Abstract
Mutations in Mucolipin 1 (MCOLN1) have been linked to mucolipidosis type IV (MLIV), a lysosomal storage disease characterized by several neurological and ophthalmological abnormalities. It has been proposed that MCOLN1 might regulate transport of membrane components in the late endosomal–lysosomal pathway; however, the mechanisms by which defects of MCOLN1 function result in mental and psychomotor retardation remain largely unknown. In this study, we show constitutive activation of autophagy in fibroblasts obtained from MLIV patients. Accumulation of autophagosomes in MLIV cells was due to the increased de novo autophagosome formation and to delayed fusion of autophagosomes with late endosomes/lysosomes. Impairment of the autophagic pathway led to increased levels and aggregation of p62, suggesting that abnormal accumulation of ubiquitin proteins may contribute to the neurodegeneration observed in MLIV patients. In addition, we found that delivery of platelet-derived growth factor receptor to lysosomes is delayed in MCOLN1-deficient cells, suggesting that MCOLN1 is necessary for efficient fusion of both autophagosomes and late endosomes with lysosomes. Our data are in agreement with recent evidence showing that autophagic defects may be a common characteristic of many neurodegenerative disorders.
INTRODUCTION
Autophagy (or more specifically macroautophagy) is a process that mediates sequestration of macromolecules and organelles into specialized cytosolic vesicles for subsequent delivery and degradation in lysosomes (1–3). Activation of autophagy occurs when cells undergo stress conditions, such as starvation, accumulation of damaged organelles or oxidative stress, and serves two main purposes: the production of nutrients and the elimination of products potentially toxic for the cell (4). Recently, it has been suggested that autophagy may also play a very important role in cell homeostasis during non-stress conditions, as the basal activity of the autophagic process is required for the degradation of misfolded proteins and organelle turnover. Basal autophagy may be especially important in neurons, where accumulation of aggregated proteins often results in cell death (5). Accordingly, transgenic mice that lack specific components of the autophagic machinery exhibit accumulation of ubiquitin-rich inclusions in neurons and progressive neurodegeneration (6,7). Moreover, accumulation of autophagic vacuoles has been described in several neurodegenerative disorders in humans, including Parkinson's (8,9), Alzheimer's (10,11) and Huntington's disease (12).
The autophagic machinery is highly conserved among eukaryotes. In yeast, over 30 autophagy-related (Atg) proteins have been identified (13). Most of these proteins mediate protein–protein and protein–lipid conjugation events, although some others, such as phosphoinositide 3-kinase (PI3K) class III and beclin-1, have an important regulatory role. The process starts with the sequestration of a portion of the cytoplasm by a double membrane structure that grows and seals to form an autophagosome. LC3, the mammalian homolog of yeast Atg8, is one of the most selective markers for autophagosomes. Cleavage and subsequent attachment of a phosphatidylethanolamine (PE) group (14) allows insertion of LC3 into the membrane of the autophagosome. It is widely accepted that increased levels of PE-conjugated LC3 correlates with increased autophagy (15).
Autophagosomes can fuse with different types of vesicles, including endosomal multivesicular bodies (MVB) and lysosomes. This fusion ensures degradation of the autophagosome content by lysosomal enzymes. Recent evidence suggests that alterations on the endosomal–lysosomal pathway that impair autophagosome degradation may also play a role in a number of neurodegenerative disorders. For example, mutations in CHMP2B, a component of the ESCRT machinery that mediates sorting of proteins into the luminal vesicles of MVBs, have been described in patients with amyotrophic lateral sclerosis (16) and a rare form of fronto-temporal dementia (17); while spastin, a protein mutated in many cases of hereditary spastic paraplegia, is known to interact with CHMP1B (18). In addition, depletion of specific subunits of the ESCRT complex by siRNA results in increased autophagy and accumulation of protein aggregates containing ubiquitinated proteins (19). Therefore, efficient autophagy requires fusion with a functional endosomal–lysosomal pathway, and defects in autophagy degradation cause accumulation of ubiquitinated protein inclusions that may lead to neurodegeneration (20).
Mucolipidosis type IV (MLIV) is a lysosomal storage disorder (LSD) characterized by severe neurological and ophthalmological abnormalities due to defective transport of membrane components in the late endosomal–lysosomal pathway. Individuals with MLIV suffer from mental and psychomotor retardation, severe neurodegeneration, diminished muscle tone or hypotonia, achlorhydria and visual problems including corneal clouding, retinal degeneration, sensitivity to light and strabismus (21–23). Mucolipin-1 (MCOLN1), the protein mutated in this disease, is a cation channel with homology to the transient receptor potential channel superfamily (24–27). MCOLN1 consists of six transmembrane-spanning domains with the N- and C-terminal tails oriented within the cytosol and the pore located between transmembrane segments 5 and 6. Two acidic di-leucine sorting motifs are responsible for the transport of the protein to late endosomes/lysosomes (28–30). MCOLN1 is a non-selective channel with preference for monovalent cations (31–34) and its activity can be modulated by phosphorylation (35) and by changes in pH and Ca2+ concentration (32,33,36).
Analysis of fibroblasts obtained from MLIV patients revealed the accumulation of enlarged vacuoles that contain sphingolipids, phospholipids and acid mucopolysaccharides (24,37–39), suggesting that MCOLN1 might be necessary for the correct trafficking of protein and lipids along the late endosomal/lysosomal pathway. This idea is supported by experiments in Caenorhabditis elegans showing that knockout of cup-5, the ortholog of MCOLN1, resulted in the formation of enlarged hybrid organelles that contain both late-endosomal and lysosomal markers (40). Recently, several groups have suggested additional roles for MCOLN1 in different cellular processes, including regulation of lysosomal acidification (36), mitochondria turnover (41) and lysosomal secretion (42). However, the mechanism by which defects on MCOLN1 function result on mental and psychomotor retardation remains largely unknown.
In this study, we examined the autophagic process in fibroblasts from MLIV patients. We reasoned that the defects on the late endosomal/lysosomal pathway observed in MLIV might have a profound impact on the formation and/or degradation of autophagosomes. As predicted, we found that absence of MCOLN1 resulted in increased basal autophagy and decreased autophagosome degradation. In normal conditions, MLIV fibroblasts accumulate many more LC3-positive structures than control fibroblasts. Almost all the LC3-positive structures also contain p62, suggesting an abnormal accumulation of ubiquitinated proteins in patients. After autophagy induction by starvation, fusion of autophagosomes with late endosomes–lysosomes was less efficient in MLIV fibroblasts. We also observed delayed delivery of PDGFR to lysosomes in MCOLN1-deficient cells. Therefore, MCOLN1 seems to regulate trafficking events along the late endosomal–lysosomal pathway and we propose that defects on these trafficking events result in inefficient autophagy and accumulation of ubiquitinated proteins that may contribute to neurodegenerative process observed in MLIV patients.
