Abstract
rRNA precursors are bound throughout their length by specific proteins, as the pre-rRNAs emerge from the transcription machinery. The association of pre-rRNA with proteins as ribonucleoprotein (RNP) complexes persists during maturation of 18S, 5.8S, and 28S rRNA, and through assembly of ribosomal subunits in the nucleolus. Preribosomal RNP complexes contain, in addition to ribosomal proteins, an unknown number of nonribosomal nucleolar proteins, as well as small nucleolar RNA-ribonucleoproteins (sno-RNPs). This report describes the use of a specific, rapid, and mild immunopurification approach to isolate and analyze human RNP complexes that contain nonribosomal nucleolar proteins, as well as ribosomal proteins and rRNA. Complexes immunopurified with antibodies to nucleolin—a major nucleolar RNA-binding protein—contain several distinct specific polypeptides that include, in addition to nucleolin, the previously identified nucleolar proteins B23 and fibrillarin, proteins with electrophoretic mobilities characteristic of ribosomal proteins including ribosomal protein S6, and a number of additional unidentified proteins. The physical association of these proteins with one another is mediated largely by RNA, in that the complexes dissociate upon digestion with RNase. Complexes isolated from M-phase cells are similar in protein composition to those isolated from interphase cell nuclear extracts. Therefore, the predominant proteins that associate with nucleolin in interphase remain in RNP complexes during mitosis, despite the cessation of rRNA synthesis and processing in M-phase. In addition, precursor rRNA, as well as processed 18S and 28S rRNA and candidate rRNA processing intermediates, is found associated with the immunopurified complexes. The characteristics of the rRNP complexes described here, therefore, indicate that they represent bona fide precursors of mature cytoplasmic ribosomal subunits.
INTRODUCTION
Assembly of ribosomal subunits in eukaryotic cells takes place primarily in the nucleolus, where rRNAs are synthesized and processed and where they associate with as many as 85 different ribosomal proteins (r-proteins) and with 5S rRNA to form the nuclear precursors to cytoplasmic 40S and 60S ribosomal subunits (for reviews, see Hadjiolov, 1985; Warner, 1990). In human cells, the 18S, 5.8S, and 28S RNAs are synthesized as part of a ∼13,500 nucleotide (47S) precursor RNA (pre-rRNA). Production of mature rRNAs involves removal of long external (ETS) and internal (ITS) spacer sequences in the pre-rRNA, as well as numerous nucleotide modifications, which include pseudouridine conversion and ribose methylation (for reviews, see Maden, 1990; Eichler and Craig, 1994; Venema and Tollervey, 1995). The available evidence indicates that all pre-rRNA cleavage steps occur posttranscriptionally, after synthesis of the whole primary transcript is completed (Hadjiolov, 1985).
Association of r-proteins with rRNA begins on the nascent pre-rRNA (e.g., Chooi and Leiby, 1981), and most of the r-proteins are already bound to the rRNA before transport of ribosomal subunits to the cytoplasm (e.g., Warner and Soeiro, 1967; Kumar and Warner, 1972; Prestayko et al., 1974; Auger-Buendia and Longuet, 1978; Warner, 1979; Hadjiolov, 1985). In addition to r-proteins, small nucleolar RNA-ribonucleoprotein (sno-RNP) complexes and a number of nonribosomal proteins are also found in association with pre-rRNAs and their processing products in the form of preribosomal RNP complexes (e.g., Kumar and Warner, 1972; Prestayko et al., 1974; Auger-Buendia and Longuet, 1978; Mougey et al., 1993). 80S preribosomes, containing 45S rRNA, as well as a 55S preribosome, which matures to the large ribosomal subunit (Warner and Soeiro, 1967), have been consistently identified in a number of organisms (see Hadjiolov, 1985). Therefore, rRNP complexes (rather than naked rRNA) are the actual native cellular substrates for rRNA processing and assembly of functional ribosomal subunits.
Much information has accumulated recently on the small nucleolar RNAs, which participate in various pre-rRNA processing events, including cleavage and nucleotide modifications (for reviews, see Maxwell and Fournier, 1995; Smith and Steitz, 1997; Tollervey and Kiss, 1997). By contrast, the precise number, identity, and function of nonribosomal proteins associated with rRNA in the nucleolus is largely unknown. Sucrose density gradient analyses have yielded differing reports on the numbers of nonribosomal proteins found in preribosomes, which range from 10 to about ∼30 such proteins (see Hadjiolov, 1985, for a review). These differences can be attributed, at least in part, to difficulties in ascertaining specific associations between the RNA and the cosedimenting proteins, and in distinguishing them from other RNP complexes (e.g., heterogeneous nuclear RNP [hnRNP] complexes) that cosediment with them (Hadjiolov, 1985). Characterization of proteins associated in native RNP precursors to ribosomal particles is also complicated by the resistance of the interphase nucleolus to a variety of extraction procedures (see Warner, 1990), which can compromise RNP complex integrity.
Among the few nonribosomal, pre-rRNA–associated proteins identified to date in vertebrate cells, the best characterized is nucleolin (also known as C23), a protein of ∼93–110 kDa, which was first identified as one of the most abundant proteins in the nucleolus of growing cells (Orrick et al., 1973; see Olson, 1990; Hernandez-Verdun, 1991). Nucleolin has been shown to bind rRNA directly (e.g., Herrera and Olson, 1986; Bugler et al., 1987). Immunoelectron microscopy studies have shown that it associates with pre-rRNA early on during rRNA transcription, and that it binds throughout the length of the pre-rRNA as transcription of the full-length rRNA continues (Ghisolfi-Nieto, et al., 1996). Nucleolin is absent from mature ribosomes, implying that it dissociates from rRNA bebore ribosome accumulation in the cytoplasm. Direct evidence for a role of nucleolin in ribosome biogenesis has been provided recently by the finding that it interacts with the U3 sno-RNP, and that interaction of nucleolin with the pre-rRNA substrate is required for the first pre-rRNA processing step (Ginisty et al., 1998). Other nonribosomal, nucleolar proteins for which there is evidence of an association with rRNA include B23 and the sno-RNP-associated protein fibrillarin (Lischwe et al., 1985; Ochs et al., 1985; see Olson, 1990). Ribocharin, a variant of B23, has been reported as a major component of nuclear precursors to the large ribosomal subunit (Hügle et al., 1985).
The intracellular distribution of nucleolin and of other nucleolar components undergoes remarkable changes in mitosis, as nucleoli disassemble (see Scheer et al., 1993). Despite the cessation of RNA (including rRNA) synthesis (Taylor, 1960; Prescott, 1964; Johnson and Holland, 1965), pre-rRNA and its processing intermediates persist during mitosis and resume processing in telophase (Fan and Penman, 1971). Proteins with central roles in rDNA transcription, such as RNA polymerase I, DNA topoisomerase I, and upstream binding factor (UBF), remain associated with rDNA-containing nucleolar organizer regions (NORs; e.g., see Scheer et al., 1993; Jordan et al., 1996; Roussel et al., 1996). Other nucleolar, nonribosomal proteins, such as nucleolin, fibrillarin, and B23, associate to varying degrees with the periphery of the chromosomes, while also dispersing throughout the cytoplasm, where they colocalize to varying degrees with one another and with rRNA (e.g., Jimenez-Garcia et al., 1994; Weisenberger and Scheer, 1995; Dundr et al., 1997, 1998; but see Fan and Penman, 1971). This colocalization suggests that at least some of the preribosomal RNP components remain physically associated during M-phase.
The studies reported here describe the isolation of RNP complexes by immunoaffinity chromatography using a monoclonal antibody to nucleolin. This isolation reveals that a subset of known nucleolar proteins, as well as ribosomal proteins, are associated with one another and with RNA in rRNP complexes in interphase cell nuclear extracts. By taking advantage of the natural solubility of nucleolar components afforded by mitosis, similar complexes have been isolated from metaphase-arrested cells, under mild and rapid conditions that are more likely to retain native RNP structure. Precursor rRNA, as well as processed rRNA and candidate processing intermediates, are also present in the immunopurified complexes. The characteristics of these rRNP complexes indicate that they are likely native intermediates in and substrates for the biogenesis of ribosomal subunits.
