum tests W = 10, P = 0.26) (Fig. 3C). If DMR0 methylation levels are indicative of LOI, then we would expect most of our informative tumour samples to have LOI, since the methylation levels in these tumours were low. Instead we found normal monoallelic IGF2 expression in four tumours with low methylation levels (range 19.4–32.9%). LOI was also detected in three (20%) of the normal colon tissue biopsies. In these samples, the methylation levels were 38.8, 40.0 and 36.6% which was significantly lower than adjacent normal tissue without LOI with median methylation levels of 47.3% (range 32.4–63.3, n = 11 Wilcoxon Rank Sum test W = 3, P = 0.038) (Fig. 3C). The positive predictive value (PPV) was calculated using a cut off of ≤35 being positive and >35 being negative. This shows the probability that a patient with a methylation value ≤35% will have LOI in their tumours is PPV 66.7% (95% CI: 29.9%, 92.5%), while in non-tumour tissue the probability that a patient with a methylation value ≤35% will have loss of imprinting (LOI) is 100.0% [95% CI: 2.5%, 100.0%]. The 95% confidence intervals (CI) are Pearson–Clopper exact confidence intervals. So while all cases with LOI had hypomethylation at IGF2 DMR0, the inverse is not true and DMR0 hypomethylation is not invariably associated with LOI in tumours.
Methylation levels of IGF2 in breast cancer tumours
Methylation levels at IGF2 DMR0 were examined in 22 breast tumours and matched control tissues from the same patients. Median methylation levels in tumours were 35.5% (IQR 29.2; 43.5), compared to 48.9% (IQR 45.5; 52.1) in the normal tissue as shown in Figure 3D. Over half of the tumours (n = 13) had methylation levels that were lower than their corresponding normal tissue samples (Fig. 3E), but only seven cases (33.3%) had less than 35% methylation. Nine of the breast cancer cases were informative for the Apa1 polymorphism and showed normal imprinting, despite two of these cases having hypomethylation at IGF2. Loss of heterozygosity for IGF2 was excluded in these patients. These results reflect that in breast tumours hypomethylation is less prevalent than in colorectal tumours and in our small sample of informative cases reduced IGF2 methylation levels did not associate with LOI. Methylation levels for individual CpGs within the DMR0 were interrelated similar to controls and are as shown in Supplementary Material, Figure S1.
Hypomethylation and cancer in a prospective population study
Since hypomethylation of IGF2 DMR0 is a feature of colorectal, and to a lesser extent, breast cancer, we set out to determine whether IGF2 DMR0 hypomethylation is present constitutively prior to the onset of these cancers in 1348 DNA samples from a subgroup of The European Prospective Investigation of Cancer (EPIC-Norfolk) cohort of peripheral blood samples. Within this population, we had two nested populations consisting of individuals who went on to develop breast (n = 228) or colorectal cancer (n = 225) 2–5 years after contributing to the EPIC study. The sample set was selected so that the control and cancer samples were age matched (range 60–80 years) as shown in Table 1. The methylation levels in our EPIC population indicated that 9.5% had hypomethylation (≤35% threshold). These samples verified by clonal analysis with conventional bisulphite sequencing to have no methylation on either allele (results not shown). Within the EPIC control population, IGF2 DMR0 methylation levels decreased with age suggesting that loss of methylation is an acquired epigenetic change (Table 2 and Supplementary Material, Fig. S2). In addition to age, gender also had a significant influence on methylation levels. Females were less frequently hypomethylated than males at IGF2 (Table 2). Importantly, no evidence for an association was found with individuals with methylation levels below the 35% threshold for either colorectal or breast cancer (Table 3). Since the underlying population contained a substantial proportion of cancer cases (228 breast and 225 colorectal), it is unlikely that methylation levels at the IGF2 DMR0 could distinguish individuals who are predisposed to cancer. These data indicate that IGF2 DMR0 hypomethylation is acquired in peripheral blood with age, but does not predict future risk for breast or colorectal cancer.
Table 1.
