Summary
Vibrio cholerae is both a human pathogen and a natural inhabitant of aquatic environments. In the aquatic environment, microorganisms are found attached to surfaces in structures known as biofilms. We have identified a transcriptional repressor in V. cholerae that inhibits exopolysaccharide synthesis and biofilm development. Our studies show that this repressor is the V. cholerae homologue of Escherichia coli CytR, a protein that represses nucleoside uptake and catabolism when nucleosides are scarce. We propose that the role of CytR in V. cholerae biofilm development is to co-ordinate bacterial biofilm accumulation with the presence of nucleosides. Thus, nucleosides may be a signal to planktonic cells to join the biofilm.
Introduction
Although surface adhesion or biofilm development is critical for the persistence and proliferation of all bacterial species in their natural habitats, the environmental signals that promote the surface-adherent mode of life are poorly understood. Vibrio cholerae, a native inhabitant of diverse aquatic environments and the human intestinal pathogen that causes the severe diarrhoeal disease cholera, is an ideal model for the study of biofilm development. Surface adhesion or biofilm development by V. cholerae is instrumental in both pathogenesis and colonization of aquatic environments. In the environment, V. cholerae has been identified on the surfaces of zooplankton, phytoplankton, crustaceans, insects and plants (Huq et al., 1983; 1986; Tamplin et al., 1990; Shukla et al., 1995). Because zoo-plankton and phytoplankton blooms precede cholera outbreaks, the association of V. cholerae with these organisms is hypothesized to play a role in the epidemiology of disease (Colwell, 1996; Lobitz et al., 2000). Thus, an understanding of the regulation of V. cholerae surface adhesion will not only increase our understanding of bacterial adaptation to aquatic habitats but may also suggest methods of improving our predictive models for cholera epidemics.
Vibrio cholerae biofilm development on abiotic surfaces has been described previously (Watnick and Kolter, 1999; Watnick et al., 2001). In batch biofilm experiments, two stages in V. cholerae biofilm development are observed during the first 24 h of growth (Watnick et al., 2001). In the initial period of 4–5 h, only transient association with the surface is observed. This association requires a functional polar flagellum. In the second stage of biofilm development, transient association with the surface continues, but now permanent immobilization and microcolony formation are observed. Permanent immobilization is dependent on the vps genes, which encode enzymes required for the synthesis of VPS, an exopolysaccharide produced by V. cholerae (Watnick and Kolter, 1999; Yildiz and Schoolnik, 1999).
Studies of both V. cholerae and Pseudomonas aeruginosa suggest that increased exopolysaccharide synthesis alters both the course of biofilm development and the three-dimensional architecture of the mature biofilm (Wai et al., 1998; Mizunoe et al., 1999; Yildiz and Schoolnik, 1999; Hentzer et al., 2001). In V. cholerae, spontaneously occurring ‘rugose’ variants have been isolated (White, 1938; Crutchley, 1968; Morris et al., 1996; Wai et al., 1998; Mizunoe et al., 1999; Yildiz and Schoolnik, 1999). These variants display a rough colony morphology that is correlated with increased transcription of the vps genes and increased VPS synthesis (Wai et al., 1998; Mizunoe et al., 1999; Yildiz and Schoolnik, 1999; Yildiz et al., 2001). Increased VPS synthesis by rugose variants also results in a higher profile biofilm than that formed by the parent V. cholerae strain (Yildiz and Schoolnik, 1999). The genetic basis of increased vps transcription in spontaneous rugose variants has not yet been ascertained, and one possibility is that these rugose variants represent a genotypically heterogeneous population.
The transcription profiles of biofilm-associated cells are quite different from their planktonic counterparts. Evidence from P. aeruginosa and Escherichia coli suggests that increased transcription of genes required for exopolysaccharide synthesis is a characteristic of cells that have been incorporated into the biofilm (Davies et al., 1993; Prigent-Combaret et al., 1999). More comprehensive genomics- and proteomics-based profiling of biofilm-associated cells is under way or completed in many bacterial backgrounds, and these studies are likely to yield a deeper understanding of the physiological state of the biofilm-associated cell (Sauer and Camper, 2001; Schoolnik et al., 2001; Whiteley et al., 2001; Sauer et al., 2002; Zhu et al., 2002). Complementary studies of the environmental signals and transcription factors that are operative in biofilm development will allow us to link signal transduction cascades with changes in gene transcription and protein expression within the biofilm.
Several environmental signals that influence bacterial biofilm development have been identified. These include quorum sensing, surface sensing and the nutritional content of the aquatic environment. In P. aeruginosa, autoinducer synthesis mutants form a flat, detergent-sensitive biofilm, suggesting that quorum sensing directs normal biofilm development (Davies et al., 1998; Parsek and Greenberg, 1999). The quorum-sensing systems of Burkholderia cepacia and V. cholerae have also been implicated in biofilm development (Huber et al., 2001; Zhu et al., 2002). Another signal that activates biofilm development is contact with the substratum. In both V. cholerae and P. aeruginosa, there is evidence that exopolysaccharide synthesis is activated by mutation of the flagellar structural genes but not the flagellar motor (Garrett et al., 1999; Watnick et al., 2001). One possibility is that, as is true for the closely related organisms Vibrio parahaemolyticus and Vibrio alginolyticus, the flagellar motor of V. cholerae is able to sense and respond to changes in torque on the polar flagellum that occur when a surface is encountered (Belas et al., 1986; McCarter et al., 1988; Kawagishi et al., 1996). For diverse bacteria, the nutritional composition of the aquatic environment is a key determinant of surface attachment (Bowden and Li, 1997; O’Toole and Kolter, 1998; Pratt and Kolter, 1998; Watnick et al., 1999; Danese et al., 2001). In particular, for many organisms, glucose and related carbohydrates greatly enhance exopolysaccharide production and, thus, biofilm development (Bryan et al., 1986; Bonet et al., 1993; Abbad Andaloussi et al., 1995; Kimmel et al., 1998; Degeest and De Vuyst, 1999; Looijesteijn et al., 1999; Petryet al., 2000; Degeest et al., 2001; Mozzi et al., 2001). Although transcription factors that regulate exopolysaccharide synthesis have been identified in various organisms, the environmental signals that these regulators sense is not known (Reeve et al., 1997; Chapman and Kao, 1998; Yildiz et al., 2001). This is the case for VpsR, the only known activator of V. cholerae biofilm development and VPS synthesis (Yildiz et al., 2001). By sequence homology, VpsR is the response element in a two-component system. Although activation of vps gene transcription by VpsR has been demonstrated, the environmental signals that govern activation by VpsR have not yet been delineated.
In E. coli, CytR has been shown to repress nucleoside uptake and catabolism in nucleoside-poor environments. In this paper, we provide evidence that V. cholerae CytR plays a role not only in nucleoside catabolism but also in the control of biofilm development and, specifically, in the synthesis of the V. cholerae exopolysaccharide, VPS. Our studies link nucleoside concentrations to transcriptional regulation of exopolysaccharide synthesis and biofilm development by V. cholerae. Thus, we suggest that nucleosides may be a signal to planktonic cells to join the nascent biofilm.