MATERIALS AND METHODS
Cell Culture and Synchronization
HeLa cells were grown in monolayer culture in DMEM supplemented with 10% FCS and 1% penicillin-streptomycin. For metaphase arrest experiments, cells were partly synchronized in S-phase of the cell cycle with a single thymidine block (2 mM thymidine, ∼16 h; Bostock et al., 1971). Two hours after removal of thymidine, nocodazole (methyl(5-[2-thienylcarbonyl]-1H-benzimidazol-2-yl)carbamate; Sigma Chemical, St. Louis, MO) was added to the culture medium to a final concentration of 0.5 μg/ml, and the cells were incubated for an additional 12–14 h to arrest and accumulate cells in mitosis (Zieve et al., 1980). Metaphase-arrested cells were collected at the end of the incubation period by selective mechanical detachment as previously described (Tobey et al., 1967), at which point >90% of the cells exhibited the rounded morphology characteristic of metaphase cells. For preparation of nuclear extracts, HeLa cells grown in suspension culture were obtained from the Cell Culture Center (Minneapolis, MN).
Antibodies
Monoclonal antibody 7G2 to nucleolin was obtained by immunization of a BALB/c mouse with HeLa cell nuclear ssDNA-binding proteins, followed by fusion of splenocytes with SP2/0 mouse myeloma cells essentially as previously described (Piñol-Roma et al., 1988; Piñol-Roma and Dreyfuss, unpublished data). The reactivity of the antibody toward nucleolin was verified by immunoblotting of nuclear fractions enriched for nucleolin, and by reactivity of the antibody with recombinant nucleolin (kindly provided by Drs. M. Olson and F. Amalric, respectively; our unpublished observations). Mouse monoclonal antibody to B23 was obtained from Dr. R. Ochs (Ochs et al., 1983), and rabbit polyclonal antisera to fibrillarin, Nopp140, and ribosomal protein S6 were provided by Drs. Michael Terns, U. Thomas Meier, and R. Traut, respectively. Monoclonal antibodies 4F4 (to hnRNP C1/C2) and 4B10 (to hnRNP A1) have been described previously (Choi et al., 1984a; Piñol-Roma et al., 1988).
Cell Fractionation
Fractionation of metaphase-arrested cells was performed essentially as previously described (Piñol-Roma and Dreyfuss, 1991). Briefly, nocodazole-arrested cells collected by selective detachment were washed twice with ice-cold PBS and resuspended in an isotonic (composition) buffer (10 mM Tris-HCl, pH 7.4/100 mM NaCl/2.5 mM MgCl2; referred to hereafter as RSB100), containing 0.5% (vol/vol) Triton X-100 and 1 μg/ml each of leupeptin, pepstatin A, and aprotinin. For RNA isolation experiments, RNasin (Promega, Madison, WI) was also included at 1 U/μl. The cells were disrupted by two 5-sec exposures to sonication on ice, using a microtip sonicator (model XL2015; Heat Systems, Farmingdale, NY), set at scale 2.5. The cell lysate was layered onto a 30% (wt/vol) sucrose cushion in RSB100, centrifuged at 4000 × g for 20 min, and the supernatant fraction was used for subsequent immunopurification experiments. Interphase cell nuclear extracts were prepared as described previously (Dignam et al., 1983).
Immunopurification Analysis
Immunopurifications were carried out using monoclonal antibodies to nucleolin (7G2) or to the hnRNP C proteins (4F4), as described previously (Piñol-Roma et al., 1990). Antibodies were bound to protein A-Sepharose CL-4B beads (Pharmacia, Piscataway, NJ) in RSB100 containing 0.5% Triton X-100, by rocking at 4°C for 1 h. The antibody-coated beads were washed three times with the same buffer and incubated for 20 min with either total M-phase HeLa cell lysate or with HeLa cell nuclear extract. The beads were then washed five times with RSB100/0.5% Triton X-100, and bound complexes were eluted with either SDS-PAGE or with nonequilibrium pH gradient gel electrophoresis (NEPHGE) sample buffer, for analysis by SDS-PAGE or two-dimensional gel electrophoresis, respectively. Where indicated, the specificity of the antibodies was confirmed by immunopurification in the presence of the ionic detergent Empigen BB (Calbiochem, San Diego, CA) at 1% in PBS containing 1 mM EDTA, 0.1 mM DTT as previously described (Choi and Dreyfuss, 1984a; Piñol-Roma et al., 1988). “Mock” immunopurifications in which HeLa proteins were omitted were routinely included in these experiments, to identify polypeptides that originated from the antibody preparations used in the immunopurification.
Gel Electrophoresis and Immunoblot Analysis
SDS-PAGE and immunoblot analyses were performed as previously described (Piñol-Roma et al., 1988), with a separating gel acrylamide concentration of 12.5%. Two-dimensional gel electrophoresis was carried out as described by O’Farrell et al. (1977). Separation in the first dimension was by NEPHGE using pH 3–10 ampholites (Bio-Rad, Richmond, Ca). The second dimension was carried out by SDS-PAGE. Where indicated, proteins were visualized by silver staining (Morrissey, 1981). For immunoblot analysis, anti-B23 and anti-fibrillarin antibodies were used at a 1:100 dilution. Anti-Nopp 140, anti-ribosomal protein S6, and anti-hnRNP A1 antibodies were used at 1:500 dilution. Detection of bound antibodies was carried out with the appropriate horseradish peroxidase-conjugated secondary antibody (Organon-Teknika-Cappel, Malvern, PA) followed by chemiluminescent detection using an ECL reagent kit (Amersham, Arlington Heights, IL).
Nuclease Digestion
Samples were incubated with micrococcal nuclease (Pharmacia) at the indicated concentrations for 10 min at 30°C in the presence of 1 mM CaCl2. Reactions were then stopped by addition of EGTA, to a final concentration of 5 mM, on ice. Where indicated, EGTA was added to 5 mM at the beginning of the incubation to inactivate the micrococcal nuclease. RNase A and RNase-free DNase I (Promega) were added directly to the extracts at the indicated final concentrations, and digestions were allowed to proceed at 30°C for 10 min. The digested samples were used for immunopurification experiments as described above.
RNA Analysis
RNP complexes were immunopurified as described above. Immunopurified complexes were eluted from the protein A-Sepharose beads by heating to 65°C for 3 min in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.8) containing 1% SDS. RNA was isolated from the eluted complexes by phenol extraction and precipitated with ethanol. The isolated RNA was fractionated by agarose gel electrophoresis in the presence of formaldehyde, and Northern blotting was performed using the indicated biotinylated oligonucleotides, following standard procedures (Maniatis et al., 1989). After the last wash, the membranes were further blocked by incubation in 3% BSA in PBS for 1 h, followed by incubation with 2 μg/ml horseradish peroxidase-conjugated Avidin D (Vector Laboratories, Burlingame, CA) in PBS containing 0.05% NP-40 for 30 min. After three washes in the same buffer (15 min each at room temperature) the hybridized oligonucleotides were visualized by enhanced chemiluminescence using an ECL reagent kit (Amersham, Arlington Heights, IL). The sequences of the biotinylated oligonucleotides (obtained from GENSET) were as follows: 5′-ETS: 5′-CCGTGGGACGCTTTCC-3′ (complementary to the segment of pre-rRNA 1,805 nucleotides upstream of the 5′-end of the mature 18S rRNA sequence); 28S rRNA: 5′-TGATGAGCGTCGGCATC-3′ (complementary to the region 1,902 nucleotides downstream of the 5′-end of the mature 28S rRNA sequence).
Immunofluorescence Microscopy
HeLa cells grown on glass coverslips were fixed with 2% formaldehyde in PBS for 20 min at room temperature, followed by permeabilization with acetone at −20°C for 3 min. Fixed and permeabilized cells were incubated with 7G2 ascites fluid diluted 1:1,000 in 3% BSA in PBS for 1 h at room temperature in a humidified chamber. After a wash in PBS, the cells were incubated with FITC-labeled goat-antimouse IgG (1:50 dilution; Organon Teknika-Cappel) for 30 min, washed again in PBS, and mounted onto a glass slide. Staining was observed using a Zeiss axiophot microscope (Carl Zeiss, Thornwood, NY) equipped with a 40× Plan-Neofluar objective.