RESULTS
Monitoring autophagy in human fibroblasts
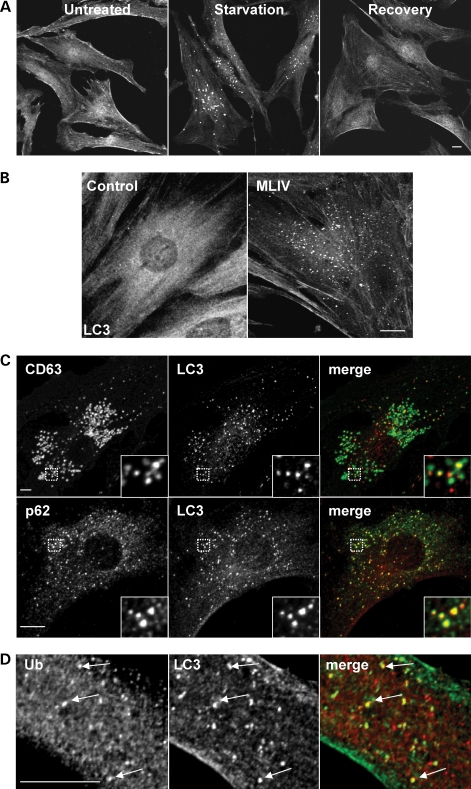
LC3 is the most commonly used marker to monitor autophagy in mammalian cells. Under normal conditions, LC3 shows a cytosolic distribution; however, following authophagy induction, it is recruited to newly formed autophagosomes and remains associated to these structures until fusion of autophagosomes with late endosomes/lysosomes. As seen in Figure 1A, starvation of fibroblasts by incubation with Earle's balanced salt solution (EBSS) for 3 h caused an evident recruitment of LC3 to discrete vesicular structures. Subsequent incubation with complete media for one additional hour (recovery) allowed autophagosome degradation and redistribution of LC3 to cytosol. Therefore, monitoring changes in the distribution of endogenous LC3 is a suitable way to follow autophagy in human fibroblasts.
Figure 1.
Increased autophagy in fibroblasts from MLIV patients. (A) Control fibroblasts grown in complete media (untreated, left), starved in EBSS media for 3 h (starvation, center) or starved for 3 h followed by 1 h of recovery in complete media (recovery, right), were fixed, permeabilized and immunostained with a polyclonal antibody to endogenous LC3. (B) LC3 staining in untreated control and MLIV fibroblasts. Note that in contrast to a diffuse staining in control fibroblasts, LC3 shows a vesicular staining in MLIV fibroblasts. (C) MLIV fibroblasts grown in complete media were fixed, permeabilized and immunostained with a polyclonal antibody to LC3 (red) and a monoclonal antibody against CD63 or p62 (green). Insets show a 3-fold magnification of the indicated region (D) Double immunostaining of untreated MLIV cells with anti-LC3 (green) and anti-ubiquitin (red). Cells were examined by confocal fluorescence microscopy. Merging the images in the red and green channels generated the third picture on each row; yellow indicates overlapping localization. Scale bars = 10 µm.
Basal levels of autophagy are increased in MLIV fibroblasts
To assess whether the autophagic process is altered in MLIV, we compared the distribution of endogenous LC3 in normal non-starvation conditions between control fibroblasts and fibroblasts obtained from MLIV patients. As expected, LC3 was mainly cytosolic in untreated control cells, although occasional LC3-labeled autophagosomes could also be detected (Fig. 1B). In contrast, numerous LC3-positive structures were observed in MLIV fibroblasts, suggesting a constitutive activation of basal autophagy in these cells (Fig. 1B). Quantification analysis revealed that the average number of autophagosomes in control cells was 12.8 (SD ± 1.3, n = 10), whereas in MLIV cells, the average number of autophagosomes increased to 54.2 (SD ± 3.2, n = 10).
We labeled MLIV cells with antibodies against LC3 and CD63, a marker for late endosomes/lysosomes. As seen in Figure 1C, some LC3-positive vesicles did not co-localize with CD63. These vesicles were probably nascent autophagosomes, because they were free from any lysosomal characteristics. We also observed many LC3-labeled puncta that co-localized with CD63, indicating fusion of autophagosomes with late endosomes/lysosomes.
Interestingly, most of the LC3-positive vesicles also contained p62 (Fig. 1C), a molecule commonly found in protein inclusions associated with neurodegenerative disorders (43). It has been suggested that the ability of p62 to interact with ubiquitinated proteins and LC3 (44) regulates the formation of protein aggregates that are removed by autophagy (20). In agreement with this idea, we found that many of the autophagosomes present in MLIV patient cells held ubiquitinated proteins (Fig. 1D). These results suggest that altered autophagy observed in MCOLN1-deficient cells results in accumulation of protein aggregates containing ubiquitinated proteins.
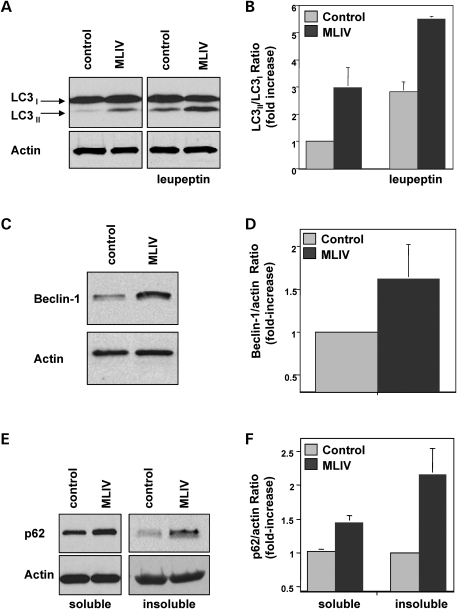
LC3 is initially synthesized in an unprocessed form that is rapidly cleaved in its C-terminal region to generate soluble LC3-I. During autophagy induction, the LC3-I isoform is converted into LC3-II by conjugation of a PE group that allows insertion of LC3 into the membrane of the autophagosome. It is well established that the amount of LC3-II correlates with the extent of autophagosome formation. To corroborate that basal autophagy is increased in MLIV, we analyzed the ratio between LC3-II and LC3-I in control and patient cells by immunoblot. As seen in Figure 2A, increased levels of LC3-II were detected in MLIV fibroblasts under basal conditions. Quantification of seven independent experiments showed a 3-fold increase (2.97; SD ± 0.3) in the LC3-II/LC3-I ratio in MCOLN1-deficient cells (Fig. 2B). It has been suggested that in some cases measurement of the LC3-II/actin ratio may be a more accurate way to monitor autophagy induction. Quantification of the LC3-II/actin ratio showed a 3.29-fold increase (SD ± 0.6; n = 7) in MLIV cells, thus confirming autophagy activation in patients (Supplementary Material, Fig. S1).
Figure 2.
Autophagy is upregulated in MLIV fibroblasts. (A) Control and MLIV fibroblasts were treated without or with 50 µg/ml leupeptin for 7 h. Cells were lysed and immunoblotted with anti-LC3 (top) or anti-actin (bottom). (B) Quantification of the experiment shown in (A). Autophagy was measured as the ratio of LC3-II/LC3-I and is represented as a fold increase of the ratio from control cells without treatment. Results are means ± SD (n = 7), P < 0.01. (C) Lysates from control fibroblasts and MLIV fibroblasts were examined by western blot for expression of beclin-1 and actin. (D) Quantification of the levels of beclin-1 normalized with actin is shown as a fold increase of control cells. Results are means ± SD (n = 9), P < 0.05. (E) Western blot showing the accumulation of p62 in the soluble and insoluble fractions of MLIV fibroblasts. (F) Quantification was performed by normalizing p62 with actin levels. Results are means ± SD (n = 4). Soluble P < 0.001; insoluble P < 0.05.