Results
Identification of a ‘super-biofilm’ mutant
A library of mini-Tn10 transposon-insertion mutants was constructed in the V. cholerae O139 strain MO10. This library was screened for mutants that formed increased or ‘super’ biofilms as described previously (Watnick and Kolter, 1999; Watnick et al., 2001). The transposon-insertion junctions of identified mutants were amplified by arbitrary polymerase chain reaction (PCR) and sequenced (O’Toole et al., 1999). Of the 6000 transposon-insertion mutants evaluated, three independent clones were found to be in the gene given the TIGR designation VC2677 (Heidelberg et al., 2000). Further evaluation demonstrated that all these mutants displayed a rugose colony morphology and formed a biofilm containing approximately twice as many cells as a wild-type V. cholerae biofilm, as determined by crystal violet staining and quantification. These mutants also displayed normal motility on swarm agar (data not shown). A transposon-insertion mutant in the same gene of V. cholerae O1 El Tor demonstrated a similar increase in biofilm development (data not shown), suggesting that this type of regulation is not strain specific.
The ‘super biofilm’ mutant is an E. coli CytR homologue
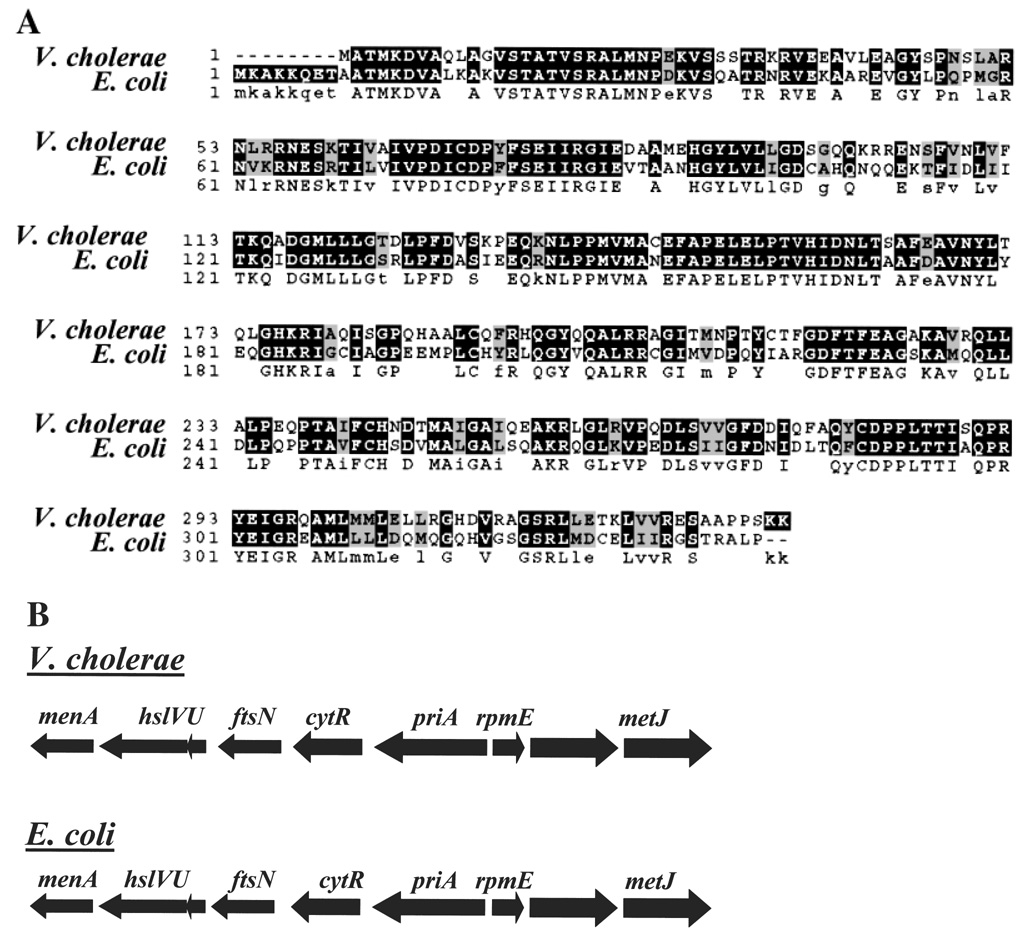
Sequence analysis demonstrated that the amino acid sequence of the gene containing the transposon insertion is 65% identical and 81% similar to the CytR protein of E. coli. No other protein in the V. cholerae genome showed this extent of similarity to E. coli CytR. As shown in Fig. 1A, this homology extends throughout the protein. Furthermore, the genomic environments of the E. coli CytR gene and the V. cholerae biofilm repressor are strikingly similar as shown in Fig. 1B. Thus, we term our biofilm repressor V. cholerae CytR and present additional evidence below that this protein is, in fact, V. cholerae CytR.
Figure 1.
A comparison of the V. cholerae O1 El Tor CytR amino acid sequence and genomic environment with that of E. coli K-12 CytR.
A. Alignment of the amino acid sequences of V. cholerae and E. coli CytR. CLUSTALW was used to produce the sequence alignment (Thompson et al., 1994).
B. Genomic environment of V. cholerae O1 El Tor cytR and E. coli K-12 cytR.
Construction of a V. cholerae cytR deletion mutant, cloning of V. cholerae cytR and complementation of the mutant phenotype
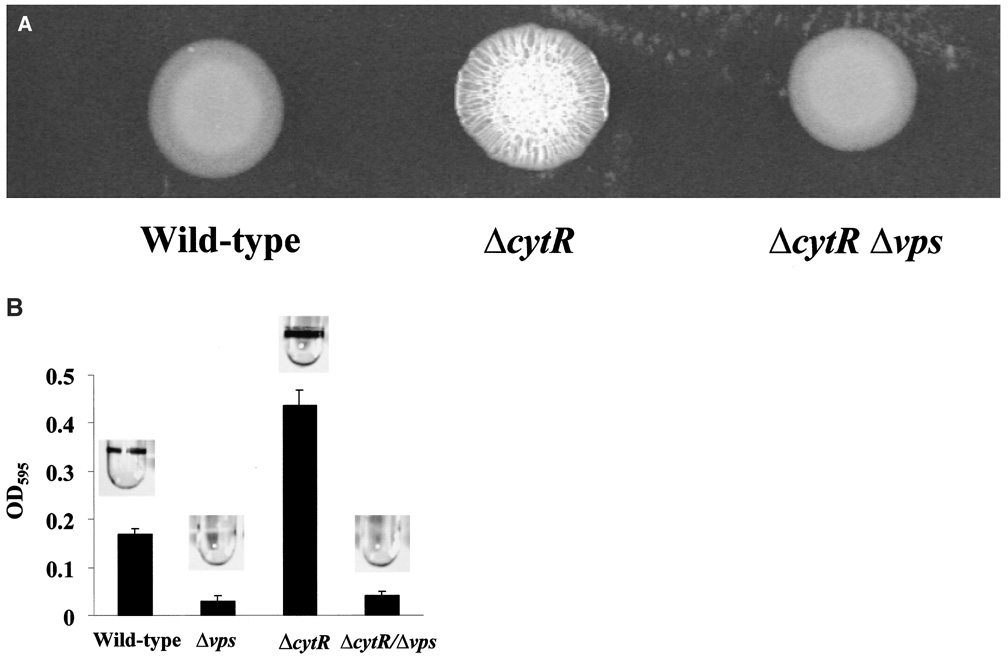
To confirm that the phenotype demonstrated by the transposon-insertion mutant was indeed the result of a mutation in cytR, a strain carrying a complete deletion of the cytR gene was constructed (strain PW324 denoted ΔcytR). The planktonic growth curves of wild-type V. cholerae and the ΔcytR mutant in LB broth were measured and found to be essentially superimposable before the onset of biofilm development (data not shown). As shown in Fig. 2, the deletion mutant demonstrated a rugose colony morphology and ‘super-biofilm’ phenotype that was similar to that of the transposon-insertion mutant. Measurements of total protein in wild-type V. cholerae and ΔcytR mutant biofilms indicated an increase in cell mass within the ΔcytR mutant biofilm that was consistent with crystal violet quantification (data not shown). Furthermore, we constructed a ΔcytR mutant harbouring a deletion in a large vps operon containing six open reading frames (ORFs) including vpsL (Yildiz et al., 2001; strain PW329 denoted ΔcytR Δvps). The ΔcytR Δvps mutant formed a smooth colony and did not accumulate on surfaces. This result suggests that the phenotype of the ΔcytR mutant results from derepression of exopolysaccharide synthesis rather than activation of a new mechanism of surface adhesion.