RESULTS
Immunolocalization of Nucleolin in Interphase and M-Phase HeLa Cells
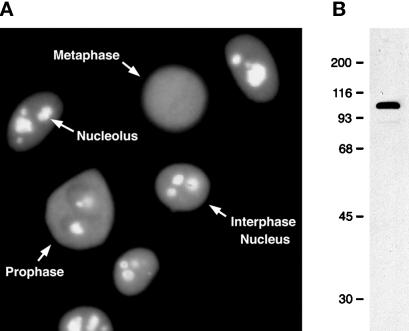
Nucleolin is one of the most abundant proteins in the nucleolus of growing vertebrate cells, where it accumulates predominantly in the dense fibrillar component (DFC) and in the granular component (GC; Olson et al., 1981; Escande et al., 1985). In agreement with these previous observations, immunofluorescence microscopy staining of HeLa cells with a new monoclonal antibody (7G2) against nucleolin shows strong staining of nucleoli, as well as weaker diffuse staining throughout the nucleoplasm (Figure 1A). Little, if any, nucleolin is detected in the cytoplasm of interphase cells at steady-state levels, although nucleolin has been shown to shuttle between the nucleus and the cytoplasm (Borer et al., 1989). Presumably, the small amount of nucleolin in transit through the cytoplasm is below the detection limit of this assay. Immunoblot analysis of total HeLa cell proteins shows that 7G2 reacts with a prominent polypeptide band of ∼100 kDa, in agreement with its identification as nucleolin, as well as a barely visible faster-migrating band, which varies among sample preparations and which is likely a proteolytic product of nucleolin (Figure 1B). As cells enter mitosis and the nucleolus disassembles, nucleolin begins to disperse throughout the cytoplasm (cell labeled ‘Prophase’ in Figure 1A), although significant staining persists in the nucleolar region. In cells in metaphase, 7G2 staining is observed throughout the cell, indicating that the bulk of nucleolin disperses throughout the cell volume during mitosis (cell labeled ‘Metaphase’ in Figure 1A). Confocal microscopy, however, shows that some of the staining remains in close association with the condensed chromosomes (Pomeranz, Henderson, and Piñol-Roma, unpublished observations), in agreement with previous reports (see Olson, 1990).
Figure 1.
Distribution of nucleolin in interphase and M-phase HeLa cells. (A) Asynchronously growing HeLa cells were fixed, permeabilized, and immunostained with the anti-nucleolin monoclonal antibody 7G2, followed by detection with FITC-conjugated secondary antibody. The position of the nuclei and nucleoli, as well as of cells in mitotic prophase and metaphase (as judged by phase microscopy, not shown here), is indicated. (B) Immunoblot analysis of total HeLa cell proteins with the anti-nucleolin monoclonal antibody 7G2. The position of molecular weight standards run in the same gel is indicated on the left (molecular masses are indicated in kilodaltons).
Immunopurification of Nucleolin from HeLa Cell Nuclear Extracts
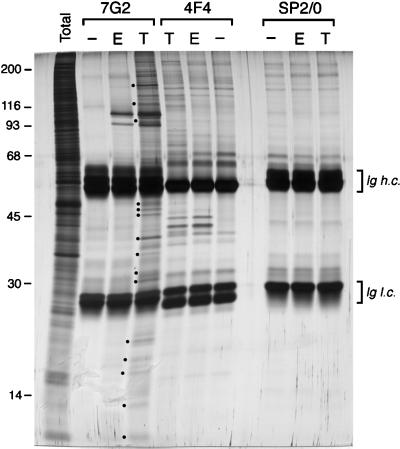
The association of nucleolin with other nuclear components in interphase nuclei was addressed by performing immunopurifications from HeLa cell nuclear extracts, using the 7G2 monoclonal antibody. When immunopurifications were carried out in the presence of the ionic detergent Empigen BB, which disrupts most protein–protein as well as protein–RNA interactions (while preserving antibody–antigen interactions; Choi and Dreyfuss, 1984a) the predominant polypeptide that was isolated corresponded to an apparent molecular mass of ∼100 kDa, which is consistent with the mobility of nucleolin, as well as a less prominent band with slightly faster migration, which is likely a proteolytic fragment of nucleolin (Figure 2, lane 7G2, E). The other predominant bands observed in this lane correspond to the heavy (h.c.) and light (l.c.) chains of the 7G2 antibody used in the immunopurification, as they are also observed when HeLa nuclear extract is omitted during the immunopurification (Figure 2, lane 7G2, −). Few, if any, additional polypeptides are immunopurified with 7G2 under these conditions, thus confirming the specificity of the 7G2 antibody toward nucleolin. By contrast, when immunopurifications were carried out under conditions expected to preserve most protein–protein as well as protein–RNA interactions (0.5% Triton X-100, 100 mM NaCl; e.g., see Piñol-Roma et al., 1988), a number of additional polypeptides were consistently found copurifying specifically with nucleolin (e.g., bands indicated with solid circles in Figure 2, lane 7G2, T). These additional polypeptides have a wide range of mobilities, including several bands of apparent molecular mass >116 kDa, at least three bands migrating just above the 45-kDa marker, a major band of ∼38 kDa, and additional less prominent bands between 30 kDa and 40 kDa. A conspicuous group of proteins is also consistently observed migrating below the 30-kDa marker. Two-dimensional gels show that most of these proteins below 30 kDa are highly basic, suggesting that they may be ribosomal proteins (see Figure 7). The proteins immunopurified with 7G2 under these conditions are only a subset of the total proteins present in the nuclear extract (compare lanes 7G2, T, and Total in Figure 2), further supporting the specificity of their association with nucleolin. The presence of these proteins results from specific antibody–antigen interactions, since no polypeptides other than those corresponding to the immunoglobulins were observed when identical immunopurifications were carried out using nonimmune parent myeloma SP2/0 antibody (lanes SP2/0).
Figure 2.
Immunopurification of nucleolin from HeLa cell nuclear extracts. Immunopurifications were carried out from interphase cell nuclear extracts using anti-nucleolin (7G2), anti-hnRNP C1/C2 (4F4), and nonimmune (SP2/0) monoclonal antibodies, in the presence of either 0.5% Triton X-100 (lanes T) or 1% Empigen BB (lanes E). The immunopurified complexes were resolved by SDS-PAGE, and proteins were visualized by silver staining. Mock immunopurifications in which nuclear extract was omitted were run in parallel to allow identification of bands that resulted from the antibody used in the isolation (lanes −). Examples of bands specific to 7G2-immunopurified complexes are indicated with •, as discussed in the text. Ig h.c. and Ig l.c. refer to the Ig heavy and light chains, respectively.
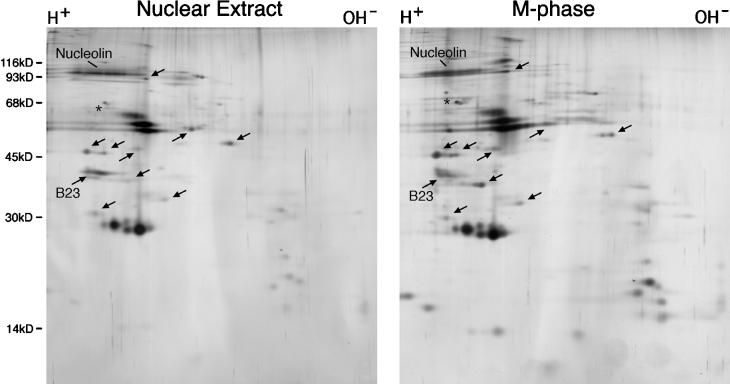
Figure 7.
Two-dimensional gel electrophoresis comparison of complexes immunopurified from interphase cell nuclear extracts and from M-phase cells. RNP complexes were immunopurified with the 7G2 monoclonal antibody from HeLa cell nuclear extracts (left panel) or from whole nocodazole-arrested cell lysates (right panel) as described for Figures 2 and 5, respectively. Proteins in each complex were resolved by two-dimensional gel electrophoresis (see legend to Figure 5) and visualized by silver staining. Arrows point to candidate nonribosomal proteins, as discussed in the text.