Increased autophagy may be due to either increased autophagosome formation or to decreased autophagosome degradation. To distinguish between these two possibilities, we incubated cells with leupeptin, an inhibitor of lysosomal proteases. If treatment with leupeptin does not cause an increase in the LC3-II levels, it is likely that autophagosome accumulation occurred due to inhibition of autophagic degradation. As seen in Figure 2A and B, the levels of LC3-II did increased in the presence of leupeptin both in control and patient cells. These results suggest that fusion of autophagosomes with lysosomes is not blocked in MLIV cells and that increased autophagy may be at least partially due to augmented autophagosome formation. However, it is important to note that leupeptin experiments do not rule out the possibility that autophagosome degradation is slower in MLIV cells (see below). Interestingly, expression of beclin-1, a component of the Class III PI3K complex that participates in autophagosome formation (45) was increased in MLIV fibroblasts (Fig. 2C and D). This upregulation of beclin-1 in response to MCOLN1 deficiency could explain the increased autophagosome formation in patient cells.
We also detected increased levels of p62 in immunoblots of cells lysates from MLIV fibroblasts (Fig. 2E and F). Importantly, we found over a 2-fold increase in the amount of insoluble p62 in patients (Fig. 2E and F). The fraction of protein that remains resistant to Triton X-100 extraction is often used as a quantitative measure of protein aggregation (46). Therefore, these data are in agreement with our immunofluorescence experiments and further demonstrate the accumulation of p62-positive inclusions in MLIV patient cells. Increased levels of LC3-II and insoluble p62 were also found in fibroblasts obtained from a different MLIV patient (see Materials and Methods), thus corroborating our results (Supplementary Material, Fig. S2).
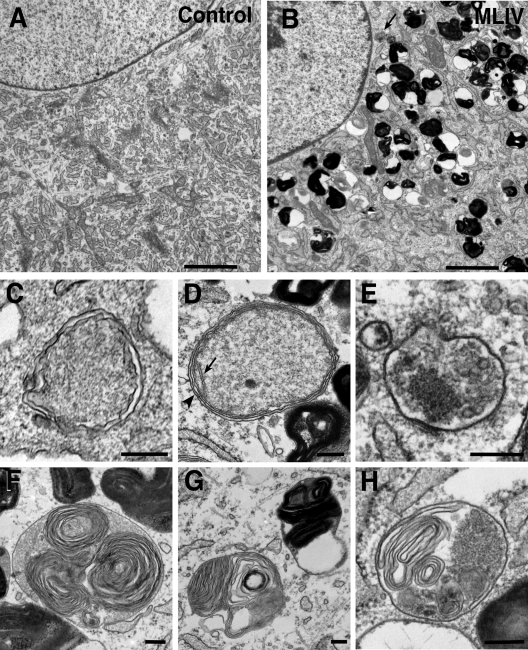
Finally, high levels of basal autophagy in MLIV fibroblasts were confirmed by transmission electron microscopy (Fig. 3). This analysis showed frequent autophagic vacuoles containing organelles and other cytoplasmic contents in mutant, but not in wild-type fibroblasts (Fig. 3A–E). In addition, we observed the characteristic concentric multi-lamellar structures and the electron-dense inclusions previously described in MLIV cells (Fig. 3F and G) (47). Vesicles containing lamellar structures and what seems to be semi-degraded material were also found (Fig. 3H) and might be the result of fusion between autophagosomes and multi-lamellar vesicles. Altogether, our results indicate constitutive enhancement of the autophagic flux in MCOLN1-depleted cells.
Figure 3.
Accumulation of autophagosomes in MLIV fibroblasts. Electron micrographs of control fibroblasts (A) and MLIV fibroblasts (B). Note the accumulation of electron-dense lysosomes and autophagosomes [arrow indicates example similar to that shown in (E)] present in MLIV cells. Scale bars = 2 µm. (C–E). High magnification images of membrane-bound structures with cytoplasmic contents, consistent with autophagosomes, in MLIV fibroblasts. In (D), the arrow indicates a crista in a mitochondrion. The arrowhead indicates an area where the mitochondrial inner and outer membranes and the surrounding double membrane of the autophagosome are clearly seen. Scale bars = 200 nm. (F and G) Characteristic lysosomes in MLIV fibroblasts containing concentric lamellar inclusions. Scale bars = 200 nm. (H) EM images of MLIV fibroblasts showing autolysosomes containing the concentric lamellar inclusions and cytoplasmatic contents. Scale bars = 200 nm.
Starvation-induced autophagy does not require MCOLN1
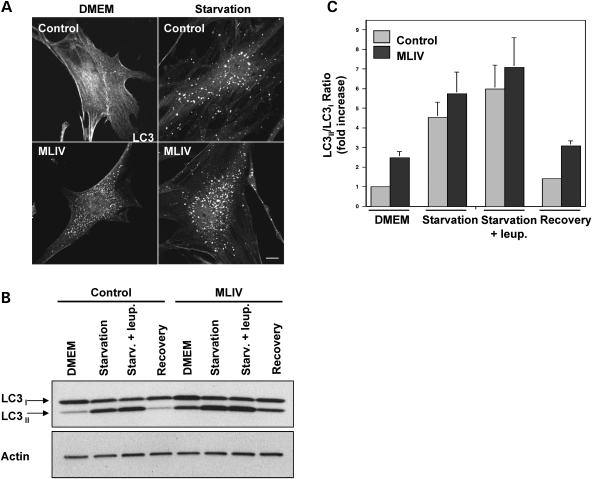
Next, we compared autophagy induction under stress conditions in control and mutant fibroblasts. Cells were incubated for 3 h in medium without amino acids (EBSS) to induce autophagy. As seen in Figure 4A, nutrient deprivation caused an increase in the number and size of LC3-positive vesicles. The average number of autophagosomes per cell under starvation conditions was 38.9 (SD ± 6.4, n = 12) in control fibroblasts and 79.88 (SD ± 9.31; n = 10) in MLIV fibroblasts.
Figure 4.
Starvation induces robust autophagy in fibroblasts from MLIV patients. (A) Control fibroblasts or MLIV fibroblasts were fixed under control conditions (DMEM) or after 3 h of starvation and immunostained with an antibody to endogenous LC3. Note that, as shown in Figure 2, in normal conditions, LC3 shows a vesicular staining in MLIV fibroblasts in contrast with the diffuse staining in control fibroblasts. After 3 hours of starvation autophagy is further increased. Scale bar = 10 µm. (B) Lysates from control fibroblasts or fibroblasts from a MLIV patient were collected from untreated cells (DMEM), cells starved for 3 hours in the absence or presence of 50 µg/ml leupeptin, or cells starved for 3 hours followed by 1 hour of recovery (recovery). Endogenous LC3 (top) and actin (bottom) were visualized by western blot. (C) Autophagy was measured as the ratio of LC3-II/LC3-I and is represented as a fold increase of the ratio from control cells in normal media (DMEM). Results are means±S.D. (n = 5). Starvation+leup P < 0.05; DMEM, Starvation P < 0.01; Recovery P < 0.001.