Baseline characteristics of cancer cases and controls by cancer site
| Cases | Controls | |
|---|---|---|
| EPIC-Norfolk, 1993–2003 (prospective) | ||
| Breast cancer, number | 228 | 460 |
| Age (years) | 60.5 (59.4–61.7) | 60.3 (59.5–61.1) |
| Smoking (pack years) | 16.3 (0.1–62.5) | 16.87 (0.05–59.5) |
| Body mass index (kg/m2) | 26.6 (26.2227.0) | 26.75 (26.2–27.3) |
| Family history of cancer (%) | 45 (19.7%) | 75 (16.3%) |
| Colorectal cancer, number | 225 | 435 |
| Age (years) | 66.1 (65.1–67.1) | 66.1 (65.3–66.8) |
| Men (%) | 56.4 | 55.2 |
| Smoking (pack years) | 22.3 (0.1–93) | 22.0 (0.4–126) |
| Body mass index (kg/m2) | 27.1 (26.6–27.7) | 26.3 (25.9–26.6) |
| Family history of cancer (%) | 34 (15.1%) | 69 (15.9%) |
| SEARCH (cross-sectional) colorectal cancer, number | 192 | 96 |
| Age (years) | 65.9 (65.1–66.8) | 43.2 (42–44.4) |
| Men (%) | 57.3 | 29.5 |
| Smoking (pack years) | 24.8 (21.4–28.2) | 13.8 (1.3–17.3) |
| Body mass index (kg/m2) | 26.7 (26.1–27.3) | 26.6 (25.1–28.1) |
| Family history of cancer (%) | N/A | N/A |
| ABC (cross-sectional) breast cancer, number | 338 | 84 |
| Age (years) | 52.6 (51.8–53.4) | 43.2 (42–44.4) |
| Body mass index (kg/m2) | 26.6 (26.2–27.1) | 26.6 (25.1–28.1) |
| Smoking (pack years) | N/A | N/A |
| Family history of cancer (%) | N/A | N/A |
Data are means and 95% CIs, unless otherwise stated. Numbers vary due to missing values for BMI. Cancer includes breast or colorectal cancer. N/A: data not available.
Table 2.
Association between loss of methylation at the IGF2 DMR0 locus in blood DNA and selected study characteristics in controls at baseline, EPIC-Norfolk, 1993–2003 (cut off 35%)
| LOM, n = 83 | Non-LOM, n = 812 | P-value | |
|---|---|---|---|
| Age (year) | 67.0 (65.1–68.8) | 62.7 (62.1–63.3) | <0.001 |
| Men (%) | 31 (37.3) | 209 (25.7) | 0.023 |
| Smoking (pack years) | 22.5 (17.7–27.2) | 19.5 (17.8–21.1) | 0.21 |
| Body mass index (kg/m2) | 26.7 (26.0–27.4) | 26.4 (26.2–26.7) | 0.53 |
| Family history of cancer (%) | 13 (15.7%) | 131 (16.1%) | 0.21 |
Data are means and 95% CIs, unless otherwise stated. Numbers vary due to missing values for BMI.
Table 3.
Association between loss of methylation at the IGF2 DMR0 locus in blood DNA and risk of future breast and colorectal cancers, EPIC-Norfolk, 1993–2003 (cut off 35%)
| Methylation status | Cases (%) | Controls (%) | Model 1a |
|---|---|---|---|
| Colorectal cancer | 225 | 435 | |
| Non-LOM | 195 (86.7) | 381 (87.6) | 1 |
| LOM | 30 (13.3) | 54 (12.4) | OR = 1.09 95% CI (0.65–1.82) P = 0.74 |
| Breast cancer | 228 | 460 | |
| Non-LOM | 213 (93.4) | 431 (93.7) | 1 |
| LOM | 15 (6.6) | 29 (6.3) | OR = 1.04 95% CI (0.54–2.0) P = 0.91 |
| All cancers | 453 | 895 | |
| Non-LOM | 408 (90.1) | 812 (90.7) | 1 |
| LOM | 45 (9.9) | 83 (9.3) | OR = 1.08 95% CI (0.72–1.61) P = 0.71 |
aModel 1: additional adjustment for possible confounders, smoking (pack years), BMI.
Frequency of IGF2 DMR0 hypomethylation in peripheral blood in a cross-sectional population study
Our data from the EPIC cohort indicated that constitutive hypomethylation at IGF2 DMR0 shows no evidence of predicting colorectal or breast cancer so we wanted to know to what extent hypomethylation is acquired during carcinogenesis. Previously published reports have described LOI in peripheral blood of colorectal cancer patients in a cross-sectional study, so we tested whether the incidence of IGF2 hypomethylation in peripheral blood would be higher in patients already diagnosed with colorectal or breast cancer. We, therefore, analysed further cohorts of peripheral blood samples from patients diagnosed with colorectal cancer (n = 192) and 96 control samples [SEARCH sample set (43)] as well as breast cancer (n = 364) samples plus 84 controls [ABC cohort (44)] (Tables 1 and 4). We found that the frequency of individuals with hypomethylation (methylation levels ≤35%) in the colorectal cancer patients was 9% which was similar to the EPIC population, but higher than a control population where the incidence was 6.4% (Table 5). These small differences were not statistically significant and can be explained by the differences in age of the case and control population. There was no evidence of a difference between breast cancer and control samples at threshold levels less than 35% (Table 5).