Figure 2.
Comparison of wild-type V. cholerae and ΔcytR mutant colony morphology and biofilm development.
A. Colony morphology of wild-type V. cholerae (MO10), a ΔcytR mutant (PW324) and a ΔcytRΔvps double mutant (PW329).
B. Biofilm accumulation by wild-type V. cholerae (MO10), a Δvps mutant (PW328), a ΔcytR mutant (PW324) and a ΔcytRΔvps double mutant (pw329). Biofilms stained with crystal violet are shown above, and quantification of crystal violet staining is shown below.
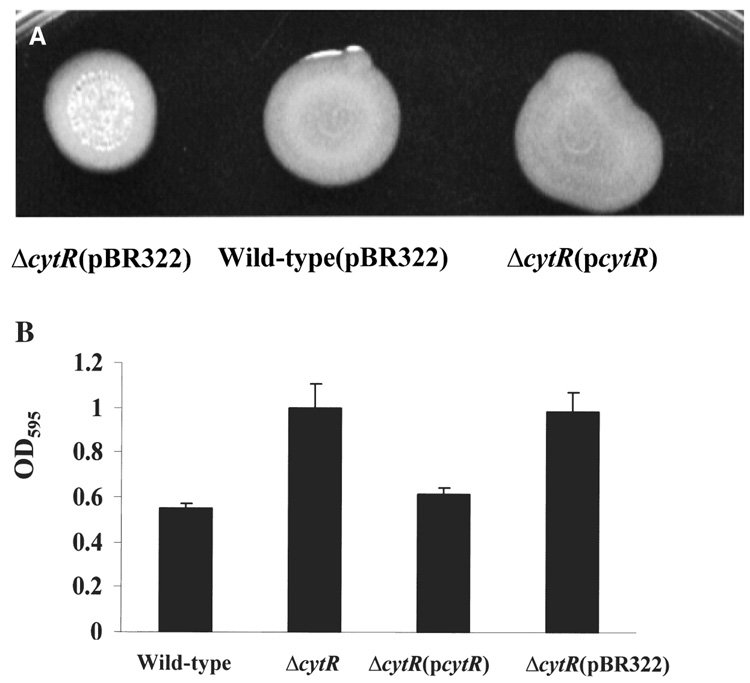
We also confirmed the correlation of the ΔcytR mutation with the ‘super-biofilm’ phenotype by complementation with the V. cholerae cytR gene in trans. We amplified the cytR gene with its native promoter by PCR, sequenced the product fully and cloned the resulting PCR product into the moderate-copy-number plasmid pBR322. The cloned cytR promoter and coding sequence from the V. cholerae O139 strain, MO10, had a nucleotide sequence identical to that of the V. cholerae O1 El Tor strain N16961. As shown in Fig. 3, when the cytR gene was provided in trans, the ΔcytR mutant displayed a smooth colony morphology and biofilm accumulation similar to that of wild-type V. cholerae.
Figure 3.
Complementation of the rugose colony morphology and increased biofilm accumulation phenotype of a V. cholerae ΔcytR mutant by the V. cholerae cytR gene provided in trans.
A. Colony morphology of the V. cholerae ΔcytR mutant (PW324) transformed with the cloning vector pBR322, wild-type V. cholerae transformed with the cloning vector pBR322 and a V. cholerae ΔcytR mutant (PW324) transformed with the pBR322-based plasmid pAJH3, denoted pcytR, carrying the promoter and coding sequence of V. cholerae cytR.
B. Quantification of biofilm accumulation by wild-type V. cholerae (MO10), a V. cholerae ΔcytR mutant (PW324), a V. cholerae ΔcytR mutant transformed with the plasmid pAJH3, denoted pcytR, and a V. cholerae ΔcytR mutant transformed with the cloning vector pBR322.
Vibrio cholerae CytR is able to regulate transcription of the E. coli udp gene, a member of the E. coli CytR regulon
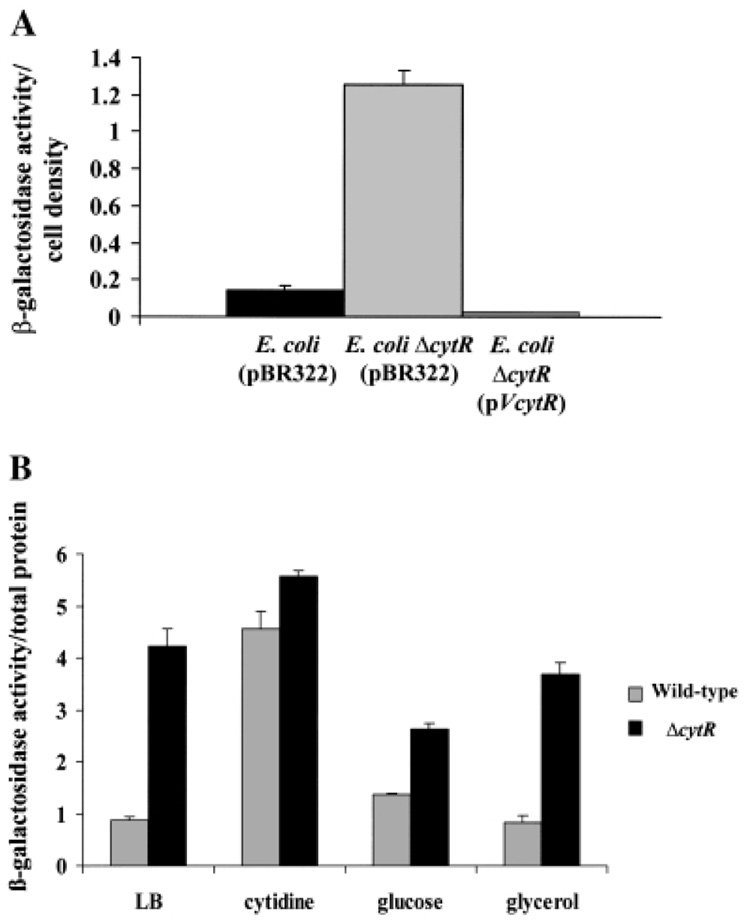
When cytidine is scarce in the environment, the E. coli CytR protein represses the transcription of genes encoding proteins involved in nucleoside uptake and catabolism. One of these genes, udp, encodes uridine dephosphorylase, an enzyme involved in the conversion of uridine to ribose-1-phosphate and uracil (Barbier and Short, 1992; Brikun et al., 1996; Neidhardt, 1996; Gavigan et al., 1999). Uridine dephosphorylase is operative in the catabolism of both uridine and cytidine. To confirm that the gene we identified as V. cholerae cytR had a function similar to that of the E. coli cytR gene, we tested whether V. cholerae cytR, provided in trans, could repress transcription of the udp gene of E. coli. We compared transcription of a udp–lacZ fusion in a parent E. coli strain containing a wild-type copy of cytR as well as in a derivative cytR mutant transformed with either a control plasmid or a plasmid carrying the wild-type V. cholerae cytR gene (Barbier and Short, 1992). As shown in Fig. 4A, udp–lacZ transcription was derepressed in the E. coli cytR mutant. When the V. cholerae cytR gene was supplied in trans to an E. coli cytR mutant, udp–lacZ transcription was repressed.