Measurement of LC3-II/LC3-I ratio immunobloting confirmed robust autophagy induction in both control and mutant cells after starvation (Fig. 4B). Quantification of four independent experiments showed a 4.54-fold increase (SD ± 0.76) in the LC3-II/LC3-I ratio after starvation in control cells and a 5.76-fold increase (SD ± 1.08) in MCOLN1-deficient cells (Fig. 4C). Similar results were obtained when the LC3-II/actin ratio was measured (Supplementary Material, Fig. S1).
Treatment of starved cells with leupeptin resulted in a further increase of the LC3-II levels in both cell types, consistent with the notion that fusion of autophagosomes and lysosomes is not blocked in MLIV cells (Fig. 4B and C). Moreover, when the cells were let to recover in complete medium for 1 h after starvation, the LC3-II/LC3-I ratio was reduced close to the level observed in untreated cells (Fig. 4B and C). Therefore, these results indicate that starvation-induced autophagy and autophagosome degradation can occur in the absence of MCOLN1.
Fusion of autophagosomes with the late endosomal/lysosomal pathway is delayed in MLIV fibroblasts
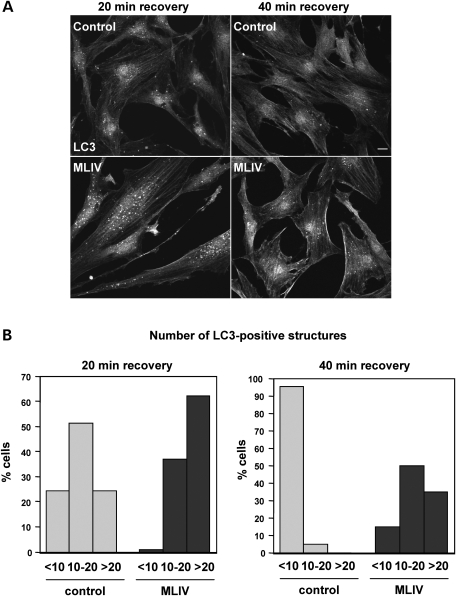
As mentioned before, the increased level of LC3-II observed after treatment with leupeptin indicates that fusion of autophagosomes with late endosomes/lysosomes is not blocked in MLIV fibroblasts (Figs 2B and 4B). However, these experiments could not disprove that the fusion was delayed at some extent in mutant cells. To test this possibility, we starved cells for 3 h in EBSS media followed by short recovery times of 20 and 40 min. As seen in Figure 5A, the number of LC3-positive vesicles was considerably higher in MCOLN1-deficient cells when compared with control cells. Quantification of the total number of autophagosomes per cell revealed that after 20 min recovery only 24.3% of control cells showed more than 20 autophagosomes and 24.3% had less than 10 (n = 37). In contrast, 62% of MCOLN1-deficient cells contained more than 20 autophagosomes and only 1% had less than 10 (n = 24) (Fig. 5B). After a 40 min recovery, there was an increase in the percent of MCOLN1-deficient cells containing less than 10 autophagosomes (from 1% at 20 min recovery to 15% at 40 min) and a decrease in the percent of cells showing more than 20 (from 62–35%) corroborating one more time that degradation of autophagosomes is not blocked in MLIV fibroblasts. However, the number of cells containing more than 20 autophagosomes after a 40 min recovery was significantly greater in mutant cells [32% in mutant cells (n = 20) versus 0% in control cells (n = 44)], whereas the percent of cells showing less than 10 autophagosomes was higher in control cells (95.45% in control cells versus 15% in mutant cells) (Fig. 5B). Similar results were obtained when we analyzed the number of autophagosomes after 20 and 40 min recovery in a different patient cell line (Supplementary Material, Fig. S3). Therefore, our data indicate that autophagosome degradation is in fact delayed in MLIV cells.
Figure 5.
Degradation of autophagosomes is delayed in MLIV fibroblasts. (A) Control and MLIV fibroblasts were starved in EBSS media for 3 h and then allowed to recover for 20 or 40 min in complete medium. Cells were then fixed and LC3 localization was examined by immunofluorescence. Scale bar = 10 µm. (B) Percentage of cells showing more than 20, between 10 and 20 or less than 10 LC3-positive structures after a 20 min recovery (left panel) or 40 min of recovery (right panel).
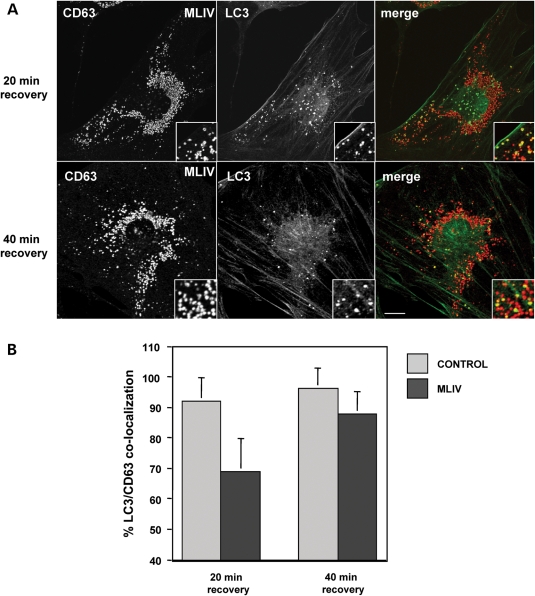
When we analyzed co-localization between CD63 and LC3 as a sign of fusion between autophagosomes and late endosomes/lysosomes, we found that in MLIV patient cells 68.8% (SD ± 11) of the LC3-positive vesicles co-localize with CD63 after 20 min recovery, and this percentage increased to 87.7% (SD ± 7) after 40 min (Fig. 6). However, the extent of LC3/CD63 co-localization was significantly reduced in MLIV fibroblasts when compared with control cells (92% in control cells versus 68.8% in mutant cells after a 20 min recovery), demonstrating that fusion of autophagosomes with late endosomes/lysosomes is less efficient in MCOLN1-deficient cells.
Figure 6.
Fusion of autophagosomes and lysosomes is delayed in MLIV fibroblasts. (A) MLIV fibroblasts were starved for 3 h and then allowed to recover 20 or 40 min in complete medium. Cells were fixed, permeabilized and immunostained with a polyclonal antibody to LC3 (green) and a monoclonal antibody to CD63 (red). Insets are 3-fold magnification. Cells were examined by confocal fluorescence microscopy. Merging images in the red and green channels generated the third picture on each row; yellow indicates overlapping localization. Scale bar = 10 µm. (B) Percentage of autophagosome–lysosome fusion events was quantified both in control and MLIV fibroblasts after 20 or 40 min of recovery, as the fraction of LC3 positive structures that co-localized with the lysosomal marker CD63. 20 min recovery, P < 0.01; 40 min recovery, P < 0.05.