Table 4.
Association between loss of methylation at the IGF2 DMR0 locus in blood DNA and selected study characteristics in SEARCH and ABC dataset
| LOM | Non-LOM | P-value | |
|---|---|---|---|
| SEARCH, n | 23 | 259 | |
| Age (year) | 60.3 (54.9–65.6) | 58.1 (56.7–59.7) | 0.44 |
| Men (%) | 47.8 | 47.5 | 0.98 |
| Smoking (pack years) | 22.7 (16.3–29.2) | 21.9 (18.9–24.9) | 0.87 |
| Body mass index (kg/m2) | 25.6 (23.6–27.5) | 26.8 (26.1–27.5) | 0.32 |
| Family history of cancer (%) | N/A | N/A | N/A |
| ABC, n | 25 | 397 | |
| Age (year) | 55.42 (52.7–58.1) | 52.44 (51.6–53.3) | 0.29 |
| Smoking (pack years) | N/A | N/A | |
| Body mass index (kg/m2) | 26.6 (24.6–27.6) | 26.8 (26.0–27.5) | 0.33 |
| Family history of cancer (%) | N/A | N/A |
Data are means and 95% CIs, unless otherwise stated. Numbers vary due to missing values for methylation. N/A, data not available.
Table 5.
Association between loss of methylation at the IGF2 DMR0 locus in blood DNA and risk of cancers in SEARCH and ABC datasets (cut off 35%)
| Methylation status | Cases (%) | Controls (%) | Model 1a | Model 2b |
|---|---|---|---|---|
| SEARCH | ||||
| Colorectal cancer | 188 | 94 | ||
| Non-LOM | 171 (91.0%) | 88 (93.6%) | 1 | 1 |
| LOM | 17 (9.0%) | 6 (6.4%) | 1.33 (0.08–22.7) P = 0.8 | 1.63 (0.06–42.5) P = 0.77 |
| ABC | ||||
| Breast cancer | 338 | 84 | ||
| Non-LOM | 319 (94.4%) | 78 (92.9%) | 1 | 1 |
| LOM | 19 (5.6%) | 6 (7.1%) | 0.93 (0.78–1.187) P = 0.65 | 0.65 (0.296–2.005) P = 0.77 |
CI, confidence interval.
aModel 1: Adjusted for age, and sex.
bModel 2: Additional adjustment for possible confounders, smoking (pack years), BMI.
DISCUSSION
IGF2 DMR0 hypomethylation has been suggested as a surrogate biomarker for LOI with the potential to predict inherent predisposition to cancer. DNA methylation studies are more robust and quantitative compared to RNA expression studies and therefore amenable to high throughput analysis or use as clinical biomarkers. In this study, we have evaluated IGF2 methylation levels at the DMR0 region in DNA from breast and colorectal cancer tissue biopsies, as well as in large cohorts of peripheral blood DNA samples from cross-sectional and prospective populations using a well validated quantitative pyrosequencing assay.
We have found that IGF2 DMR0 hypomethylation is more prevalent than LOI and 36% of our colorectal tumour tissue samples with hypomethylation were monoallelic for IGF2 expression. Interestingly, the tumours with LOI had lower methylation levels than those without LOI. Since IGF2 DMR0 is methylated on the paternally inherited allele (36), it is unlikely that the loss of methylation at this region would have a direct role in reactivation of the silent maternal allele. This is supported by our finding that IGF2 DMR0 hypomethylation and LOI are not tightly linked. However, IGF2 DMR0 hypomethylation was found in 80% of colorectal and 33% of breast cancer tissues with most of the colorectal and breast cancer patients having significantly lower methylation in their tumours compared to adjacent non-tumour tissues. These results suggest that hypomethylation at IGF2 DMR0 in tumour tissue should not be viewed as a surrogate biomarker for LOI but as a frequently identifiable attribute of colorectal cancer.