As a control for the specificity of the complexes isolated with 7G2, and for comparison purposes, the protein composition of complexes isolated with the 4F4 antibody to the pre-mRNA–binding (hnRNP) proteins C1/C2 (Choi and Dreyfuss, 1984a) was analyzed in parallel. As shown in Figure 2 (lane 4F4, T), in the absence of ionic detergent 4F4 isolates a group of polypeptides in the apparent molecular mass range of 35 to 120 kDa, consistent with the protein composition previously reported for the hnRNP complex (Choi and Dreyfuss, 1984b; Piñol-Roma et al., 1988). At the level of resolution afforded by this analysis, the number and apparent molecular mass of these polypeptides appear different from those observed in association with nucleolin (compare lanes 7G2, T and 4F4, T). These differences were confirmed by two-dimensional gel electrophoresis, which showed little, if any, overlap in the protein composition of the complexes isolated with 7G2 and with 4F4 (Pomeranz and Piñol-Roma, unpublished observations; see Figure 5). A few polypeptides with similar mobilities in the 7G2- and 4F4-purified complexes are apparent above the 116-kDa marker (see Figure 2). Whether these are the same or different proteins in both complexes is not known, since attempts at comparing these proteins by two-dimensional gel electrophoresis were not successful because these proteins did not enter the first dimension gel (our unpublished observations). The specificity of the 4F4 antibody toward the C proteins, determined also by immunopurification in the presence of the ionic detergent Empigen BB (lane 4F4, E), has been shown previously (Choi and Dreyfuss, 1984a). It is noteworthy that the stoichiometry observed for the hnRNP proteins in the hnRNP complexes immunopurified here from nuclear extracts (prepared by high-salt extraction from nuclei, followed by dialysis) is markedly different from that previously reported for hnRNP complexes isolated under similar conditions from soluble nucleoplasm at moderate (100 mM NaCl) ionic strength (e.g., see Piñol-Roma et al., 1988), suggesting that there is considerable disruption or rearrangement of the complexes during nuclear extract preparation. This is in agreement with the known sensitivity of immunopurified hnRNP complexes to nuclease digestion and ionic strength conditions (see Choi and Dreyfuss, 1984b; Dreyfuss et al., 1993, and references therein). However, attempts at immunopurification of nucleolin from soluble nucleoplasm were unsuccessful, since little nucleolin could be recovered in the soluble fraction, and most of the protein was associated with the insoluble nuclear fraction (Wu and Piñol-Roma, unpublished observations).
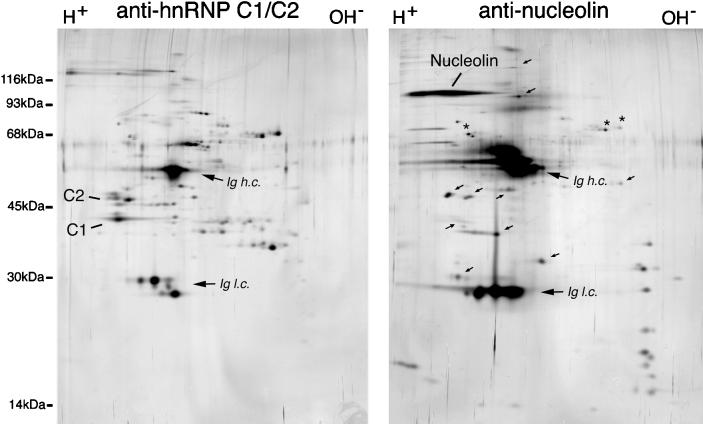
Figure 5.
Immunopurification of nucleolin and hnRNP C proteins from M-phase cells. Whole cell lysates were prepared from metaphase-arrested HeLa cells in RSB-100 containing 0.5% Triton X-100. Immunopurifications were then carried out from these lysates using antinucleolin (right panel) or antihnRNP C (left panel) monoclonal antibodies. The antibody-antigen complexes were eluted with NEPHGE sample buffer, resolved by two-dimensional gel electrophoresis (NEPHGE in the first dimension, left to right, followed by SDS-PAGE in the second dimension; top to bottom), and proteins visualized by silver staining. Analysis of immunoglobulins incubated similarly but in the absence of cell lysate was carried out in parallel gels (not shown) to identify those polypeptides resulting from the antibody and not from the HeLa cells. Arrows indicate some of the proteins specifically associated in complexes with nucleolin. Ig h.c. and Ig l.c. refer to the Ig heavy and light chains respectively.
Association of Nonribosomal Nucleolar Proteins B23 and Fibrillarin, and Ribosomal Protein S6, with Nucleolin-containing Complexes
Few nucleolar proteins have been implicated directly or indirectly in pre-rRNA packaging and metabolism in vertebrate cells. Among these are, in addition to nucleolin, the nucleolar protein B23 and the box C/D sno-RNA-associated protein fibrillarin (for reviews, see Olson, 1990; Hernandez-Verdun, 1991; Shaw and Jordan, 1995). Therefore, it was of interest to determine whether these proteins are among those that copurify with nucleolin. This was addressed by immunoblot analysis of complexes immunopurified with 7G2. Immunopurified hnRNP complexes were again used in these experiments as a control for specificity of any observed associations.
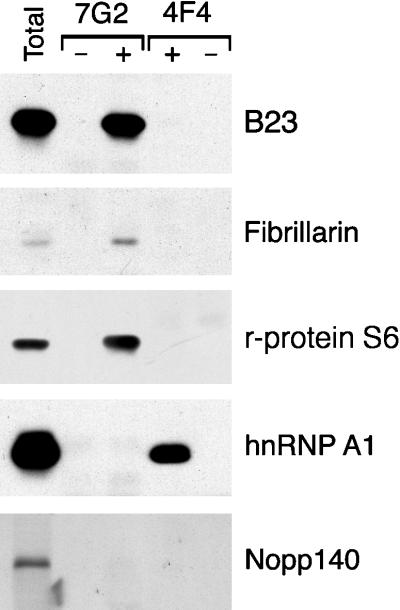
As shown in Figure 3, B23 is readily detectable by immunoblot analysis of complexes isolated with the anti-nucleolin antibody (lane 7G2, +), whereas no detectable B23 is observed copurifying with hnRNP complexes (lane 4F4, +). The immunoreactive band is due to nuclear extract proteins, rather than to reactivity of the anti-B23 antibody or the secondary antibody with immunoglobulins used for immunopurification, as verified by the lack of reactive bands in mock immunopurifications carried out in the absence of nuclear extract (lane 7G2, −). This association of B23 with nucleolin is consistent with a previous report by Li et al. (1996). Similarly, immunoblotting with antibodies to fibrillarin shows that it is also found specifically in complexes immunopurified with 7G2 (Figure 3, lane 7G2, +), whereas little, if any, detectable fibrillarin was observed in the 4F4-immunopurified complexes (Figure 3, lane 4F4, −). In addition to B23 and fibrillarin, immunoblot analysis shows that ribosomal protein S6, which is also a component of the mature ribosome, is specifically associated with nucleolin but not with the hnRNP complexes (Figure 3, panel r-protein S6). By contrast, immunoblotting with the 4B10 monoclonal antibody shows that while hnRNP A1 (an abundant component of hnRNP complexes; e.g., Piñol-Roma et al., 1988) is readily detectable in 4F4-immunopurified hnRNP complexes (Figure 3, panel hnRNP A1, lane 4F4,+), no hnRNP A1 is detectable in the complexes immunopurified with 7G2 (Figure 3, panel hnRNP A1, 7G2, +). This shows that 4F4 does indeed immunopurify hnRNP complexes in the nuclear extracts used here and underscores the specificity of the interactions observed among the various components examined here.
Figure 3.
Immunoblot analysis of RNP complexes immunopurified from nuclear extracts. Proteins from complexes immunopurified with 7G2 (lanes 7G2) or with 4F4 (lanes 4F4) monoclonal antibodies (to nucleolin and hnRNP C1/C2 respectively) were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with antibodies to B23, fibrillarin, ribosomal protein S6 (r-protein S6), hnRNP A1, or Nopp140 (lanes +), as indicated on the right. As a control for reactivity with the antibodies used in the immunopurification, mock immunopurifications were also included in which HeLa cell proteins were omitted (lanes -).
Finally, the possible association of another major nucleolar protein of unknown function, Nopp140 (Meier and Blobel, 1992), with either 7G2- or 4F4-immunopurified complexes, was also addressed. As shown in Figure 3 (panel Nopp 140), whereas Nopp140 can be detected in the unfractionated nuclear extract using anti-peptide antisera, no significant levels of this protein are found in association with either of the immunopurified complexes. Therefore, only a subset of the nucleolar proteins are found in association with nucleolin, and the interaction of these proteins appears to be specific in that they are not found in hnRNP complexes (nor are hnRNP proteins detected to any significant levels in complexes with nucleolin; see also Figure 5, below).