Overexpression of MCOLN1 in NRK cells induces accumulation of autophagosomes
It has been reported that the expression of certain proteins that alter the endosomal/lysosomal pathway may affect the efficiency of the autophagic process. For example, overexpression of a mutant version of CHPM2B, one of the components of the ESCRT-III complex, induces accumulation of LC3-positive vesicles presumably due to defects in the fusion of autophagosomes with MVBs (19). We have previously described that overexpression of MCOLN1 affects the endosomal/lysosomal pathway and induces enlargement of late endosomes/lysomes (28). Therefore, in this study, we examined whether MCOLN1 overexpression might cause autophagosome accumulation.
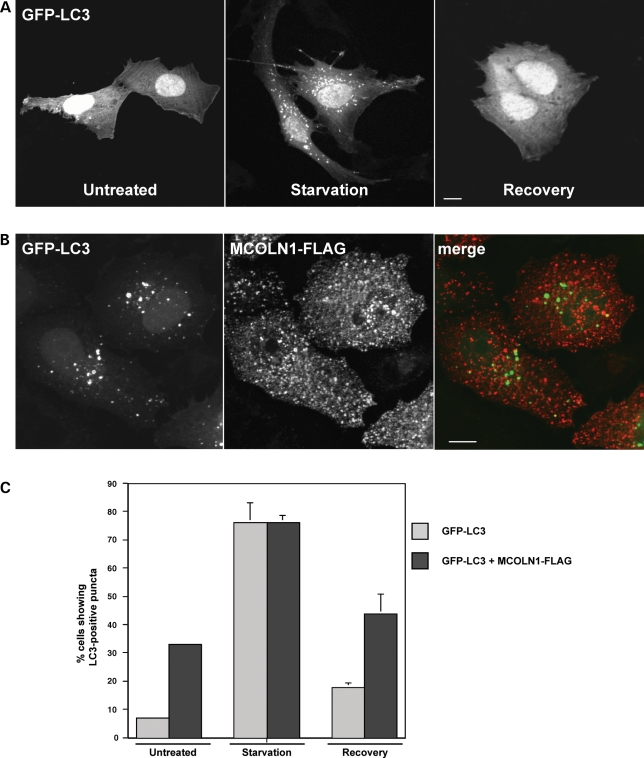
To monitor autophagic activity, we followed changes in the distribution of GFP-tagged LC3 transiently expressed in NRK cells. Similar to endogenous LC3, GFP-LC3 was largely cytosolic in untreated NRK cells, and an obvious increase of punctate vesicular structures labeled with GFP-LC3 was observed after 3 h of starvation in EBSS (Fig. 7A). Interestingly, overexpression of MCOLN1 caused a considerable accumulation of GFP-LC3-labeled autophagosomes (Fig. 7B). Quantification of 300 cells per experimental condition showed that under nutrient rich conditions, <8% (7.21%, SD ± 0.58) of NRK cells expressing GFP-LC3 alone displayed GFP-LC3 puncta. In contrast, 33% (SD ± 0.86) of cells co-expressing GFP-LC3 and MCOLN1 showed GFP-LC3-positive vesicles (Fig. 7C). Starvation-induced autophagy did not seem to be affected by MCOLN1 expression, as the percentage of cells accumulating GFP-LC3-labeled puncta after 3 h of incubation in EBSS was very similar between cells expressing GFP-LC3 alone or GFP-LC3 plus MCOLN1 (76.13%, SD ± 7.04 and 76.33%, SD ± 2.3, respectively) (Fig. 7C). Finally, the number of GFP-LC3 positive vesicles decreased significantly in both cell types after 1 h recovery (Fig. 7C). Taken together, our results indicate that alterations in the late endosomal/lysosomal pathway induced by MCOLN1 overexpression result in abnormal accumulation of autophagosomes in basal conditions.
Figure 7.
Overexpression of MCOLN1 increases autophagy. (A) NRK cells were transfected with GFP-LC3 for 24 h. Untreated cells, cells starved in EBSS media for 3 h (starvation, center) or starved for 3 h followed by 1 h of recovery in complete media (recovery, right), were fixed and examined by confocal fluorescence microscopy. Scale bar = 10 µm. (B) NRK cells were co-transfected with GFP-LC3 and MCOLN1-FLAG, fixed and stained with an antibody to FLAG. Scale bar = 10 µm. (C) Percentage of cells expressing GFP-LC3 in a vesicular pattern in normal conditions (untreated), after 3 h starvation or after 1 h of recovery, in the absence or presence of MCOLN1-FLAG. Results are means ± SD (n = 300), P < 0.001.
Absence of MCOLN1 causes delayed transport of cargo along the endosomal/lysosomal pathway
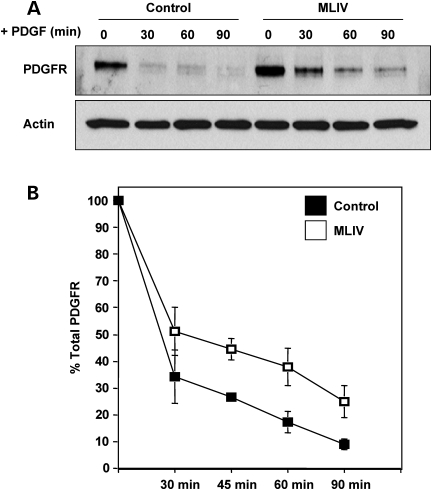
As mentioned above, fusion of autophagosomes with lysosomes is delayed in MLIV fibroblasts, so we hypothesized that fusion between late endosomes and lysosomes might also be impaired in the absence of MCOLN1. To test this possibility, we followed delivery of cargo from plasma membrane to lysosomes. Platelet-derived growth factor receptor (PDGFR) is a prototypical member of the receptor tyrosine kinases family in which activation and trafficking have been exhaustively characterized (48). Ligand-induced dimerization of the PDGF receptor leads to autophosphorylation of the receptor and the subsequent binding and phosphorylation of downstream signaling proteins. Termination of PDFGR signaling occurs via endocytosis and delivery of the receptor to lysosomes for degradation. We therefore stimulated cells with PDGF for different periods of time and determined the amount of PDGFR by immunoblot analysis. Interestingly, we observed that ligand-induced PDGFR degradation was reduced in MCOLN1-deficient cells when compared with control cells (Fig. 8A). Quantitative analysis of three independent experiments revealed that in MLIV fibroblasts the remaining amount of PDGFR at 60 and 90 min after ligand stimulation was increased by more than 50% when compared with control cells (Fig. 8B). These results indicate that MCOLN1 is required for efficient transport of PDGFR to lysosomes. These data combined are indicative that MCOLN1 is required for the efficient delivery of material from both late endosomes and autophagosomes to lysosomes.
Figure 8.