Figure 4.
Functional similarity of E. coli CytR and V. cholerae CytR.
A. Normalized β-galactosidase activity of an E. coli strain containing a chromosomal fusion of lacZ to the E. coli udp promoter (SS6005) transformed with pBR322, an E. coli ΔcytR mutant (SS6018) transformed with pBR322 and the same E. coli ΔcytR mutant transformed with the plasmid pAJH3, denoted pVcytR, carrying the promoter and coding sequence of V. cholerae cytR.
B. Normalized β-galactosidase activity of wild-type V. cholerae (PW387) and V. cholerae ΔcytR mutant (PW386) cells containing a chromosomal fusion of lacZ to the V. cholerae udp promoter grown in LB broth, LB broth supplemented with 0.3% cytidine, LB broth supplemented with 0.3% glucose and LB broth supplemented with 0.3% glycerol.
Vibrio cholerae CytR regulates the V. cholerae udp gene in response to cytidine
We questioned whether CytR regulates nucleoside catabolism in V. cholerae as it does in E. coli. We identified an ORF in the V. cholerae genome (TIGR locus VC1034) whose putative protein product is 75% identical and 89% similar to the Udp protein of E. coli K-12. We constructed a lacZ fusion to the V. cholerae udp promoter and inserted this fusion into the lacZ locus of both wild-type V. cholerae and a ΔcytR mutant. We then assayed β-galactosidase activity in extracts of planktonic cells grown in LB broth alone as well as in LB broth with added cytidine, glucose and glycerol. In all the media used, the growth curves of wild-type V. cholerae and the ΔcytR mutant were similar (data not shown). As shown in Fig. 4B, when these strains were grown in LB broth, udp–lacZ transcription was greater in the V. cholerae ΔcytR mutant than in wild-type V. cholerae. As predicted, the addition of cytidine to LB broth increased udp–lacZ transcription in wild-type cells almost to the level of the V. cholerae ΔcytR mutant. The addition of glucose to the growth medium decreased the difference in udp–lacZ transcription between wild-type V. cholerae and a ΔcytR mutant slightly, whereas the addition of glycerol to the growth medium had no significant effect on udp–lacZ transcription in wild-type V. cholerae and ΔcytR mutant cells.
Phase-contrast microscopy of wild-type V. cholerae and ΔcytR mutant biofilms
Once the biofilm repressor was identified as the V. cholerae CytR protein, the mechanism of biofilm repression and its role in biofilm development was explored further. It has been shown previously that biofilm development by V. cholerae O139 consists of an initial phase of several hours when only transient surface association is seen. After this initial phase, immobilization and accumulation of the cells in a biofilm is observed. This step requires exopolysaccharide synthesis (Watnick et al., 2001).
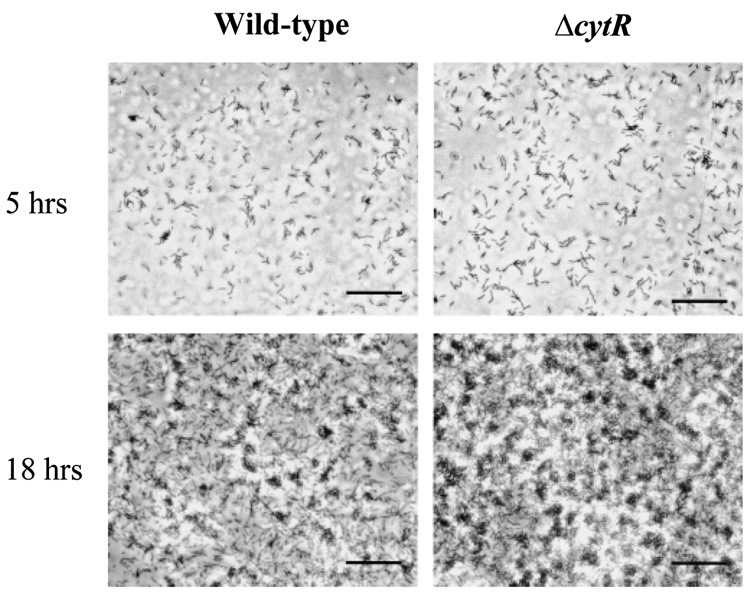
The colony morphology and biofilm phenotype of the V. cholerae ΔcytR mutant suggests that exopolysaccharide synthesis is derepressed in this mutant. We were interested in determining how the course of biofilm development was altered by this derepression. We hypothesized that increased exopolysaccharide synthesis might (i) shorten or eliminate the initial phase of transient attachment; (ii) increase deposition of bacteria on the surface during the biofilm accumulation phase; and/or (iii) alter the mature biofilm architecture. Video and time-lapse microscopy of the early stages of biofilm development demonstrated that the initial phase of transient attachment was indistinguishable from that of wild-type V. cholerae both in duration and in the types of interactions observed between the bacterium and the surface. In Fig. 5A, we show representative micrographs of early surface immobilization by wild-type V. cholerae and a ΔcytR mutant. These micrographs represent simultaneous experiments in which wells were incubated with similar numbers of either wild-type V. cholerae or ΔcytR mutant cells for 5 h at 27°C. These micrographs demonstrate that the early stages of biofilm development by wild-type V. cholerae and a ΔcytR mutant were indistinguishable. However, as shown in Fig. 5B, after 18 h of incubation with a surface, the biofilm formed by the V. cholerae ΔcytR mutant was confluent and dense, whereas the wild-type V. cholerae biofilm had not yet reached confluence. Thus, biofilm development by the ΔcytR mutant deviates from that by wild-type V. cholerae in the surface accumulation phase but not in the initial phase of transient association.
Figure 5.
Phase-contrast micrographs comparing batch biofilm development in LB broth by wild-type V. cholerae (MO10) and a V. cholerae ΔcytR mutant (PW324) after 5 h and 18 h of exposure to a polystyrene surface. Bar = 10 µm.
Three-dimensional architecture of V. cholerae ΔcytR mutant biofilms
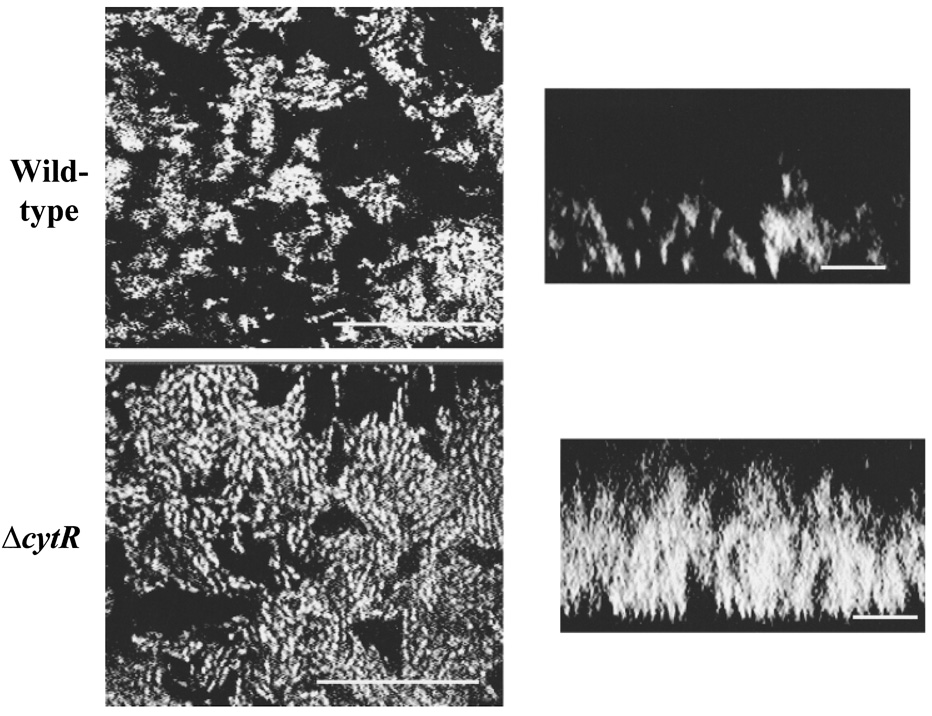
Previous studies have suggested that bacterial variants or mutants that exhibit increased exopolysaccharide production may form three-dimensional biofilms with an altered architecture (Hentzer et al., 2001). To determine whether the architecture of a V. cholerae ΔcytR mutant biofilm is grossly different from that of wild-type V. cholerae, we used confocal microscopy to visualize the three-dimensional structures of wild-type V. cholerae and ΔcytR mutant biofilms after 2 days of growth. As shown in Fig. 6, horizontal and vertical sections through both wild-type V. cholerae and ΔcytR mutant biofilms demonstrate pillars of bacteria with water channels between. In contrast to the wild-type V. cholerae biofilm, however, the water channels observed in the ΔcytR mutant biofilm are much narrower, and the bacterial pillars are almost confluent. Thus, the density of cells in the ΔcytR mutant biofilm is much greater than that in wild-type biofilms.