Our second finding was that in our prospective studies using the EPIC population samples only 9.5% of samples had ≤35% methylation. Based on previous reports in which IGF2 DMR0 hypomethylation in peripheral blood was found to be associated with colorectal cancer and the perception that loss of methylation is likely to be an early event in tumorigenesis, we expected to find a higher incidence of colorectal cancer and possibly also breast cancer in the 10% percentile proportion of the population with hypomethylation. However, this was not the case and hypomethylation of IGF2 DMR0 was present with similar incidences in controls and persons who later went on to develop cancer. This observation indicates that constitutive hypomethylation of IGF2 is actually rare and not a predetermining factor for colorectal or breast cancer.
It has recently been shown that Dnmt3b-mediated methylation in cancer targets specific genes rather than being a stochastic random event (45). It is possible that demethylation is also targeting specific loci during cancer and that IGF2 DMR0 is one such region. Demethylation in cancer may be passively mediated by failure to maintain methylation during replication or active via DNA repair mechanisms (46,47). Age-related DNA methylation changes at the IGF2/H19 locus have been previously reported (48) and there are also indications that heritable factors may determine methylation levels at this locus (49). Within the EPIC sample set we found a strong correlation with age and hypomethylation (Table 2). In our study, IGF2 DMR0 methylation decreased after 60 years of age. Thus, if genetic factors initially predispose to hypomethylation, environmental or age-related factors probably augment hypomethylation. We have recently reported that the methylation profiles in congenital growth disorders with IGF2 LOI and Wilms tumour cases with IGF2 LOI are not the same (36). This would suggest that even if similar epigenetic reprogramming mechanisms operate in the germ-line and in cancer tissues the IGF2 and H19 DMR sequences respond differently.
The results of our study in the EPIC samples contradict reports that show an increased frequency of LOI in peripheral blood samples of colorectal cancer patients (13–15). The peripheral blood samples in these authors' studies came from individuals who were known to have LOI in colon tissue. The difference in our study is firstly that our prospective study utilized blood samples that did not come from the same patients as our tumour samples. Secondly, we looked at IGF2 DMR0 methylation levels as an indicator for cancer predisposition, whereas other studies examined LOI in mRNA. IGF2 expression is very low in peripheral blood samples. RNA analysis by nested PCR in peripheral blood could enrich for a subset of IGF2 expressing cells with LOI that is beyond the range of detection of quantitative pyrosequencing methylation analysis.
Overall, our results show that hypomethylation of IGF2 DMR0 is an acquired cancer-specific epigenetic event. Hypomethylation of IGF2 DMR0 is more frequent than LOI and may be a consequence of LOI as well as other cancer-specific epigenetic-mediated events such as chromosomal and chromatin rearrangements at the locus. It is plausible that the detection of lower levels of methylation in cancer tissue samples reflects a relative sampling effect in tumours with less-differentiated cell populations. Routine single cell analysis of methylation in situ is not yet feasible, but if loss of methylation at the DMR0 is a feature of less differentiated cells, then pyrosequencing assays for methylation may prove a valuable alternate tool for quantifying less differentiated cells in heterogeneous tumour samples. Because IGF2 DMR0 hypomethylation is so highly prevalent in colon tissue from colorectal cancer patients and rarely present constitutively, it has a value as a diagnostic indicator of colorectal cancer and should be added to the modest list of discriminant methylation markers suitable for cancer screening (50–53). Currently, there is no national UK screening programme aimed at reducing colorectal cancer-related deaths. The Danish (54) and the Nottingham (55) randomized controlled trials and the colorectal screening pilot study (56) have shown the efficacy of screening with faecal-occult-blood test (FOBT) in reducing colorectal cancer-related mortality. The PPV of FOBT ranges from 9 to 17% with a sensitivity of ∼52% (56). Such low sensitivity and PPV result in a large number of unnecessary colonoscopies and highlight the need for a test with better predictive value. Our IGF2 DMR0 hypomethylation assay is robust and requires only 100 ng of DNA which makes it suitable as a screening test to complement FOBT. While the clinical application of these results with regard to diagnostic value is clear, detailed analyses of hypomethylation and IGF2 expression levels in subgroups of colorectal tumours, and normal tissue at different stages of disease are still required in order to evaluate the prognostic value of IGF2 hypomethylation.
MATERIALS AND METHODS
Patient and population samples
Colorectal and breast tumour samples
DNA and RNA from Colorectal tumour samples and adjacent controls were extracted from tissue obtained from the Addenbrooke's hospital (NIHR Cambridge Biomedical Research Centre, LREC 06/Q0108/307).