Down-regulation of PDGFR is impaired in MLIV fibroblasts. (A) Control fibroblasts and MLIV fibroblasts were starved for 16 h in DMEM containing 0.1% BSA and incubated with 100 ng/ml of PDGF-BB for the indicated times. Cells were then lysed, subjected to SDS–PAGE and immunoblotted with antibodies against PDGFR. Lysates were also blotted for actin as a loading control. (B) Quantification of the optical densities for the corresponding PDGFR bands from three independent experiments.
DISCUSSION
In this paper, we report constitutive activation of autophagy in fibroblasts obtained from MLIV patients. This activation was observed under basal conditions and was the result of increased autophagosome formation and delayed autophagosome degradation. To monitor autophagy, we analyzed recruitment of LC3 from the cytosol to vesicular structures, as well as an increase in the levels of LC3-II. We found that the number of LC3-positive puncta was dramatically increased in MCOLN1-deficient cells when compared with control fibroblasts. Interestingly, most of the LC3-labeled structures also contained p62, suggesting accumulation of ubiquitinated proteins in MLIV fibroblasts. Increased levels of LC3-II and p62 were also confirmed by immunoblot analysis. Treatment with leupeptin and co-localization experiments between CD63 and LC3 showed that fusion of autophagosomes with late endosomes/lysosomes is not blocked in MLIV cells. These data, together with the increased levels of beclin-1 observed in mutant fibroblasts, suggest that autophagosome accumulation must be at least partially due to increased autophagosome formation. However, we also observed that autophagosome maturation is less efficient in MLIV patient cells. Therefore, we suggest that both augmented autophagosome formation and delayed autophagosome fusion with late endosomes/lysosomes account for the autophagy induction reported in MLIV cells.
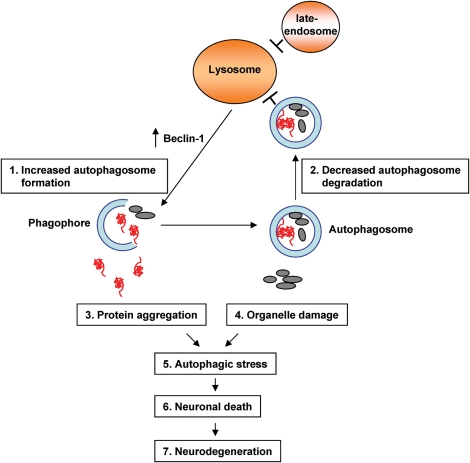
MLIV is characterized by severe neurological abnormalities; however, the underlying mechanism of neurodegeneration is poorly understood. We propose that trafficking defects along the endosomal/lysosomal pathway due to the absence of MCOLN1 impairs lysosomal function and results in inefficient autophagosome degradation. This in turn causes accumulation of ubiquitinated aggregates that generate cellular stress and further induces autophagosome formation and accumulation (Fig. 9). Defects in autophagy may not be especially detrimental in fibroblasts, where the rapid division of the cells helps to prevent the accumulation of misfolded or aggregated proteins. However, the accumulation of these products in neurons may result in cell death, and as the neurons cannot be replaced, neurodegeneration occurs.
Figure 9.
A proposed model of mechanism of defective autophagy induced neurodegeneration in MLIV patients. Trafficking along the late endosomal/lysosomal pathway is perturbed in MCOLN1-deficient cells. As a result, fusion between autophagosomes and lysosomes is delayed and de novo formation of autophagosomes is increased. Dysfunction of the autophagic pathway leads to accumulation of protein aggregates and to defects in organelle turnover thus rendering cells, especially neurons, more susceptible to cell death.
In addition, autophagic stress may cause accumulation of damage organelles, such as mitochondria, making the cells more susceptible to pro-apoptotic signals (49). In support of this idea, a recent study described that MLIV fibroblasts accumulate fragmented mitochondria and exhibit increased sensitivity to apoptosis induced by Ca2+-mobilizing agonists (41).
Defects in autophagy caused by lysosomal malfunction may not be an exclusive characteristic of MLIV. Recent evidence shows that in many LSDs such as multiple sulfatase deficiency and mucopolysaccharidosis type IIIA, accumulation of undegraded products in lysosomes impairs autophagosome maturation, resulting in accumulation of ubiquitinated protein aggregates and defective mitochondria (50). In addition, factors which reduce lysosomal activity, for instance, aging or oxidative stress, could reduce the ability of autophagy to eliminate toxic or aggregated proteins thus exacerbating age-related neurodegenerative disorders such as Parkinson's, Alzheimer's and Huntington's disease (5).
Accumulation of autophagosomes has also been described in Niemann-Pick type C disease (NPC), a sphingolipid storage disorder characterized by progressive neurodegeneration (51). Interestingly, increased formation of autophagosomes rather than defective degradation was observed in NPC. These results led to the suggestion that accumulation of cholesterol in late endosomes/lysosomes increases the levels of beclin-1 resulting in elevated basal autophagy (52). Abnormal accumulation of cholesterol in late endosomes has also been reported in MLIV fibroblasts (36). Therefore, disrupted cholesterol trafficking could be the cause of the increased autophagosome formation associated with elevated beclin-1 levels that we observe in MLIV cells, while defective lysosomal function would account for the inefficient autophagosome degradation.
We cannot dismiss the possibility that MCOLN1 plays a specific role in the fusion between autophagosomes and late endosomes/lysosomes. Transient expression of MCOLN1 in NRK cells induced accumulation of GFP-LC3-labeled vesicles. As it is improbable that many lipids accumulate within lysosomes a short time after transfection, it is plausible that the alteration of MCOLN1 function due to overexpression is responsible for impaired autophagosome maturation. It has also been suggested that MCOLN1 might regulate fusion/fission events between late endosomes and lysosomes (40,53). Consistent with this idea, we found that delivery of PDGFR to lysosomes is delayed in MLIV fibroblasts (Fig. 8). In addition, it has recently been shown that transport of fluid-phased markers to lysosomes is slower in macrophages treated with MCOLN1 siRNA (54). Since some of the proteins required for late endosome–lysosome fusion are likely to be the same as those required for autophagosome–lysosome fusion, it is possible that MCOLN1 participates in both fusion events.
Our results suggest that modulation of autophagy might be an effective strategy for treatment of MLIV. We found robust induction of autophagy after starvation in both control and MLIV cells, indicating that the autophagy machinery is functional in MCOLN1-deficient cells. Therefore, therapies designed to induce autophagy activation, such as caloric restriction or treatment with rapamycin, might help to overcome the delayed autophagosome degradation seen in basal conditions. This strategy has been suggested for the treatment of other neurodegenerative disorders (55,56). For example, autophagy induction by treatment with rapamycin has been shown to attenuate neuronal death in animal models of Huntington's disease (57).
In summary, our data demonstrate the impact of lysosomal function on autophagic activity and suggest that, analogous to other LSDs, accumulation of ubiquitinated protein inclusions due to defective autophagy may contribute to the neurodegeneration observed in MLIV patients.