Figure 6.
Confocal micrographs of 2-day-old wild-type V. cholerae (MO10) and V. cholerae ΔcytR mutant (PW324) biofilms. XY-sections through the biofilms are shown on the left (bar = 25 µm), and vertical sections through the biofilms are shown on the right (bar = 10 µm).
Reporter measurements of vps gene transcription
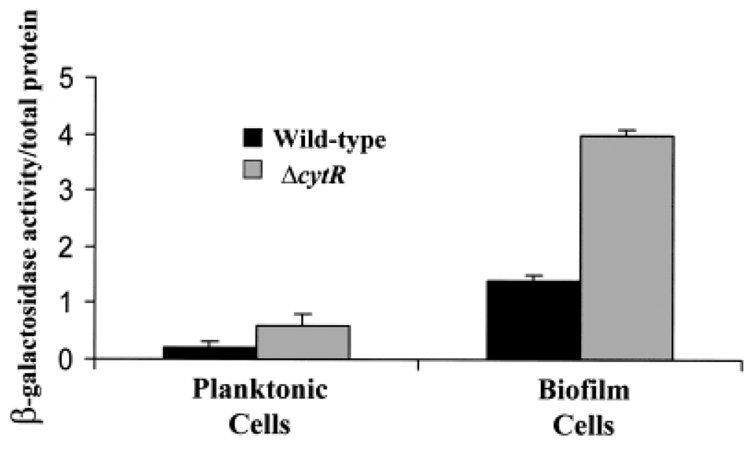
To investigate further the observation that a cytR mutation affects the surface accumulation phase of biofilm development, we measured transcription of one of the identified vps operons in the planktonic and biofilm-associated phases. We inserted a lacZ fusion to the vpsL promoter into the chromosomal lacZ gene of wild-type V. cholerae and a ΔcytR mutant. vpsL is the first gene in one of two large putative operons involved in VPS synthesis (Yildiz et al., 2001). vpsL–lacZ transcription was measured in planktonic and biofilm-associated wild-type V. cholerae and ΔcytR mutant cells. As shown in Fig. 7, transcription of vpsL–lacZ in both wild-type V. cholerae and ΔcytR mutant planktonic cells was low. However, vpsL–lacZ transcription in biofilm-associated cells was seven and 14 times, respectively, that in planktonic cells with the same genetic background. Furthermore, vpsL–lacZ transcription in the V. cholerae ΔcytR biofilm was three times that of vpsL–lacZ transcription in the wild-type V. cholerae biofilm.
Figure 7.
Normalized β-galactosidase activity of biofilm-associated and planktonic wild-type V. cholerae (PW357) and V. cholerae ΔcytR mutant (PW358) cells carrying a chromosomal fusion of the vpsL promoter to lacZ. Black bars represent β-galactosidase measurements of wild-type V. cholerae, and grey bars represent β-galactosidase measurements of V. cholerae ΔcytR mutants.
Discussion
We have identified a repressor of exopolysaccharide synthesis and biofilm development in V. cholerae and have shown that this repressor is the V. cholerae CytR protein. The E. coli CytR protein is a member of the LacI family of repressors. These repressors respond to increased concentrations of effector molecules by lifting repression of the catabolic pathways relevant to these effector molecules. The previously defined function of CytR in E. coli and other Gram-negative bacteria was repression of the transcription of genes encoding proteins involved in nucleoside uptake and catabolism in response to low nucleoside concentrations (Neidhardt, 1996; Thomsen et al., 1999). Our experiments demonstrate that V. cholerae CytR functions in a manner similar to that of E. coli CytR. When provided in trans, V. cholerae CytR is able to complement an E. coli cytR mutant. The udp gene product, encoding uridine dephosphorylase, is involved in cytidine and uridine catabolism and is regulated by CytR in E. coli. We have shown that V. cholerae CytR represses transcription of the V. cholerae udp gene in response to low environmental cytidine concentrations. Thus, we provide strong evidence that the mechanism of regulation and binding site specificity of V. cholerae CytR is similar to that of E. coli CytR and that V. cholerae CytR is also involved in the repression of nucleoside catabolism in response to low nucleoside concentrations.
We draw two conclusions from our studies of vpsL transcription in wild-type V. cholerae and ΔcytR mutant planktonic and biofilm-associated cells. The first is that, in both planktonic and biofilm-associated cells, V. cholerae CytR regulates VPS synthesis at the level of vps gene transcription. Furthermore, for wild-type V. cholerae, vpsL transcription in the biofilm is greater than vpsL transcription in the planktonic phase. We hypothesize that, as has been observed for other Gram-negative bacteria, surface contact activates transcription of exopolysaccharide synthesis genes in wild-type V. cholerae (Davies et al., 1993; Prigent-Combaret et al., 1999; Sauer and Camper, 2001; Whiteley et al., 2001; Sauer et al., 2002). Because surface contact activation of vpsL is also observed in ΔcytR mutant cells, we conclude that CytR is not a component of the surface-sensing signal transduction cascade. Instead, an independent regulatory circuit must be operative in surface sensing.
Previous studies suggest that wild-type V. cholerae biofilm development includes a period of transient surface association followed by surface immobilization and accumulation (Watnick and Kolter, 1999; Watnick et al., 2001). The initial period of transient surface association by the V. cholerae ΔcytR mutant is indistinguishable from that of wild-type V. cholerae in both its character and duration. Transient surface association has previously been postulated to be important for surface sensing (Watnick et al., 2001). The observation that transient surface association is unaltered in biofilm development by the V. cholerae ΔcytR mutant is consistent with the conclusion stated above that surface contact activation, which occurs during the period of transient surface activation, is functional in the V. cholerae ΔcytR mutant. The period of biofilm accumulation is altered, however, in a V. cholerae ΔcytR mutant. Once biofilm accumulation by the V. cholerae ΔcytR mutant begins, it proceeds rapidly and produces a structure in which cells are much more densely packed together than they are in the wild-type V. cholerae biofilm. We hypothesize therefore that signals that maintain water channels between the pillars of the biofilm, such as those involved in quorum sensing, are over-ridden in the ΔcytR mutant.