Colorectal tissue was collected from surgically resected colectomy specimens where normal tissue, ‘near’ and ‘further away’ from the tumour as well as carcinoma tissue samples were available. Sections from tissue immediately adjacent to the analysed samples were examined to confirm their histological nature.
DNA and RNA from breast tumour samples and adjacent controls were extracted from tissue that had been collected during surgery from patients treated at Colchester General Hospital (Essex, UK), with written consent taken before surgery (OREC ref MH363). The samples were immediately frozen and stored at −80°C. Breast cancers were invasive ductal carcinomas (n = 12); invasive adenocarcinomas (n = 6); tumours with mixed phenotypes (n = 2); DCIS (n = 1) and mucinous carcinomas (n = 1).
Placenta samples
Placental tissues were collected from term pregnancies at Queen Charlotte's and Chelsea Hospital, London under the guidance of Hammersmith and Queen Charlotte's and Chelsea Hospital Trust Research Ethics Committee (ref 2001/6029). From each placenta, five 1 cm3 biopsies were sectioned from the foetal side and washed three times in 1xPBS to remove maternal blood, and DNA extraction was performed as previously described (26).
Peripheral blood samples
DNA from peripheral blood samples was obtained from the following cohorts EPIC—Norfolk population (40); SEARCH population (43); Breast Cancer Consortium (ABC) (44). All cancers in these cohorts were invasive, i.e. no polyps or DCIS were included. Information on grade and stage of cancers was not made available.
The cases and controls were randomly allocated and methylation assays were done blind to case–control status. The nested study design utilizing the EPIC samples uniquely capitalises on the fact that DNA was extracted from individuals who were healthy and subsequently went on to develop disease. This approach provides an exceptional opportunity to examine whether the loss of methylation at IGF2 DMR0 occurs prior to disease onset. The cases within the SEARCH and Breast Cancer Consortium populations had been diagnosed with colorectal and breast cancer at the time of blood collection. Number of samples required for statistical power of 80% with an alpha of 0.01 were calculated to be 200–250 cases and 400–500 based on an assumption that the true relative risk is two, which would be consistent with a simplistic exposure-risk models (in this case, normal methylation exposure to hypomethylated exposure), and that based on earlier literature (57,58), 10% of controls were expected to have hypomethylation.
Analysis of imprinting status
IGF2 imprinting status was determined by assaying for Apa1 polymorphism (rs680) within the IGF2 exon 9 by restriction digestion or sequencing of PCR products obtained as described previously (14). We called LOI when we could clearly see two visible bands on a gel which was at least in a 1:4 ratio. Sequence traces were called biallelic when two peaks above background could be discerned.
Pyrosequencing assay
Our pyrosequencing assay for IGF2 DMR0 region (Fig. 1) is as described previously (36) and included six CpGs, three of which were previously reported to be hypomethylated in colorectal patients with LOI at IGF2 (15). For high through-put analysis, the following controls were included on each 96 well plate: DNA from a cell line SUM159 which has 0–20% methylation at IGF2 DMR0 as well as commercially obtained pooled peripheral blood leucocyte DNA (BD Biosciences, Clonetech range) which had methylation levels of 42–43%. These samples enabled us to check our inter-assay variation. Using a minimum of 100 ng of DNA as template for bisulphite conversion eliminated experimental error due to allele drop-out and the inter- and intra-assay variability was 5 and 2%, respectively. Linearity of the assay was checked by adding unmethylated template sequences (obtained by PCR amplification of target sequence, and then bisulphite converting the PCR products) in varying concentrations to bisulphite converted genomic DNA.
The methylation quantification was analysed by PSQ HS 96A SNP and Pyro Q-CpG Software (Biotage, Uppsala, Sweden) and other than excluding samples that did not meet quality control checks the data was not normalized.
In the EPIC cohort, we verified allele-specific loss of methylation by conventional bisulphite sequencing of cloned PCR products in sample of 12 hypomethylated cases and similarly confirmed allele-specific methylation in a further five random cases from the cohort.
Statistical analysis
GraphPad Prism software was used to determine Medians and IQR for tumour and control samples and for D' Agostino and Pearson omnibus normality tests which returned skewness and Kurtosis values on methylation profiles of peripheral blood and placenta samples. Free software from R Development Core Team http://www.R-project.org was used for Wilcoxon Rank Sum test to determine whether there were differences in methylation levels in tumours and adjacent non-tumour tissues, as well as in cases with and without LOI. PPVs were calculated in Microsoft Excel using Pearson Clopper Exact Confidence intervals because of the small sample size (59).