MATERIALS AND METHODS
Antibodies and reagents
The primary antibodies used were: mouse monoclonal anti-ubiquitin (FK2; MBL, Woburn, MA, USA), mouse monoclonal anti-CD63 (H5C6) and mouse monoclonal anti-p62 lck ligand (BD Pharmigen, San Jose, CA, USA), mouse monoclonal anti-actin (BD Transduction, San Jose, CA, USA), rabbit polyclonal anti-LC3 (Sigma, St Louis, MO, USA), rabbit polyclonal anti-Beclin-1 (Cell Signaling Technology, Beverly, MA, USA) and rabbit polyclonal anti-PDGFR (Upstate/Millipore, Bellerica, MA, USA). The secondary antibodies used were: goat anti-mouse or goat anti-rabbit conjugated to Alexa Fluor 488 or 555 (Molecular Probes, Eugene, OR, USA). HRP-conjugated anti-mouse or anti-rabbit IgG (GE Healhcare/Amersham Bioscience, Piscataway, NJ, USA). Leupeptin, mammalian protease inhibitor cocktail and NP-40 were obtained from Sigma. PDGF-BB was obtained from Upstate/Millipore.
Cell lines and transfections
Skin fibroblasts from a MLIV patient (clone WG0909), the corresponding heterozygous non-diseased parent skin fibroblasts (clone WG0988) and unrelated non-diseased skin fibroblasts (clone MCH065) were obtained from the Repository for Mutant Human Cell Strains of Montreal Children's Hospital (Montreal, Canada). MLIV fibroblast clone MG02527 was from Coriell Cell Repositories (Boston, MA, USA). Fibroblasts and NRK cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Life technologies, Invitrogen/Gibco, Carlsbad, CA, USA) supplemented with 100 U/ml penicillin (Gibco) and 10% (w/v) fetal bovine serum (FBS). Transfection of NRK cells was performed using FuGENE-6 (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's recommendations. Analyses were conducted 24 hours post-transfection.
Starvation of cells
Cells were starved by being washed three times with PBS and incubated in EBSS (EBSS, Sigma) at 37°C for 3 h. To examine the recovery of cells from starvation, cells incubated in EBSS for 3 h were transferred to DMEM–10% FBS at 37°C for the indicated times.
Plasmids
GFP-LC3 chimera was a kind gift of Dale Hailey (NIH, Bethesda, MD, USA). MCOLN1-FLAG was cloned as described previously (35).
Immunofluorescence microscopy
For immunofluorescence, skin fibroblasts or NRK cells were grown on cover slips and fixed in methanol/acetone (1:1, v/v) for 10 min at −20°C. Incubation with primary antibodies diluted in PBS, 0.1% (wt/vol) saponin and 0.1% BSA, was carried out for 1 h at room temperature. Unbound antibodies were removed by rinsing with PBS for 5 min, and cells were subsequently incubated with a secondary antibody (Alexa555 or Alexa488-conjugated goat anti-rabbit or anti-mouse Ig) diluted in PBS, 0.1% (wt/vol) saponin and 0.1% BSA, for 30–60 min at room temperature. After a final rinse with PBS, cover slips were mounted onto glass slides with Fluoromount G (Southern Biotechnology Associates, Birmingham, AL, USA). Fluorescence images were acquired on an LSM 510 confocal microscope (Carl Zeiss, Thornwood, NY, USA). For co-localization analysis in recovery conditions, six cells per cell line per experiment were imaged and analyzed as follows: LC3 positive vesicles were counted and marked by switching off the GFP channel. Then, the GFP channel was switched back on and the number of co-localized vesicles was counted. From these two values, the fraction of co-localized vesicles was then determined.
Electron microscopy
Cells were cultured in 60 mm Permanox dishes (Nunc Nalgene) and processed for transmission electron microscopy. In brief, cultures were fixed with glutaraldehyde and paraformaldehyde, post-fixed with osmium tetroxide, stained en bloc with uranyl acetate, ethanol dehydrated and Epon embedded. Chemicals were from Electron Microscopy Sciences. Sections 60 nm thick were cut parallel to the adherent surface on a Sorvall MT2 ultramicrotome, stained with uranyl acetate and lead citrate and viewed with a JEM 1200 EXII electron microscope (JEOL USA) equipped with an AMT XR-60 digital camera (Advanced Microscopy Techniques).
Western blot
Fibroblasts were washed with ice-cold PBS, extracted in ice-cold lysis buffer (25 mm Tris–HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% NP-40 supplemented with protease inhibitor cocktails), centrifuged (16 000 g) for 10 min and the supernatants (soluble fraction) were collected. To analyze protein levels in the insoluble fractions, the remaining protein pellets were washed with lysis buffer, centrifuged and extracted with 2% SDS containing sample buffer. Samples were analyzed by SDS–PAGE (4–20% gradient gels) under reducing conditions and transferred on to nitrocellulose. The membrane was then blocked with 1× PBS, 0.05% Tween-20, 10% non-fat milk and incubated with the appropriate antibodies. Enhanced chemiluminescence reagent (Amersham Biosciences) was used for protein detection. The amounts of the different proteins were quantified by using the public domain ImageJ program (1.38), normalized with the actin content in the same sample and represented as fold increase of the amount present in control fibroblasts.
Degradation of PDGFR
Skin fibroblasts were serum starved for 16 h and stimulated with PDGF-BB (100 ng/ml) for the indicated times. Total cell lysates were subjected to immunoblotting with antibodies against PDGFR. The amounts of PDGFR were quantified at each time point by using the public domain ImageJ program (1.38) and are represented as the percentage of remaining PDGFR when compared with unstimulated fibroblasts in the same experiment.
SUPPLEMENTARY MATERIAL
FUNDING
This project was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute (NHLBI).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dale Hailey for helpful discussions and suggestions. We also appreciate the editorial assistance of the NIH Fellows Editorial Board.
Conflict of Interest statement. The authors have no conflicts of interest to declare.