Sensitivity of Immunopurified Nucleolin-containing Complexes to RNase Digestion
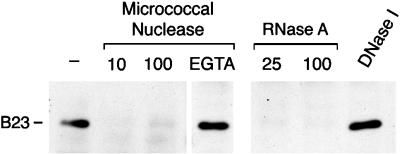
Nucleolin, B23, and fibrillarin all have sequence elements characteristic of RNA-binding proteins, and all three proteins are known or surmised to associate with pre-rRNA and/or processed rRNA (e.g., Bugler et al., 1987; Lapeyre et al., 1987; Dumbar et al., 1989; Lapeyre et al., 1990; see Shaw and Jordan, 1995). The association of these proteins with nucleolin, therefore, could be explained by direct interactions among these proteins, by their binding to common RNA molecules, or both. To distinguish among these possibilities, nuclear extracts were digested with a variety of nucleases before immunopurification with 7G2. Proteins in the immunopurified complexes were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to various proteins. Results obtained for the B23 protein are shown in Figure 4. Digestion of nuclear extract with micrococcal nuclease at either 10 or 100 U/ml resulted in dissociation of B23 from the complexes (Figure 4, lanes ‘micrococcal nuclease’). As a control for possible nonspecific effects unrelated to the nuclease activity, similar digestions were carried out, in the presence of EGTA, to inactivate the nuclease. Under these conditions, B23 continued to copurify with nucleolin (Figure 4, lane EGTA). Because micrococcal nuclease digests both RNA and DNA under the digestion conditions used here, similar analyses were also carried out using nuclear extracts that had been predigested with either with RNase A or with DNase I. As shown also in Figure 4, digestion with RNase A, but not with high concentrations of DNase I, resulted in the dissociation of the B23 from the complexes, confirming that dissociation of B23 from the complexes results from digestion of RNA rather than of DNA (Figure 4, lanes RNase A and DNase I). Silver staining of identical gels indicates that this is not due to loss of nucleolin during the digestion, and that other proteins dissociate upon digestion of the RNA in a way similar to B23 (our unpublished observations). Therefore, RNA integrity is necessary for the association of these proteins with nucleolin. This, in turn, indicates that the complexes isolated with the 7G2 antibody to nucleolin are indeed RNP complexes.
Figure 4.
Sensitivity of nucleolin complexes to RNase. Nuclear extracts were incubated with micrococcal nuclease, RNase A, or DNase I, as indicated, and immunopurifications were then carried out with the 7G2 monoclonal antibody in RSB-100 containing 0.5% Triton X-100. The proteins in the immunopurified complexes were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with the antiB23 monoclonal antibody. Micrococcal nuclease digestions were carried out using either 10 or 100 units/ml, as indicated, in the presence of 2 mM CaCl2. lane EGTA: incubation with 100 u/ml micrococcal nuclease in the presence of both CaCl2 and 5 mM EGTA. RNase A digestions were carried out using either 25 or 100 μg/ml, as indicated. DNase I incubations were carried out in the presence of 100 units RNase-free DNase I/ml. lane -: complexes immunopurified from nuclear extract incubated similarly in the absence of added nuclease.
Immunopurification of Nucleolin Complexes from Metaphase-arrested HeLa Cells
Some of the difficulties in the study of native preribosomal RNP complexes arise from the resistance of the nucleolus to a variety of extraction procedures that are likely to result in at least partial disassembly of RNP structures (see INTRODUCTION). In addition, the results shown in Figure 2 suggested that, at least in the case of hnRNP complexes, some rearrangements or disruptions had occurred during nuclear extract preparations, raising the possibility that similar disruptions may have occurred with the 7G2-immunopurified complexes. To minimize this problem, and to pursue an alternative experimental approach to isolate complexes containing nucleolin, similar immunopurifications to those shown in Figure 2 were carried out from metaphase-arrested HeLa cells. The rationale for this approach is as follows: 1) rRNA precursors are found in metaphase-arrested cells in similar relative abundance to that observed in interphase cells (Fan and Penman, 1971); 2) a different abundant RNP complex, the hnRNP complex, which contains premRNA and its associated (hnRNP) proteins, has sedimentation properties and protein composition in mitosis that are virtually indistinguishable from those in interphase (Lahiri and Thomas, 1985; Piñol-Roma and Dreyfuss, 1991). Thus, there is precedent for the preservation of RNP complex integrity in mitosis; 3) nucleoli disassemble in mitosis and most of the pre-rRNA–associated proteins disperse throughout the cytoplasm (Scheer et al., 1993; see Figure 1), from which they can be recovered in a soluble fraction obtained by simple lysis in isotonic buffers (see below), and are thus amenable to isolation without resorting to lengthy or stringent extraction procedures that can compromise the integrity of the complexes.
When using the monoclonal antibody 7G2 to immunopurify nucleolin from whole-cell lysate prepared from cells arrested in metaphase, in the presence of isotonic buffer conditions and nonionic detergent, a number of proteins were seen to copurify with nucleolin (Figure 5). Analysis of the complexes by two-dimensional gel electrophoresis is shown here to provide a clearer picture of the number and characteristics of these proteins. The proteins that are found associated with nucleolin have a wide range of apparent molecular masses and isoelectric points. They include a prominent group of very basic proteins with apparent molecular masses between 14 kDa and 35 kDa; proteins with molecular masses in the range of 14 kDa-20 kDa and with a wide range of isoelectric points; and additional proteins above ∼30 kDa, at least some of which do not have mobilities consistent with any of the known ribosomal proteins (indicated with arrows in Figure 5). Importantly, the specificity of the association of the observed proteins with nucleolin is underscored upon comparison with hnRNP complexes immunopurified similarly using the anti-hnRNP C proteins antibody 4F4 (Figure 5, panel “antihnRNP C1/C2”). Very few of the polypeptides observed in the nucleolin-containing complexes comigrate with hnRNP proteins, and vice versa (few of the hnRNP proteins are observed in the complexes immunopurified with 7G2). Only minor components, indicated with asterisks in Figure 5, overlap between the two complexes.
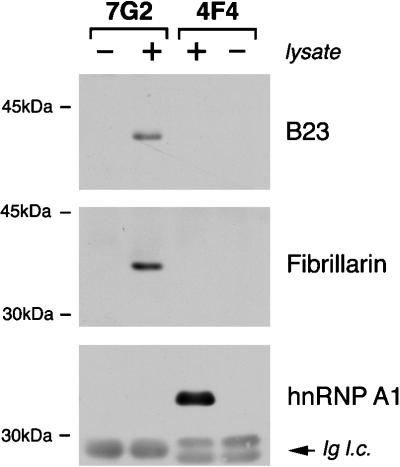
To address whether B23 and fibrillarin remain in RNP complexes with nucleolin in mitosis, their association with complexes isolated from metaphase-arrested cells was assessed by immunoblot analysis, using a similar approach to that shown above with nuclear extracts (see Figure 3). Again, 4F4-immunopurified hnRNP complexes were used as a control for the specificity of the association. Figure 6 shows that, in a manner similar to what is seen in interphase nuclear extracts, both B23 and fibrillarin remain associated in complexes with nucleolin in metaphase-arrested cells (lanes 7G2, +). Furthermore, this interaction is specific, since these two proteins are not detected in hnRNP complexes (lanes 4F4, +), nor is hnRNP A1 detected in association with nucleolin (Figure 6, panel hnRNP A1).
Figure 6.
Immunoblot analysis of RNP complexes immunopurified from metaphase-arrested HeLa cells. RNP complexes were immunopurified with either monoclonal antibody 4F4 or 7G2 from metaphase-arrested HeLa cells (lanes +). The proteins in the respective complexes were resolved by SDS-PAGE and blotted onto nitrocellulose. Identical filters were probed with antiserum to fibrillarin, a monoclonal antibody to B23, or 4B10 monoclonal antibody to hnRNP A1, as indicated. Lanes - contain mock immunopurifications carried out without HeLa cell lysate, to ascertain whether any of the signal observed is due to the antibodies used in the immunopurification. Ig l.c. refers to the Ig light chains originating from the antibody used for immunopurification, and which are detected by the secondary antibody used in the immunoblot.