For larger data sets such as the EPIC, ABC and SEARCH cohorts, baseline characteristics of cancer cases and controls by cancer site were calculated using means and 95% confidence intervals. Loss of methylation (LOM) was defined as level of methylation ≤35% and non loss of methylation (non-LOM) is equivalent to a methylation status >35%. Mean age, smoking pack per year and BMI (continuous variables) were analysed in LOM and non-LOM groups using the t test statistic. We used a χ2 test to compare proportions such as family history of cancer and sex ratio with LOM and non-LOM. Because this study is a match case and control study, association between case/control status and LOM was examined using conditional logistic regression (fixed effect). At first, we tested the association for breast and colorectal cancer separately; we then pooled both cancers and tested the association. Conditional logistic regression models were adjusted for BMI, number of packet smocked per year and family history of cancer.
P-values less than 0.05 were considered to be statistically significant. We used the STATA statistical package, version 8.2 (Stats Corp., College Station, TX, USA) for all analyses. All P-values were two-sided.
SUPPLEMENTARY MATERIAL
FUNDING
Funding for this work was from Cancer Research United Kingdom, Senior Cancer Research Fellowship (A.M.), Association for International Cancer Research (A.M), Wenner-Gren Foundation (M.J.) and Wellcome Trust (S.B.). Cancer Research United Kingdom; Medical Research Council; British Heart Foundation; Food Standards Agency; the Department of Health; Academy of Medical Sciences (EPIC Norfolk Study). Funding to pay the Open Access Publication charges for this article was provided by Cancer Research United Kingdom.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the support of the NIHR Cambridge Biomedical Research Centre, The University of Cambridge, Cancer Research UK and Hutchison Whampoa Limited.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.van Roozendaal C.E., Gillis A.J., Klijn J.G., van Ooijen B., Claassen C.J., Eggermont A.M., Henzen-Logmans S.C., Oosterhuis J.W., Foekens J.A., Looijenga L.H. Loss of imprinting of IGF2 and not H19 in breast cancer, adjacent normal tissue and derived fibroblast cultures. FEBS Lett. 1998;437:107–111. doi: 10.1016/s0014-5793(98)01211-3. [DOI] [PubMed] [Google Scholar]
- 3.Kim K.S., Lee Y.I. Biallelic expression of the H19 and IGF2 genes in hepatocellular carcinoma. Cancer Lett. 1997;119:143–148. doi: 10.1016/s0304-3835(97)00264-4. [DOI] [PubMed] [Google Scholar]
- 4.Kim H.T., Choi B.H., Niikawa N., Lee T.S., Chang S.I. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am. J. Med. Genet. 1998;80:391–395. doi: 10.1002/(sici)1096-8628(19981204)80:4<391::aid-ajmg16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 5.Murphy S.K., Huang Z., Wen Y., Spillman M.A., Whitaker R.S., Simel L.R., Nichols T.D., Marks J.R., Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 2006;4:283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 6.Byun H.M., Wong H.L., Birnstein E.A., Wolff E.M., Liang G., Yang A.S. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 7.Liou J.-M., Wu M.-S., Lin J.-T., Wang H.-P., Huang S.-P., Chiu H.-M., Lee Y.-C., Lin Y.-B., Shun C.-T., Liang T. Loss of imprinting of insulin-like growth factor II is associated with increased risk of proximal colon cancer. Eur. J. Cancer. 2007;43:1276–1282. doi: 10.1016/j.ejca.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki J., Konishi F., Kawamura Y.J., Kai T., Takata O., Tsukamoto T. Clinicopathological characteristics of colorectal cancers with loss of imprinting of insulin-like growth factor 2. Int. J. Cancer. 2006;119:80–83. doi: 10.1002/ijc.21741. [DOI] [PubMed] [Google Scholar]
- 9.Maenaka S., Hikichi T., Imai M.A., Minamoto T., Kawahara E. Loss of imprinting in IGF2 in colorectal carcinoma assessed by microdissection. Oncol. Rep. 2006;15:791–795. [PubMed] [Google Scholar]
- 10.Kaneda A., Feinberg A.P. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 11.Woodson K., Flood A., Green L., Tangrea J.A., Hanson J., Cash B., Schatzkin A., Schoenfeld P. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J. Natl Cancer Inst. 2004;96:407–410. doi: 10.1093/jnci/djh042. [DOI] [PubMed] [Google Scholar]