REFERENCES
- 1.Seglen P.O., Berg T.O., Blankson H., Fengsrud M., Holen I., Stromhaug P.E. Structural aspects of autophagy. Adv. Exp. Med. Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- 2.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 3.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuervo A.M. Autophagy: many paths to the same end. Mol. Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Vicente M., Cuervo A.M. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 6.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J.H., Guo F., Shelburne J., Watkins S., Chu T. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anglade P., Vyas S., Javoy-Agid F., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 10.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo M.A. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 11.Yu W.H., Cuervo A.M., Kumar A., Peterhoff C.M., Schmidt S.D., Lee J.H., Mohan P.S., Mercken M., Farmery M.R., Tjernberg L.O., et al. Macroautophagy, a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kegel K.B., Kim M., Sapp E., McIntyre C., Castaño J.G., Aronin N., DiFiglia M. Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z., Klionsky D.J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky D.J., Cuervo A.M., Seglen P.O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson N., Ince P.G., Smith M.O., Highley R., Skibinski G., Andersen P.M., Morrison K.E., Pall H.S., Hardiman O., Collinge J., et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 17.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 18.Reid E., Connell J., Edwards T.L., Duley S., Brown S.E., Sanderson C.M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 19.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J.I., Ezakim J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Amir N., Zlotogora J., Bach G. Mucolipidosis type IV: clinical spectrum and natural history. Pediatrics. 1987;79:953–959. [PubMed] [Google Scholar]
- 22.Bach G. Mucolipidosis type IV. Mol. Genet. Metab. 2001;73:197–203. doi: 10.1006/mgme.2001.3195. [DOI] [PubMed] [Google Scholar]
- 23.Altarescu G., Sun M., Moore D.F., Smith J.A., Wiggs E.A., Solomon B.I., Patronas N.J., Frei K.P., Gupta S., Kaneski C.R., et al. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59:306–313. doi: 10.1212/wnl.59.3.306. [DOI] [PubMed] [Google Scholar]
- 24.Slaugenhaupt S.A., Acierno J.S., Jr, Helbling L.A., Bove C., Goldin E., Bach G., Schiffmann R., Gusella J.F. Mapping of the mucolipidosis type IV gene to chromosome 19p and definition of founder haplotypes. Am. J. Hum. Genet. 1999;65:773–778. doi: 10.1086/302549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bargal R., Avidan N., Ben Asher E., Olender Z., Zeigler M., Frumkin A., Raas-Rothschild A., Glusman G., Lancet D., Bach G. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 26.Bassi M.T., Manzoni M., Monti E., Pizzo M.T., Ballabio A., Borsani G. Cloning of the gene encoding a novel integral membrane protein mucolipin, and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M., Goldin E., Stahl S., Falardeau J.L., Kennedy J.C., Acierno J.S., Jr, Bove C., Kaneski C.R., Nagle J., Bromley M.C., et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 28.Vergarajauregui S., Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–353. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miedel M.T., Weixel K.M., Bruns J.R., Traub L.M., Weisz O.A. Posttranslational cleavage and adaptor protein complex-dependent trafficking of mucolipin-1. J. Biol. Chem. 2006;281:12751–12759. doi: 10.1074/jbc.M511104200. [DOI] [PubMed] [Google Scholar]
- 30.Pryor P.R., Reimann F., Gribble F.M., Luzio J.P. Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic. 2006;7:1388–1398. doi: 10.1111/j.1600-0854.2006.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaPlante J.M., Falardeau J., Sun M., Kanazirska M., Brown E.M., Slaugenhaupt S.A., Vassilev P.M. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 2002;532:183–187. doi: 10.1016/s0014-5793(02)03670-0. [DOI] [PubMed] [Google Scholar]
- 32.Raychowdhury M.K., Gonzalez-Perrett S., Montalbetti N., Timpanaro G.A., Chasan B., Goldmann W.H., Stahl S., Cooney A., Goldin E., Cantiello H.F. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum. Mol. Genet. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 33.Cantiello H.F., Montalbetti N., Goldmann W.H., Raychowdhury M.K., Gonzalez Perrett S., Timpanaro G.A., Chasan B. Cation channel activity of mucolipin-1: the effect of calcium. Pflugers. Arch. 2005;451:304–312. doi: 10.1007/s00424-005-1448-9. [DOI] [PubMed] [Google Scholar]
- 34.Kiselyov K., Chen J., Rbaibi Y., Oberdick D., Tjon-Kon-Sang S., Shcheynikov N., Muallem S., Soyombo A. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 2005;280:43218–43223. doi: 10.1074/jbc.M508210200. [DOI] [PubMed] [Google Scholar]
- 35.Vergarajauregui S., Oberdick R., Kiselyov K., Puertollano R. Mucolipin 1 channel activity is regulated by protein kinase A-mediated phosphorylation. Biochem. J. 2008;410:417–425. doi: 10.1042/BJ20070713. [DOI] [PubMed] [Google Scholar]
- 36.Soyombo A.A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 2006;281:7294–7301. doi: 10.1074/jbc.M508211200. [DOI] [PubMed] [Google Scholar]
- 37.Berman E.R., Livni N., Shapira E., Merin S., Levij I.S. Congenital corneal clouding with abnormal systemic storage bodies: a new variant of mucolipidosis. J. Pediatr. 1974;84:519–526. doi: 10.1016/s0022-3476(74)80671-2. [DOI] [PubMed] [Google Scholar]
- 38.Riedel K.G., Zwaan J., Kenyon K.R., Kolodny E.H., Hanninen L., Albert D.M. Ocular abnormalities in mucolipidosis IV. Am. J. Ophthalmol. 1985;99:125–136. doi: 10.1016/0002-9394(85)90220-x. [DOI] [PubMed] [Google Scholar]
- 39.Goldin E., Cooney A., Kaneski C.R., Brady R.O., Schiffmann R. Mucolipidosis IV consists of one complementation group. Proc. Natl Acad. Sci. USA. 1999;96:8562–8566. doi: 10.1073/pnas.96.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treusch S., Knuth S., Slaugenhaupt S.A., Goldin E., Grant B.D., Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl Acad. Sci. USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jennings J.J., Jr, Zhu J.H., Rbaibi Y., Luo X., Chu C.T., Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 42.Laplante J.M., Sun M., Falardeau J., Dai D., Brown E.M., Slaugenhaupt S.A., Vassilev P.M. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 2006;89:339–348. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Talbot K., Ansorge O. Recent advances in the genetics of amyotrophic lateral sclerosis and frontotemporal dementia: common pathways in neurodegenerative disease. Hum. Mol. Genet. 2006;15:R182–R187. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- 44.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 45.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanemura K., Murayama M., Agagi T., Hashikawa T., Tominaga T., Ichikawa M. Formation of filamentous tau aggregations in transgenic mice expressing V337M human tau. J. Neurosci. 2002;22:133–141. [Google Scholar]
- 47.Tellez-Nagel I., Rapin I., Iwamoto T., Johnson A.B., Norton W.T., Nitowsky H. Mucolipidosis IV. Clinical, ultrastructural, histochemical, and chemical studies of a case, including a brain biopsy. Arch. Neurol. 1976;33:828–835. doi: 10.1001/archneur.1976.00500120032005. [DOI] [PubMed] [Google Scholar]
- 48.Haglund K., Di Fiore P.P., Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 50.Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D., de Pablo R., Tacchetti C., Rubinsztein D.C., Ballabio A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 51.Higgins J.J., Patterson M.C., Dambrosia J.M., Pikus A.T., Pentchev P.G., Sato S., Brady R.O., Barton N.W. A clinical staging classification for type C Niemann–Pick disease. Neurology. 1992;42:2286–2290. doi: 10.1212/wnl.42.12.2286. [DOI] [PubMed] [Google Scholar]
- 52.Pacheco C.D., Kunkel R., Lieberman A.P. Autophagy in Niemann–Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 53.Piper R.C., Luzio J.P. CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 2004;14:471–473. doi: 10.1016/j.tcb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Thompson E.G., Schaheen L., Dang H., Fares H. Lysosomal trafficking functions of mucolipin-1 in murine macrophages. BMC Cell Biol. 2007;8:54. doi: 10.1186/1471-2121-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergamini E., Cavallini G., Donati A., Gori Z. The role of macroautophagy in the ageing process, anti- ageing intervention and age-associated diseases. Int. J. Biochem. Cell Biol. 2004;36:2392–2404. doi: 10.1016/j.biocel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Ingram D.K., Zhu M., Mamczarz J., Zou S., Lane M.A., Roth G.S., deCabo R. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 57.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.