To gain a better appreciation of the similarities in protein composition between complexes immunopurified from M-phase cells (Figure 5, right panel) and those isolated from interphase nuclear extracts (Figure 2, lane 7G2, T), a side-by-side comparison was performed by two-dimensional gel electrophoresis (Figure 7). This analysis reveals that most (if not all) of the apparent nonribosomal proteins (marked with arrows in Figure 7) are present in both complexes, as are most of the candidate ribosomal proteins. The observed slight differences in relative migration of some of the proteins can be accounted for, at least in part, by mitosis-specific phosphorylation of nucleolar proteins (Belenguer et al., 1990; Peter et al., 1990). Furthermore, quantitative differences could be due in part to dissociation of proteins during nuclear extract preparation, as was seen to be the case with proteins of the hnRNP complex (see Figure 2). B23, which is noted in Figure 7, was identified by immunoblot analysis of identical gels. Its relative amount in two-dimensional gels of mitotic complexes varies from preparation to preparation (e.g., compare Figures 7 and 5) although immunoblot analyses show a consistent association with the immunopurified complexes. Therefore, it is likely that variations observed on two-dimensional gels are due to inefficient solubilization of the protein in mitotic samples, or to failure of the protein to enter the gel due to its lower isoelectric point resulting from phosphorylation. Attempts at identifying fibrillarin and ribosomal protein S6 in the two-dimensional gels shown above were not successful. It is likely that these proteins migrated beyond the gel in the first dimension, due to their highly basic isoelectric points. This would suggest that there may also be additional proteins in the complexes missed by this two-dimensional gel analysis.
Analysis of RNA Associated with Nucleolin-containing RNP Complexes
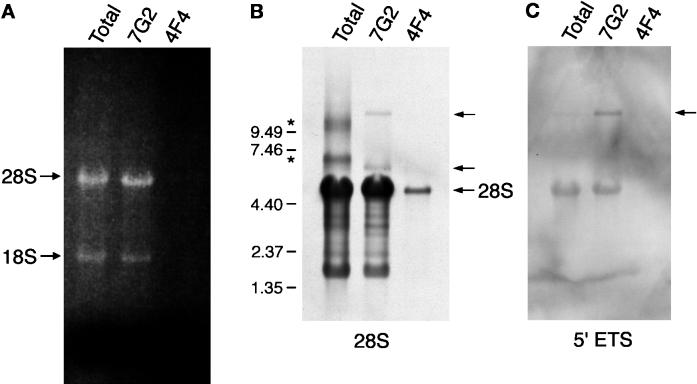
As shown in Figure 4, the association of B23 with nucleolin in complexes immunopurified from nuclear extracts is sensitive to RNase digestion, indicating this association is mediated by binding to RNA to which nucleolin is also bound. Similar results were obtained with metaphase-arrested cells (our unpublished observations). To examine what RNA is present in the nucleolin-containing complexes, RNA was extracted from complexes immunopurified from metaphase-arrested cells and analyzed by formaldehyde-agarose gel electrophoresis (RNA recovered from nuclear extract complexes was found to be too fragmented for appropriate analysis; our unpublished observations). As a reference, RNA was extracted under similar conditions from whole-cell lysates prepared from metaphase-arrested cells. Upon visualization with ethidium bromide staining, predominant bands were visible in the complexes immunopurified with 7G2 (Figure 8A, lane 7G2), which comigrate with the 18S and 28S rRNA observed in the whole cell RNA (Figure 8A, lane ‘Total’). Little, if any, discrete RNA species were observed copurifying with the hnRNP C proteins by this analysis (Figure 8A, lane 4F4). Northern blot analysis of these RNA preparations, using specific oligonucleotide probes, confirmed that 28S rRNA (Figure 7B) and 18S rRNA (our unpublished observations) are specifically copurified with nucleolin. A small amount of 28S rRNA is observed in the 4F4-immunopurified sample with this much more highly sensitive detection method (Figure 8B, lane 4F4). This level of 28S rRNA is much smaller that that observed copurifying with nucleolin and probably reflects background contamination during the immunopurification.
Figure 8.
Analysis of RNA immunopurified with antibodies to nucleolin. (A) RNP complexes were immunopurified from metaphase-arrested HeLa cells using monoclonal antibodies to nucleolin (7G2) or to hnRNP C1/C2 (4F4). RNA was eluted from the complexes as described in the main text and resolved in a 1% agarose gel containing formaldehyde. For comparison, total RNA isolated from whole cell lysates was also included (lane Total). The RNA was visualized by staining with ethidium bromide. The position of 18S and 28S rRNA is indicated on the left. (B) Northern blot analysis of the RNA shown in panel A. The RNA was transferred to a nitrocellulose membrane and probed with biotinylated oligonucleotide probes complementary to 28S rRNA. Detection of hybridized oligonucleotides was carried out with HRP-conjugated avidin, followed by ECL detection. The arrows point to the position of the mature 28S rRNA (bottom) and of precursor (top) and candidate 32S intermediate (middle) rRNA, as described in the text. Asterisks on the left indicate the position of likely RNA aggregates. The position of RNAs of known length run on the same gel is also indicated on the left. (C) Northern blot analysis carried out under identical conditions to those shown in panel B, but using an oligonucleotide complementary to the 5′-pre-rRNA external transcribed spacer. The arrow points to the precursor rRNA.
In addition to apparently mature 28S rRNA, two additional RNA species associated with nucleolin hybridize with the 28S probe, as indicated by arrows in Figure 8B. These correspond to lengths of ∼13,000 nucleotides (top band) and slightly >6000 nucleotides (middle band). The length of the longer RNA species is consistent with the length of the 45–47S precursor rRNA. Hybridizing bands above the mature 28S rRNA are also seen in the total RNA sample (marked with asterisks in Figure 8B). The diffuse appearance of these bands, together with their great variability among samples (not shown), suggests that they may be due to aggregation. To address directly whether precursor rRNA is indeed associated with the complexes immunopurified with 7G2, an identical blot was probed with an oligonucleotide complementary to the 5′-external transcribed spacer of pre-rRNA, downstream of the primary processing site (see MATERIALS AND METHODS). Figure 8C shows that this probe hybridizes with a band that comigrates precisely with the top band seen in panel B (lane 7G2) but not with the middle band. This oligonucleotide cross-hybridizes very weakly with the mature 28S rRNA, and this is visible in this figure due to the substantially larger amounts of mature 28S rRNA in the sample. Nonetheless, the relative signals between the two bands indicate that the contribution of this nonspecific cross-hybridization to the signal in the top band is negligible. Furthermore, the signal for the precursor is enriched in the 7G2-immunopurified sample as compared with the total RNA, where the signal is barely visible. No signal is detected with this oligonucleotide in the 4F4-immunopurified sample, thus underscoring the specificity of the enrichment with the nucleolin-associated RNA. Altogether, this demonstrates that in addition to mature 18S and 28S rRNA, precursor rRNA is associated with the immunopurified complexes. It is noted that these results do not address the possible presence of mature 5.8S rRNA, since at this level of sensitivity it would not be expected to be detectable by ethidium bromide staining. Furthermore, this analysis is likely to grossly underestimate the amount of precursor rRNA associated in the immunopurified complexes, since the need to carry out the isolation in the absence of strong denaturing agents results in substantial degradation of the RNA, as evidenced by the multiple bands below the 28S rRNA that hybridize with the 28S rRNA probe in Figure 8B.
DISCUSSION
This report describes the isolation of a RNP complex or set of complexes that contain nucleolin, a major nucleolar RNA-binding protein, and its associated proteins and RNA. The approach used here is based on rapid immunopurification using a novel monoclonal antibody, 7G2, to isolate nucleolin under conditions that preserve most protein–protein as well as protein-RNA interactions (e.g., see Choi and Dreyfuss, 1984b; Piñol-Roma et al., 1988). Therefore, the expectation was that proteins that associate with nucleolin either by direct protein–protein interactions or by binding to a common RNA will be coisolated with nucleolin. Similar immunopurifications have been used in the past for the specific isolation of hnRNP complexes, which contain hnRNA (including pre-mRNA) and its associated proteins (Choi and Dreyfuss, 1984b; Piñol-Roma et al., 1988). This approach has several advantages when compared with sedimentation analysis of preribosomal RNP complexes (e.g., see Warner and Soeiro, 1967; Kumar and Warner, 1972; Prestayko et al., 1974; Hadjiolov, 1985) in that it is highly specific, by virtue of the monoclonal antibody probes used, can be carried out relatively rapidly under mild buffer conditions, without resorting to lengthy sedimentations through sucrose density gradients, and is not complicated by the cosedimentation of other cellular structures, including RNP complexes (such as hnRNP complexes) that make it difficult to ascertain specific associations between cosedimenting components (for a discussion, see Hadjiolov, 1985).