- 12.Sasamoto H., Nagasaka T., Notohara K., Ozaki K., Isozaki H., Tanaka N., Matsubara N. Allele-specific methylation analysis on upstream promoter region of H19 by methylation-specific PCR with confronting two-pair primers. Int. J. Oncol. 2004;25:1273–1278. [PubMed] [Google Scholar]
- 13.Cruz-Correa M., Cui H., Giardiello F.M., Powe N.R., Hylind L., Robinson A., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., et al. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126:964–970. doi: 10.1053/j.gastro.2003.12.051. [DOI] [PubMed] [Google Scholar]
- 14.Cui H., Cruz-Correa M., Giardiello F.M., Hutcheon D.F., Kafonek D.R., Brandenburg S., Wu Y., He X., Powe N.R., Feinberg A.P. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 15.Cui H., Onyango P., Brandenburg S., Wu Y., Hsieh C.L., Feinberg A.P. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 16.Arslan A.A., Gold L.I., Mittal K., Suen T.-C., Belitskaya-Levy I., Tang M.-S., Toniolo P. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: new evidence and a systematic review. Hum. Reprod. 2005;20:852–863. doi: 10.1093/humrep/deh698. [DOI] [PubMed] [Google Scholar]
- 17.Bertucci F., Salas S., Eysteries S., Nasser V., Finetti P., Ginestier C., Charafe-Jauffret E., Loriod B., Bachelart L., Montfort J., et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 18.Giordano T.J., Thomas D.G., Kuick R., Lizyness M., Misek D.E., Smith A.L., Sanders D., Aljundi R.T., Gauger P.G., Thompson N.W., et al. Distinct transcriptional profiles of adrenocortical tumors uncovered by DNA microarray analysis. Am. J. Pathol. 2003;162:521–531. doi: 10.1016/S0002-9440(10)63846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwienbacher C., Angioni A., Scelfo R., Veronese A., Calin G.A., Massazza G., Hatada I., Barbanti-Brodano G., Negrini M. Abnormal RNA expression of 11p15 imprinted genes and kidney developmental genes in Wilms’ tumor. Cancer Res. 2000;60:1521–1525. [PubMed] [Google Scholar]
- 20.Cooper W.N., Luharia A., Evans G.A., Raza H., Haire A.C., Grundy R., Bowdin S.C., Riccio A., Sebastio G., Bliek J., et al. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 2005;13:1025–1032. doi: 10.1038/sj.ejhg.5201463. [DOI] [PubMed] [Google Scholar]
- 21.Reik W., Constancia M., Dean W., Davies K., Bowden L., Murrell A., Feil R., Walter J., Kelsey G. Igf2 imprinting in development and disease. Int. J. Dev. Biol. 2000;44:145–150. [PubMed] [Google Scholar]
- 22.Weksberg R., Nishikawa J., Caluseriu O., Fei Y.L., Shuman C., Wei C., Steele L., Cameron J., Smith A., Ambus I., et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum. Mol. Genet. 2001;10:2989–3000. doi: 10.1093/hmg/10.26.2989. [DOI] [PubMed] [Google Scholar]
- 23.Thorvaldsen J.L., Duran K.L., Bartolomei M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constancia M., Dean W., Lopes S., Moore T., Kelsey G., Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 25.Lopes S., Lewis A., Hajkova P., Dean W., Oswald J., Forne T., Murrell A., Constancia M., Bartolomei M., Walter J., et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 26.Monk D., Sanches R., Arnaud P., Apostolidou S., Hills F.A., Abu-Amero S., Murrell A., Friess H., Reik W., Stanier P., et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 27.Murrell A., Heeson S., Bowden L., Constancia M., Dean W., Kelsey G., Reik W. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 29.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 30.Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 31.Szabo P., Tang S.H., Rentsendorj A., Pfeifer G.P., Mann J.R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 32.Lewis A., Murrell A. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 2004;14:R284–R286. doi: 10.1016/j.cub.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrell A., Heeson S., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 35.Moore T., Constancia M., Zubair M., Bailleul B., Feil R., Sasaki H., Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl Acad. Sci. USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murrell A., Ito Y., Verde G., Huddleston J., Woodfine K., Silengo M., Spreafico F., Perotti D., De Crescenzo A., Sparago A., et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PlosONE. 2008;3:e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan M.J., Taniguchi T., Jhee A., Kerr N., Reeve A.E. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene. 1999;18:7527–7534. doi: 10.1038/sj.onc.1203096. [DOI] [PubMed] [Google Scholar]