To the well-characterized roles of the CytR protein in repression of nucleoside uptake and catabolism, we have now added a novel role for V. cholerae CytR in the repression of an anabolic process, namely synthesis of the V. cholerae exopolysaccharide VPS. The co-regulation of a catabolic process and an anabolic process is particularly thought provoking. One possibility is that derepression of exopolysaccharide synthesis gene transcription in a V. cholerae ΔcytR mutant is simply the indirect result of increased uptake and catabolism of nucleosides, leading to increased carbon and energy stores within the cell. This seems unlikely given the growth medium used in our experiments. The primary carbon and energy sources in LB broth are amino acids, and nucleosides represent ≈ = 0.1% of the dry weight of amino acids in LB. Thus, the carbon and energy stores of a bacterium growing in LB broth would not be significantly altered by derepression of nucleoside uptake and catabolism. Rather, we propose that elevated intracellular cytidine levels may be a signal for surface immobilization and that this signal is transduced by the V. cholerae CytR protein.
Our studies build a bridge between increased levels of cytidine and synthesis of an extracellular polysaccharide matrix leading to surface attachment. These findings are particularly intriguing in view of a recent report that DNA is an important component of the extracellular matrix of P. aeruginosa biofilms and may actually be required for initial biofilm development (Whitchurch et al., 2002). Although it seems improbable that nucleosides would be present in abundance in the aqueous phase of an aquatic environment, we hypothesize that DNA and its catabolites may accumulate within the extracellular matrix of the biofilm. Thus, nucleoside concentrations in and around biofilms may be increased, providing an environmental signal that recruits new bacteria into the biofilm.
Experimental procedures
Bacterial strains and media
The bacterial strains and plasmids used in this study are listed in Table 1. All experiments used Luria–Bertani (LB) broth. Cytidine, glucose and glycerol were used at concentrations of 3 g l−1 where noted. The wild-type V. cholerae strain, MO10, is resistant to streptomycin, as are all mutants derived from this strain. Thus, most biofilm experiments were performed in LB broth supplemented with 100 µg ml−1 streptomycin. Furthermore, biofilm experiments involving strains harbouring a plasmid encoding ampicillin resistance were performed in LB broth supplemented with 150 µg ml−1 ampicillin. Procedures used to construct strains and plasmids for this study are detailed below.
Table 1.
Bacterial strains and plasmids.
| Strains or plasmids | Genotype | Reference/source |
|---|---|---|
| E. coli | ||
| β2155 | ThrB1004 pro thi strA hsdS lacZΔM15 (F' lacZΔM15 lacIq trajD36 proA+ proB+) ΔdapA::erm pir.:RP4, Ermr, Kmr | Dehio and Meyer (1997) |
| SM10λpir | Thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpirR6K; Kmr | Miller and Mekalanos (1988) |
| SS6005 | F− thi leu rpsL Δ(argF-lac)U169 Φ(udp-lac)8 (λRS45) | Barbier and Short (1992) |
| SS6018 | SS6005, cytR::Tn10dTet | Barbier and Short (1992) |
| V. cholerae | ||
| MO10 | 1992 clinical isolate of V. cholerae O139 from India, Smr | Waldor and Mekalanos (1994) |
| PW324 | MO10ΔcytR, Smr | This study |
| PW328 | MO10Δvps, Smr | This study |
| PW329 | MO10ΔcytRΔvps, Smr | This study |
| PW357 | MO10lacZ::vpsLp→lacZ, Smr | This study |
| PW358 | PW324lacZ::vpsLp→lacZ, Smr | This study |
| PW386 | PW324lacZ::udp1p→lacZ, Smr | This study |
| PW387 | MO10lacZ::udp1p→lacZ, Smr | This study |
| Plasmids | ||
| pBSL180 | OriR6K mobRP4 lacI pTac tnp mini-Tn10Km; Kmr, Apr | Alexeyev and Shokolenko (1995) |
| pWM91 | OriR6K OriT sacB lacZα, Apr | Metcalf et al. (1996) |
| pAJH1 | pWM91 carrying a fragment of cytR harbouring an internal, unmarked deletion | This study |
| pAJH5 | pWM91 carrying a fragment of a vps operon harbouring an internal, unmarked deletion | This study |
| pAJH3 | A pBR322 derivative containing the promoter and coding sequence of cytR; Tcr Apr | This study |
| p6891MCS | pBR327 with 8 kbp Sau3A fragment of V. cholerae lacZ gene interrupted by a multiple cloning site | Butterton et al. (1995) |
| PAJH8 | p6891MCS derivative with udp1p→lacZ inserted in the multiple cloning site | This study |
| pAJH2 | p6891MCS derivative with vpsLp→lacZ inserted in the multiple cloning site | This study |
| pSMC2 | pKEN carrying a bright mutant of gfp and 1.8 kb stabilizing fragment from pUC181.8; Apr | Bloemberg et al. (1997) |
Construction of Δvps and ΔcytR mutants
A VPS locus containing two large operons required for VPS synthesis has been identified. (Yildiz et al., 2001). One of these, containing vpsL as well as five additional putative ORFs, was selected for deletion. A 400 bp fragment located just upstream of the start codon of vpsL and a 346 bp fragment located 184 bp downstream from the start codon of the sixth ORF were amplified by PCR using the primer pairs P62, P63 and P64, P65 respectively (see Table 2). Primers P63 and P64 included a complementary 15 bp sequence at their 3′ and 5′ ends respectively. These two fragments were joined using the splicing by overlap extension (SOE) technique, resulting in the construction of a fragment with a 6 kb deletion in the vpsL operon (Horton et al., 1990; Lefebvre et al., 1995). The fragment containing the deletion was purified and ligated into pCR2.1TOPO (Invitrogen). This fragment was then removed from pCR2.1TOPO by digestion with SpeI and XhoI and ligated into pWM91 to create the suicide plasmid pAJH5. This plasmid was used to create vpsL operon deletions in the relevant strains by double homologous recombination and sucrose selection as described previously (Donnenberg and Kaper, 1991).
Table 2.
Primers used for PCR.
| Primer | Location in chromosome I | Sequence |
|---|---|---|
| Construction of V. cholerae vps deletion | ||
| P62 | 998815–998834 | GGCTGGTCTATGTGG CTTGT |
| P63 | 999215–999195 | TTACGAGCGGCCGCATCCCGAAGAATAATTGGAATG |
| P64 | 1005215–1005235 | TGCGGCCGCTCGTAATGGTTGGTGATTGAACATCTG |
| P65 | 1005561–1005542 | CAACGTCAGCAACATGGCTA |
| Construction of V. cholerae cytR deletion | ||
| P38 | 2843448–2843465 | CGGATATGGCTGCTGATG |
| P39 | 2844012–2843994 | TTACGAGCGGCCGCACTTGAAAGGTTTGAACTCG |
| P40 | 2845064–2845081 | TGCGGCCGCTCGTAACTCTATCGAGTGATGTTG |
| P41 | 2845571–2845554 | GCTGCAATCCTTGGTTTG |
| Cloning of V. cholerae cytR | ||
| P55 | 2845374–2845354 | ATTGATCCGCAAGATTTGAGC |
| P56 | 2843961–2843978 | TCTCAGCGCTGCGACAGG |
| Construction of V. cholerae vpsLp→lacZ | ||
| P75 | 998382–998403 | TAGTTCAATCGCGGCTTGTACCACA |
| P76 | 999402–999382 | TAGACCCCTAGCAAGGCAACCGAA |
| Construction of V. cholerae udp1p→lacZ | ||
| P95 | 1099912–1099931 | TAGCGTCTGCTTAAA TTCCGCTC |
| P96 | 1100416–1100435 | TAGCACCAAGGTGGA ATACGGTT |
ΔcytR mutants were constructed in a manner similar to that described for the construction of Δvps mutants. Briefly, the primer pairs P38, P39 and P40, P41 were used to amplify a 564 bp fragment immediately upstream of the cytR gene and a 507 bp fragment beginning 23 bp downstream from the putative cytR stop codon. These fragments were ligated using the SOE technique and used to create a deletion in the V. cholerae cytR gene, resulting in strain PW324.