The results shown here reveal a considerable number of proteins associated with nucleolin in HeLa cell nuclear extracts, including the previously identified nonribosomal proteins, B23 and fibrillarin (Figure 3). Several additional proteins of unknown identity and ranging in apparent molecular mass between 14 kDa and >200 kDa are also in complexes with nucleolin (Figure 2). Among these is a prominent set of relatively small, basic proteins, both of which are characteristic properties of ribosomal proteins (e.g., see Wool, 1979). In agreement with this characterization, immunoblot analysis has shown that ribosomal protein S6 is associated with nucleolin in the immunopurified complexes (Figure 3). The precise identification of the other candidate ribosomal proteins is currently being pursued. The identity of the remaining proteins, which do not have electrophoretic mobilities consistent with any of the known ribosomal proteins, is currently unknown (see Figure 5 and Wool, 1979). Their association with the immunopurified complexes is in agreement with previous sedimentation analyses that showed nonribosomal proteins (in addition to ribosomal proteins) associated in RNP complexes with precursor rRNA and processing intermediates (e.g., Kumar and Warner, 1972; Prestayko et al., 1974; Auger-Buendia and Longuet, 1978). The specificity of the association of these proteins with nucleolin is strongly supported by comparing the complexes isolated using 7G2, with hnRNP complexes immunopurified under identical conditions. Silver staining and immunoblot analyses presented here show that the predominant proteins that associate with nucleolin are distinct from those found in hnRNP complexes, and vice versa. This is particularly relevant considering that both hnRNP complexes and the complexes isolated with 7G2 contain proteins that have a general affinity for single-stranded nucleic acids and therefore could potentially interact with the RNA in either of the complexes (e.g., Piñol-Roma et al., 1988; Dumbar et al., 1989; Ghisolfi-Nieto et al., 1996, and references therein). The fact that the proteins that associate with nucleolin are not found in hnRNP complexes, therefore, makes it highly unlikely (although it has not been completely ruled out) that they are merely binding nonspecifically to the RNA in the complex during the isolation. Gel filtration chromatography studies also indicate that most (if not all) of the nucleolin is in complexes that are distinct from 80S ribosomes and polysomes (Kim, Bassit, Perez, and Pinol-Roma, unpublished data), indicating that, while nucleolin binds rRNA sequences (Ghisolfi-Nieto et al., 1996; Herrera and Olson, 1986; Serin et al., 1996), it is not binding to cytoplasmic ribosomes during sample preparation, and therefore it is unlikely that mature ribosomes are significantly contaminating the immunopurified complexes. The number of proteins found here in association with nucleolin is much smaller than the number of proteins observed in isolated nucleoli (e.g., see Orrick et al., 1973). Therefore, the nucleolin-associated proteins constitute only a subset of the total nucleolar proteins. This is further illustrated by the absence of Nopp140, an abundant nucleolar protein (Meier and Blobel, 1992), from the 7G2-immunopurified complexes (Figure 3). Using antisera raised against immunopurified pre-rRNP complexes, we found that those proteins to which we obtained an immunological response are enriched in nucleoli of interphase cells, and exhibit single-stranded nucleic acid-binding activity, probably reflecting intrinsic rRNA-binding activity of each protein (Pomeranz, Marcu, and Pinol-Roma, unpublished data).
Importantly, nuclease digestion experiments indicate that the association of B23 (Figure 4) as well as of most of the other proteins in the complex (data not shown) with nucleolin is dependent on the presence of RNA. This indicates that the copurification of these proteins with nucleolin is due to their being bound to a common RNA (or RNAs) and implies that the complexes isolated here are indeed RNP complexes. This does not rule out the likely possibility of important protein–protein interactions among the proteins in the immunopurified complexes. In fact, direct interactions between nucleolin and B23 have been reported by Li et al. (1996). Nonetheless, such interactions would appear to not be sufficient, under the conditions used here, for the stable association of these proteins with nucleolin in the absence of RNA during the immunopurification of the complex. In addition, the sensitivity of the complexes to even low RNase or micrococcal nuclease activity (see Figure 4) indicates that the RNA in the complexes is accessible to nucleases, suggesting that, in vivo, the same RNA may be readily accessible to cellular factors involved in its metabolism. This situation would therefore be akin to that of hnRNP complexes, in which the RNA appears to be exposed while bound by the hnRNP proteins (see Dreyfuss et al., 1993).
The results presented here also show that fibrillarin and B23 remain in RNP complexes with nucleolin during mitosis (Figure 6), despite the cessation of rRNA synthesis and processing that occurs in M-phase (Taylor, 1960; Prescott, 1964; Johnson and Holland, 1965). Furthermore, the overall protein composition of complexes immunopurified from M-phase cells is very similar to those isolated from nuclear extracts (Figure 7). This is only an approximation, and precise comparisons are difficult to make by such analyses due primarily to mobility differences in individual proteins, at least some of which probably result from mitosis-specific phosphorylation (Belenguer et al., 1990; Peter et al., 1990). In addition, the likely disruption of RNP complexes during nuclear extract preparation is expected to result in alterations of the relative amount of the different proteins in the complexes as compared with their native stoichiometry. Nonetheless, it appears that most of the proteins that associate in RNP complexes with nucleolin in interphase remain associated during mitosis. This is consistent with immunolocalization studies that show that nucleolin, B23, and fibrillarin colocalize during mitosis (see Dundr et al., 1996, and references therein) and with more recent studies showing that partially processed pre-rRNA also colocalizes with these proteins (Dundr and Olson, 1998). In addition, Nopp140 is not detected in the complexes immunopurified with 7G2 (Figure 3, and our unpublished data) which is consistent with previous observations that Nopp140 does not colocalize with B23, nucleolin, and fibrillarin in nucleolus-derived foci in mitosis (Dundr et al., 1996).
The finding of pre-rRNA in association with the complexes described here is in agreement with the known association of nucleolin with pre-rRNA, determined through other lines of evidence (see INTRODUCTION). It is also consistent with previous studies that showed that rRNA precursors persist during mitosis at relative levels similar to those found in interphase cells. In addition, our experiments also revealed the presence of a nucleolin-associated RNA species that has characteristics of the 32S pre-rRNA processing intermediate (see Figure 8B, lane 7G2) based on its size (slightly >6000 nucleotides) and its hybridization to 28S rRNA sequences. Efforts are currently underway to examine in detail the identity of this species, as well as of other intermediates that may be present in the complexes. The association of mature 18S and 28S rRNA with nucleolin in the immunopurified complexes is also consistent with previous experiments that mapped binding sites for nucleolin on 18S and 28S sequences (Herrera and Olson, 1986; Bugler et al., 1987; Ghisolfi-Nieto et al., 1996; Serin et al., 1996). Therefore, taken together, these results support a continued association of nucleolin with 18S and 28S rRNA sequences all the way from their emergence from the transcription machinery, as part of the primary transcript, to their final processing into mature 18S and 28S rRNA molecules.
The precise relationship of the complexes of rRNA with its associated proteins in mitosis versus interphase cells is unknown. The results shown here, together with previous studies (e.g., Fan and Penman, 1971), suggest that the gross structure of these rRNP complexes, in terms of overall organization and protein composition, is retained during mitosis. This is akin to the situation with pre-mRNA-containing hnRNP complexes, which retain their overall protein composition and sedimentation properties in mitosis as compared with interphase cells (Lahiri and Thomas, 1985; Piñol-Roma and Dreyfuss, 1991). This maintenance of stable interactions between pre-rRNA and its associated proteins may also be relevant to the stability of rRNA precursors during mitosis (Fan and Penman, 1971). Therefore, taken together, these features suggest that studies of mitotic pre-rRNPs will provide important insights into the properties of these complexes in interphase, when they are less stable and, importantly, when they are much less amenable to study in their native conformation due to the extraction conditions required for their release from the interphase nucleolus.