- 38.Xu W., Fan H., He X., Zhang J., Xie W. LOI of IGF2 is associated with esophageal cancer and linked to methylation status of IGF2 DMR. J. Exp. Clin. Cancer Res. 2006;25:543–547. [PubMed] [Google Scholar]
- 39.Ulaner G.A., Vu T.H., Li T., Hu J.F., Yao X.M., Yang Y., Gorlick R., Meyers P., Healey J., Ladanyi M., et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 2003;12:535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 40.Day N., Oakes S., Luben R., Khaw K.T., Bingham S., Welch A., Wareham N. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br. J. Cancer. 1999;80(Suppl. 1):95–103. [PubMed] [Google Scholar]
- 41.Colella S., Shen L., Baggerly K.A., Issa J.P., Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 42.Tost J., Gut I.G. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol. Biol. 2007;373:89–102. doi: 10.1385/1-59745-377-3:89. [DOI] [PubMed] [Google Scholar]
- 43.Cebrian A., Lesueur F., Martin S., Leyland J., Ahmed S., Luccarini C., Smith P.L., Luben R., Whittaker J., Pharoah P.D., et al. Polymorphisms in the initiators of RET (rearranged during transfection) signaling pathway and susceptibility to sporadic medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2005;90:6268–6274. doi: 10.1210/jc.2004-2449. [DOI] [PubMed] [Google Scholar]
- 44.Easton D.F., Bishop D.T., Ford D., Crockford G.P. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 45.Linhart H.G., Lin H., Yamada Y., Moran E., Steine E.J., Gokhale S., Lo G., Cantu E., Ehrich M., He T., et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barreto G., Schafer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Doderlein G., Maltry N., Wu W., Lyko F., et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 47.Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 48.Ahuja N., Li Q., Mohan A.L., Baylin S.B., Issa J.P. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 49.Heijmans B.T., Kremer D., Tobi E.W., Boomsma D.I., Slagboom P.E. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 50.Chen W.-D., Han Z.J., Skoletsky J., Olson J., Sah J., Myeroff L., Platzer P., Lu S., Dawson D., Willis J., et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 51.Kann L., Han J., Ahlquist D., Levin T., Rex D., Whitney D., Markowitz S., Shuber A. Improved marker combination for detection of de novo genetic variation and aberrant DNA in colorectal neoplasia. Clin. Chem. 2006;52:2299–2302. doi: 10.1373/clinchem.2007.070896. [DOI] [PubMed] [Google Scholar]
- 52.Koinuma K., Kaneda R., Toyota M., Yamashita Y., Takada S., Choi Y.L., Wada T., Okada M., Konishi F., Nagai H., et al. Screening for genomic fragments that are methylated specifically in colorectal carcinoma with a methylated MLH1 promoter. Carcinogenesis. 2005;26:2078–2085. doi: 10.1093/carcin/bgi184. [DOI] [PubMed] [Google Scholar]
- 53.Zou H., Harrington J.J., Shire A.M., Rego R.L., Wang L., Campbell M.E., Oberg A.L., Ahlquist D.A. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol. Biomarkers Prev. 2007;16:2686–2696. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 54.Kronborg O., Fenger C., Olsen J., Jorgensen O.D., Sondergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467–1471. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 55.Hardcastle J.D., Chamberlain J.O., Robinson M.H., Moss S.M., Amar S.S., Balfour T.W., James P.D., Mangham C.M. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472–1477. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 56.Alexander F., Weller D. Evaluation of the UK Colorectal Cancer Screening Pilot. Final Report. 2003 http://www.cancerscreening.nhs.uk/bowel/pilot.html . [Google Scholar]
- 57.Sakatani T., Wei M., Katoh M., Okita C., Wada D., Mitsuya K., Meguro M., Ikeguchi M., Ito H., Tycko B., et al. Epigenetic heterogeneity at imprinted loci in normal populations. Biochem. Biophys. Res. Commun. 2001;283:1124–1130. doi: 10.1006/bbrc.2001.4916. [DOI] [PubMed] [Google Scholar]
- 58.Sandovici I., Leppert M., Hawk P.R., Suarez A., Linares Y., Sapienza C. Familial aggregation of abnormal methylation of parental alleles at the IGF2/H19 and IGF2R differentially methylated regions. Hum. Mol. Genet. 2003;12:1569–1578. doi: 10.1093/hmg/ddg167. [DOI] [PubMed] [Google Scholar]
- 59.Clopper C.J., Pearson E. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.