Construction of pAJH3 for complementation studies
In order to determine whether we could complement the phenotype of our ΔcytR mutant with the cytR gene provided in trans, we cloned cytR into the cloning vector pBR322. The cytR gene is 309 bp downstream of the priA gene. Thus, it is most probably transcribed by a promoter positioned directly upstream of its own coding sequence. The primers P55 and P56, which are positioned 20 bp upstream of the stop codon of priA and 80 bp downstream of the stop codon of cytR, respectively, were used to amplify the cytR gene promoter and coding sequence. The amplification product was ligated into pCR2.1-TOPO (Invitrogen) to yield pCR2.1-TOPO:: cytR. This plasmid was digested with EcoRI, and the fragment containing the cytR gene was purified and ligated into pBR322.
Construction of chromosomally based operon fusions of the vpsL promoter (vpsLp)and the udp1 promoter (udp1p) to lacZ
The intergenic region between the divergently transcribed genes, VC0933 and VC0934, was amplified by PCR using the primer pair P75 and P76. This produced a DNA fragment (vpsLp) including 31 bp of the coding region of VC0933 and 140 bp of the coding region of VC0934. The primers were designed with stop codons at either end of the fragment to avoid the generation of chimeric proteins. vpsLp was recovered by cloning into pCR2.1-TOPO (Invitrogen) to create PCR2.-TOPO::vpsLp. PCR2.-TOPO::vpsLp and pUJ10 were digested with XhoI and BamHI. The resulting vpsLp promoter fragment and the pUJ10 fragment harbouring a promoterless lacZ gene were purified and ligated to yield a plasmid-based fusion of vpsLp to the lacZ gene. The resulting plasmid and p6891 multiple cloning site (MCS) were digested with NotI. The liberated vpsLp–lacZ fusion was purified and ligated with p6891MCS to yield the vpsL–lacZ fusion inserted between two homologous fragments of the V. cholerae lacZ gene and oriented opposite to the direction of transcription of the wild-type V. cholerae lacZ gene. The promoter region of this construct was fully sequenced to confirm the absence of mutations introduced by PCR or cloning. This fusion was crossed into the chromosomal locus of the V. cholerae lacZ gene of relevant strains as described previously (Watnick et al., 2001).
The chromosomal udp1p–lacZ fusion was constructed as described for the vpsLp–lacZ fusion except that the region between the putative ORFs with the TIGR designation VC1033 and VC1034 was amplified by PCR using the primer pair P95 and P96.
Transposon mutagenesis and screen for biofilm-altered mutants
Transposon mutagenesis and the screen for biofilm-altered mutants were performed as described previously (Watnick and Kolter, 1999). Briefly, wild-type V. cholerae and the E. coli strain β2155(pBSL180) were crossed on an LB agar plate supplemented with 0.3 mM diaminopimelic acid for 2 h at 37°C. Transposon-insertion mutants were selected on LB agar plates containing kanamycin. The resulting V. cholerae transposon-insertion mutants were replica plated onto LB agar plates and then transferred to LB-filled polyvinylchloride microtitre dish wells using a multipronged device. The bacteria were incubated in these microtitre dish wells at 27°C for 18 h. Wells were then rinsed and stained with crystal violet. Wells that demonstrated an enhanced crystal violet ring were stored at −80°C in LB broth supplemented with 15% glycerol.
Biofilm assays
For observation and quantification of biofilms by crystal violet staining, biofilms were formed either in wells of sterile polystyrene microtitre dishes or in borosilicate glass tubes. Very little difference in bacterial surface accumulation was observed between these two surfaces. Again, biofilms were incubated at 27°C for 18 h. Quantification of surface-adherent cells was measured by crystal violet staining as described previously (Watnick et al., 2001). Briefly, biofilms were stained by incubation with a 1 mg ml−1 aqueous solution of crystal violet for 6 min. Dimethyl sulphoxide (DMSO) was then added immediately to disrupt the biofilm, and an OD595 of the resulting solution was measured.
Phase-contrast and confocal microscopy
Biofilms intended for observation by time-lapse phase-contrast microscopy were formed in 24-well polystyrene microtitre dishes. Wells were filled with 300 µl of LB broth, which allowed adequate aeration of the base of the well and, thus, the formation of a robust biofilm. Biofilm development on the bottoms of wells was recorded either at discrete times, by video microscopy, or at intervals by time-lapse microscopy using an Eclipse TE-200 inverted phase-contrast microscope (Nikon) equipped with an Orca digital CCD camera (Hamamatsu) and VVM-D1 shutter drivers (Uniblitz). A computer equipped with METAMORPH imaging software (Universal Imaging) was used for image acquisition and processing.
For confocal microscopy, bacteria harbouring the plasmid pSMC2 were incubated with coverslips placed vertically in Falcon tubes containing LB broth supplemented with ampicillin as described previously (Watnick et al., 2001). For these experiments, the medium was changed daily. Confocal micrographs were obtained using an Odyssey confocal microscope (Noran) equipped with a graphics work station.
β-Galactosidase measurements
For measurements of gene transcription by the vpsLp promoter in both planktonic and biofilm-associated cells, strains of interest were incubated in 96-well plates at 27°C for 18 h. The planktonic cells from four wells were combined in a separate tube, wells were rinsed with LB broth, and biofilms from these four wells were removed by mechanical disruption and pooled. All experiments were done in triplicate. Planktonic and biofilm-associated cells were gently pelleted and then resuspended in 1.5 ml of Z-buffer (Miller, 1992). Borosilicate beads (1 mm; BioSpec) were added to the cell suspension, and this suspension was agitated using a mini-Beadbeater (BioSpec) for 10 s at 2500 r.p.m. to separate cell aggregates. Although this agitation step was specifically included to disperse biofilm-associated cells, planktonic and biofilm-associated cells were treated similarly. Cells were then lysed by three freeze–thaw cycles at −80°C. A sample of this lysate (200 µl) was set aside for subsequent protein determination using the Coomassie Plus protein assay (Pierce), and ONPG (Sigma), a colorimetric substrate for β-galactosidase, was added to 1 ml of the remaining mixture. All samples were incubated for 18.5 h at 37°C to allow a yellow colour to develop, samples were centrifuged to remove cell debris, and the OD405 of each sample was measured (model 550 microplate reader; Bio-Rad). To obtain relative β-galactosidase activity measurements for each sample, the OD405 was multiplied by 1000 and divided by the calculated protein concentration.
For all measurements of udp–lacZ transcription in E. coli and V. cholerae strains, relevant cells were grown overnight in LB broth at 37°C. Cells were lysed by three freeze–thaw cycles at −80°C. Subsequent measurements of β-galactosidase activity followed the protocol above.
Acknowledgements
We would like to thank Drs Matthew Waldor and Carol Kumamoto for many stimulating discussions and for careful reading of this manuscript. We would also like to acknowledge Dr Anne Kane of the GRASP Center and her staff for expert advice and the preparation of many reagents, which greatly accelerated the course of these experiments. We would like to thank Dr Stephen Short for the generous gift of several E. coli strains and plasmids. Confocal micrographs were obtained at the Tufts Confocal Microscope Facility with the assistance of Dr Robert Willson. This work was supported by NIH K08 AI01588 and an award from the Ellison Medical Foundation to P.I.W., and by the New England Medical Center GRASP Center NIH/NIDDK, P30 DK34928.