Altogether, the characteristics of the complexes reported here indicate that these complexes are bona fide ribosome precursors and, furthermore, that they are likely to represent the entire nuclear pathway in the assembly of ribosomal subunits from the precursor rRNA to late stages in the maturation process (as evidenced by the presence of both precursor and mature 18S and 28S rRNA). The identification and characterization of the proteins observed here in RNP complexes with nucleolin, the determination of their function, and the production of additional specific probes for these proteins should facilitate the study of native ribosomal subunit precursors, including those the analysis of which has been complicated by their similar sedimentation properties to those of mature cytoplasmic ribosomal subunits. This should lead to a better picture of the cellular events leading to ribosome assembly and maturation. These studies are also of potential relevance to other RNP complexes (e.g., hnRNP complexes) for which detailed analyses have been made more difficult because of the heterogeneity of associated RNA species and the lower relative abundance of any given specific RNAs.
ACKNOWLEDGMENTS
I thank Audrey Marcu and Lisa Pomeranz for excellent technical assistance, Dr. Gillian Small and members of the Small laboratory for help with the initial Northern blot analyses; Drs. Robert Ochs, Michael and Rebecca Terns, U. Thomas Meier, and Robert Traut for gifts of antibodies to B23, fibrillarin, Nopp140, and S6, respectively; and Avrom Caplan, Jeannie Hirsch, Paul Wassarman, and members of my laboratory for helpful discussions throughout the course of this work. This research was supported by a grant from the National Institutes of Health.
REFERENCES
- Auger-Buendia M-A, Longuet M. Characterization of proteins from nucleolar preribosomes of mouse leukemia cells by two-dimensional polyacrylamide gel electrophoresis. Eur J Biochem. 1978;85:105–114. doi: 10.1111/j.1432-1033.1978.tb12217.x. [DOI] [PubMed] [Google Scholar]
- Belenguer P, Caizergues-Ferrer M, Labbé J-C, Dorée M, Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol Cell Biol. 1990;10:3607–3618. doi: 10.1128/mcb.10.7.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- Bostock CJ, Prescott DM, Kirkpatrick JB. An evaluation of the double thymidine block for synchronizing mammalian cells at the G1-S border. Exp Cell Res. 1971;68:163–168. doi: 10.1016/0014-4827(71)90599-4. [DOI] [PubMed] [Google Scholar]
- Bugler B, Bourbon H, Lapeyre B, Wallace MO, Chang J-H, Amalric F, Olson MOJ. RNA binding fragments from nucleolin contain the ribonucleoprotein consensus sequence. J Biol Chem. 1987;262:10922–10925. [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984a;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. Isolation of the heterogeneous nuclear ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci USA. 1984b;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi WY, Leiby KR. An electron microscopic method for localization of ribosomal proteins during transcription of ribosomal DNA: a method for studying protein assembly. Proc Natl Acad Sci USA. 1981;78:4823–4827. doi: 10.1073/pnas.78.8.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dumbar TS, Gentry GA, Olson MOJ. Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry. 1989;28:9495–9501. doi: 10.1021/bi00450a037. [DOI] [PubMed] [Google Scholar]
- Dundr M, Meier UT, Lewis N, Rekosh D, Hammarskjöld M-L, Olson MOJ. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Dundr M, Olson MOJ. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol Biol Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- Escande ML, Gas N, Stevens BJ. Immunolocalisation of the 100K nucleolar protein in CHO cells. Biol Cell. 1985;53:99–109. doi: 10.1111/j.1768-322x.1985.tb00359.x. [DOI] [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of synthesis and processing of nucleolar components in metaphase-arrested cells. J Mol Biol. 1971;59:27–42. doi: 10.1016/0022-2836(71)90411-6. [DOI] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov AA. Cell Biology Monographs. Vol. 12. New York: Springer-Verlag; 1985. The nucleolus and ribosome biogenesis; pp. 1–263. [Google Scholar]
- Hernandez-Verdun D. The nucleolus today. J Cell Sci. 1991;99:465–471. doi: 10.1242/jcs.99.3.465. [DOI] [PubMed] [Google Scholar]
- Herrera AH, Olson MOJ. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986;25:6258–6263. doi: 10.1021/bi00368a063. [DOI] [PubMed] [Google Scholar]
- Hügle B, Scheer U, Franke WW. Ribocharin: a nuclear Mr 40,000 protein specific to precursor particles of the large ribosomal subunit. Cell. 1985;41:615–627. doi: 10.1016/s0092-8674(85)80034-9. [DOI] [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez M de L, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TC, Holland JJ. Ribonucleic acid and protein synthesis in mitotic HeLa cells. J Cell Biol. 1965;27:565–574. doi: 10.1083/jcb.27.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Warner JR. Characterization of ribosomal precursor particles from HeLa cell nucleoli. J Mol Biol. 1972;63:233–246. doi: 10.1016/0022-2836(72)90372-5. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Thomas JO. The fate of heterogeneous nuclear ribonucleoprotein complexes during mitosis. J Biol Chem. 1985;260:598–603. [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Mariottini P, Matthiey C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- Lischwe M, Ochs R, Reddy R, Cook R, Yeoman L, Tan E, Reichlin M, Busch H. Purification and partial characterization of a nucleolar schleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG, NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- Maden BEH. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes & Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe M, O’Leary P, Busch H. Localization of nucleolar phosphoproteins B23 and C23 during mitosis. Exp Cell Res. 1983;146:139–149. doi: 10.1016/0014-4827(83)90332-4. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–134. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- O’Farrell PZ, Goodman HM, O’Farrell PH. High resolution two-dimensional gel electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Olson MOJ, Guetzow K, Busch H. Localization of phosphoprotein C23 in nucleoli by immunological methods. Exp Cell Res. 1981;135:259–265. doi: 10.1016/0014-4827(81)90161-0. [DOI] [PubMed] [Google Scholar]
- Olson MOJ. The role of proteins in nucleolar structure and function. In: Straus PR, Wilson SH, editors. The Eukaryotic Nucleus: Molecular Biochemistry and Macromolecular Assemblies. Vol. 2. Caldwell, NJ: The Telford Press; 1990. pp. 519–559. [Google Scholar]
- Orrick LR, Olson MOJ, Busch H. Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc Natl Acad Sci USA. 1973;70:1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes & Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Choi YD, Dreyfuss G. Immunological methods for purification and characterization of heterogeneous nuclear ribonucleoprotein particles. Methods Enzymol. 1990;181:317–325. doi: 10.1016/0076-6879(90)81132-e. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- Prescott DM. Cellular sites of RNA synthesis. Prog Nucleic Acid Res. 1964;3:33–57. doi: 10.1016/s0079-6603(08)60738-2. [DOI] [PubMed] [Google Scholar]
- Prestayko AW, Klomp GR, Schmoll DJ, Busch H. Comparison of proteins of ribosomal subunits and nucleolar preribosomal particles from Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Biochemistry. 1974;13:1945–1951. doi: 10.1021/bi00706a026. [DOI] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Thiry M, Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Taylor JH. Nucleic acid synthesis in relation to the cell division cycle. Ann NY Acad Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- Tobey RA, Anderson EC, Peterson DG. Properties of mitotic cells prepared by mechanically shaking monolayer cultures of Chinese hamster cells. J Cell Physiol. 1967;70:63–68. doi: 10.1002/jcp.1040700109. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- Warner JR. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Warner JR, Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA. 1967;58:1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. Distribution of newly formed ribosomal proteins in HeLa cell fractions. J Cell Biol. 1979;80:767–772. doi: 10.1083/jcb.80.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D, Scheer U. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J Cell Biol. 1995;129:561–575. doi: 10.1083/jcb.129.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool IG. The structure and function of eukaryotic ribosomes. Annu Rev Biochem. 1979;48:719–754. doi: 10.1146/annurev.bi.48.070179.003443. [DOI] [PubMed] [Google Scholar]
- Zieve GW, Turnbull D, Mullins JM, McIntosh JR. Production of large numbers of mitotic mammalian cells by the use of the reversible microtubule inhibitor nocodazole. Exp Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]