References
- Abbad Andaloussi S, Talbaoui H, Marczak R, Bonaly R. Isolation and characterization of exocellular polysaccharides produced by Bifidobacterium longum. Appl Microbiol Biotechnol. 1995;43:995–1000. doi: 10.1007/BF00166915. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- Barbier CS, Short SA. Amino acid substitutions in the CytR repressor which alter its capacity to regulate gene expression. J Bacteriol. 1992;174:2881–2890. doi: 10.1128/jb.174.9.2881-2890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg GV, O’Toole GA, Lugtenberg BJ, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet R, Simon-Pujol MD, Congregado F. Effects of nutrients on exopolysaccharide production and surface properties of Aeromonas salmonicida. Appl Environ Microbiol. 1993;59:2437–2441. doi: 10.1128/aem.59.8.2437-2441.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden GH, Li YH. Nutritional influences on biofilm development. Adv Dent Res. 1997;11:81–99. doi: 10.1177/08959374970110012101. [DOI] [PubMed] [Google Scholar]
- Brikun I, Suziedelis K, Stemmann O, Zhong R, Alikhanian L, Linkova E, et al. Analysis of CRP-CytR interactions at the Escherichia coli udp promoter. J Bacteriol. 1996;178:1614–1622. doi: 10.1128/jb.178.6.1614-1622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan BA, Linhardt RJ, Daniels L. Variation in composition and yield of exopolysaccharides produced by Klebsiella sp. strain K32 and Acinetobacter calcoaceticus BD4. Appl Environ Microbiol. 1986;51:1304–1308. doi: 10.1128/aem.51.6.1304-1308.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterton JR, Beattie DT, Gardel CL, Carroll PA, Hyman T, Killeen KP, et al. Heterologous antigen expression in Vibrio cholerae vector strains. Infect Immun. 1995;63:2689–2696. doi: 10.1128/iai.63.7.2689-2696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Kao CC. EpsR modulates production of extracellular polysaccharides in the bacterial wilt pathogen Ralstonia (Pseudomonas) solanacearum. J Bacteriol. 1998;180:27–34. doi: 10.1128/jb.180.1.27-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- Crutchley MJ. Rugose forms of an El Tor vibrio. J Gen Microbiol. 1968;50:vii. [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Biofilm formation as a developmental process. Methods Enzymol. 2001;336:19–26. doi: 10.1016/s0076-6879(01)36574-6. [DOI] [PubMed] [Google Scholar]
- Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Degeest B, De Vuyst L. Indication that the nitrogen source influences both amount and size of exopolysaccharides produced by Streptococcus thermophilus LY03 and modelling of the bacterial growth and exopolysaccharide production in a complex medium. Appl Environ Microbiol. 1999;65:2863–2870. doi: 10.1128/aem.65.7.2863-2870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degeest B, Janssens B, De Vuyst L. Exopolysaccharide (EPS) biosynthesis by Lactobacillus sakei 0–1: production kinetics, enzyme activities and EPS yields. J Appl Microbiol. 2001;91:470–477. doi: 10.1046/j.1365-2672.2001.01404.x. [DOI] [PubMed] [Google Scholar]
- Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;58:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett ES, Perlegas D, Wozniak DJ. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavigan SA, Nguyen T, Nguyen N, Senear DF. Role of multiple CytR binding sites on cooperativity, competition, and induction at the Escherichia coli udp promoter. J Biol Chem. 1999;274:16010–16019. doi: 10.1074/jbc.274.23.16010. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate over-production affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, Givskov M, et al. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq A, Huq SA, Grimes DJ, O’Brien M, Chu KH, Capuzzo JM, Colwell RR. Colonization of the gut of the blue crab (Callinectes sapidus) by Vibrio cholerae. Appl Environ Microbiol. 1986;52:586–588. doi: 10.1128/aem.52.3.586-588.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol. 1996;20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- Kimmel SA, Roberts RF, Ziegler GR. Optimization of exopolysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus RR grown in a semidefined medium. Appl Environ Microbiol. 1998;64:659–664. doi: 10.1128/aem.64.2.659-664.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Formstecher P, Lefebvre P. Improvement of the gene splicing overlap (SOE) method. Biotechniques. 1995;19:186–188. [PubMed] [Google Scholar]
- Lobitz B, Beck L, Huq A, Wood B, Fuchs G, Faruque ASG, Colwell R. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc Natl Acad Sci USA. 2000;97:1438–1443. doi: 10.1073/pnas.97.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijesteijn PJ, Boels IC, Kleerebezem M, Hugenholtz J. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremoris by the sugar source. Appl Environ Microbiol. 1999;65:5003–5008. doi: 10.1128/aem.65.11.5003-5008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of Vibrio parahaemolyticus. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunoe Y, Wai SN, Takade A, Yoshida SI. Isolation and characterization of rugose form of Vibrio cholerae O139 strain MO10. Infect Immun. 1999;67:958–963. doi: 10.1128/iai.67.2.958-963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JG, Jr, Sztein MB, Rice EW, Nataro JP, Losonsky GA, Panigrahi PO, et al. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J Infect Dis. 1996;174:1364–1368. doi: 10.1093/infdis/174.6.1364. [DOI] [PubMed] [Google Scholar]
- Mozzi F, Rollan G, de Giori GS, Font de Valdez G. Effect of galactose and glucose on the exopolysaccharide production and the activities of biosynthetic enzymes in Lactobacillus casei CRL 87. J Appl Microbiol. 2001;91:160–167. doi: 10.1046/j.1365-2672.2001.01367.x. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, editor. Escherichia coli and Salmonella. Washington, DC: American Society for Microbiology Press; 1996. [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- Parsek MR, Greenberg EP. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 1999;310:43–55. doi: 10.1016/s0076-6879(99)10005-3. [DOI] [PubMed] [Google Scholar]
- Petry S, Furlan S, Crepeau MJ, Cerning J, Desmazeaud M. Factors affecting exocellular polysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus grown in a chemically defined medium. Appl Environ Microbiol. 2000;66:3427–3431. doi: 10.1128/aem.66.8.3427-3431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve WG, Dilworth MJ, Tiwari RP, Glenn AR. Regulation of exopolysaccharide production in Rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology. 1997;143:1951–1958. doi: 10.1099/00221287-143-6-1951. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolnik GK, Voskuil MI, Schnappinger D, Yildiz FH, Meibom K, Dolganov NA, et al. Whole genome DNA microarray expression analysis of biofilm development by Vibrio cholerae O1, E1 Tor. Methods Enzymol. 2001;336:3–18. doi: 10.1016/s0076-6879(01)36573-4. [DOI] [PubMed] [Google Scholar]
- Shukla BN, Singh DV, Sanyal SC. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol Med Microbiol. 1995;12:113–120. doi: 10.1111/j.1574-695X.1995.tb00182.x. [DOI] [PubMed] [Google Scholar]
- Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56:1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen LE, Pedersen M, Norregaard-Madsen M, Valentin-Hansen P, Kallipolitis BH. Protein-ligand interaction: grafting of the uridine-specific determinants from the CytR regulator of Salmonella typhimurium to Escherichia coli CytR. J Mol Biol. 1999;288:165–175. doi: 10.1006/jmbi.1999.2668. [DOI] [PubMed] [Google Scholar]
- Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. Absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in V. cholerae O139. Mol Microbiol. 2001;39:223–235. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- White PB. The rugose variant of vibrios. J Pathol Bacteriol. 1938;46:1–6. [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS (ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]