Abstract
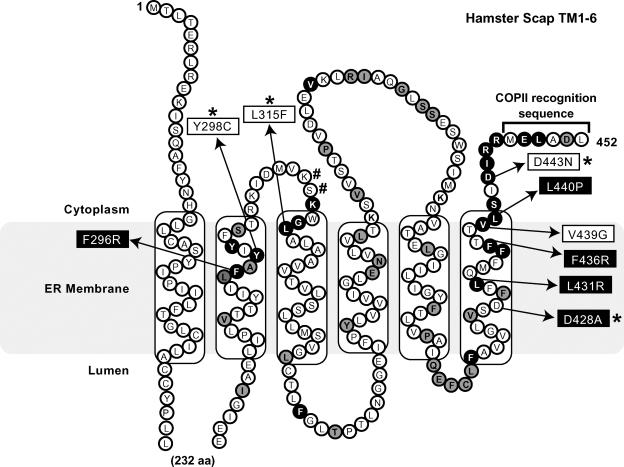
The endoplasmic reticulum membrane protein SREBP cleavage-activating protein (Scap) senses sterols and regulates activation of sterol-regulatory element binding proteins (SREBPs), membrane-bound transcription factors that control lipid homeostasis in fission yeast and mammals. Transmembrane segments 2–6 of Scap function as a sterol-sensing domain (SSD) that recognizes changes in cellular sterols and facilitates activation of SREBP. Previous studies identified conserved mutations Y298C, L315F, and D443N in the SSD of mammalian Scap and fission yeast Scap (Scp1) that render cells insensitive to sterols and cause constitutive SREBP activation. In this study, we utilized fission yeast genetics to identify additional functionally important residues in the SSD of Scp1 and Scap. Using a site-directed mutagenesis selection, we sampled all possible amino acid substitutions at 50 conserved residues in the SSD of Scp1 for their effects on yeast SREBP (Sre1) activation. We found mutations at 23 different amino acids in Scp1 that rendered Scp1 insensitive to sterols and caused constitutive activation of Sre1. To our surprise, the majority of the homologous Scap mutants displayed wild-type function, and only one mutation, V439G, caused constitutive activation of SREBP in mammals. These results suggest that the sterol-sensing mechanism of Scap and the functional requirements for SREBP activation are different between fission yeast and mammals.
Keywords: sterol-regulatory element binding protein, cholesterol, SREBP cleavage-activating protein, sterol, oxygen, Sre1, ergosterol
SREBP cleavage-activating protein (Scap) is a central regulator of lipid homeostasis in mammals (1, 2). Scap is a polytopic endoplasmic reticulum (ER) membrane protein that binds to and facilitates activation of sterol-regulatory element binding proteins (SREBPs), a family of ER membrane-bound transcription factors that control expression of over 30 genes required for cellular uptake of LDL and the synthesis of cholesterol, fatty acids, phospholipids, and triglycerides (3). Scap contains eight N-terminal transmembrane segments and a cytosolic C terminus that binds directly to the C terminus of SREBP. Transmembrane segments 2–6 of Scap comprise the sterol-sensing domain (SSD) that allows Scap to regulate activation of SREBP in response to changes in cellular cholesterol levels (4).
In cells replete with cholesterol, the SSD of Scap binds to the ER resident membrane protein insulin-induced gene (Insig) and retains SREBP in an inactive state in the ER (5). In cells depleted of cholesterol, Scap undergoes a conformational change that disrupts Insig binding and exposes a hexapeptide sequence (MELADL) on the cytosolic face of Scap (6, 7). Exposure of MELADL causes the COPII cargo receptor Sec23/24 to bind to Scap and incorporate Scap and SREBP into transport vesicles. SREBP and Scap are transported to the Golgi apparatus, where the N-terminal basic helix-loop-helix leucine zipper transcription factor domain of SREBP is released from the membrane by two proteolytic cleavage events mediated by the Site-1 and Site-2 proteases (8). The SREBP transcription factor domain enters the nucleus and increases transcription of genes that restore lipid homeostasis.
The sterol-regulated interaction between Scap and Insig is a central event in the control of cellular lipid homeostasis (9). ER retention of Scap is regulated by a synergistic relationship of Insig and sterols. High amounts of cholesterol are sufficient to retain Scap in the ER, but retention is enhanced in the presence of Insig (7). Moreover, overexpression of Insig, even in the absence of sterols, causes Scap ER retention. Although evidence suggests Scap binds cholesterol directly, it is not clear how Scap interacts with sterols and Insig in the ER membrane (10, 11). Mutations Y298C, L315F, and D443N in the SSD of Scap disrupt Insig binding and activate SREBP even in the presence of sterol (12). In addition, a sterol- and Insig-sensitive mutant of Scap (D428A) was described that binds Insig and inhibits activation of SREBP even in the absence of sterol (13). Other than the MELADL sequence, these four amino acids are the only functionally described residues to date in the SSD of Scap.
Recently, our lab has characterized an orthologous SREBP pathway in the fission yeast Schizosaccharomyces pombe that functions as a sterol-dependent oxygen-sensing system (14). In the presence of oxygen, the synthesis rate of the fungal sterol ergosterol is high and fission yeast SREBP (Sre1) is maintained in an inactive state. In the absence of oxygen, ergosterol synthesis is inhibited and this causes Scap (Scp1)-dependent proteolytic activation of Sre1. The cleaved N-terminal transcription factor domain of Sre1 enters the nucleus and increases transcription of genes in ergosterol synthesis as well as other genes required for low oxygen growth (15). As in mammals, strong evidence suggests that Scp1 functions as a sterol sensor and regulates the activation of Sre1 in response to changes in cellular sterols. Sre1 activation is abolished in the absence of Scp1, and homologous mutations Y247C, L264F, and D392N in the SSD of SCP1 cause constitutive activation of Sre1 even in the presence of oxygen and sterols (16). Interestingly, fission yeast Insig (Ins1) is not required for inhibition of Sre1 activation in the presence of sterols, suggesting that Scp1 has an intrinsic ability to sense sterol levels and regulate Sre1 activation (14). Consistent with this, overexpression of Scp1 in fission yeast does not cause constitutive activation of Sre1 (16).
In this study, we perform a genetic selection with fission yeast Scp1 to enhance our understanding of SSD function and to identify new residues required for the function of both fission yeast and mammalian Scap. Through site-directed screening and selection of mutations at residues conserved in the SSDs of Scap and Scp1, we identify single mutations at 23 amino acids in the SSD of Scp1 that cause constitutive activation of Sre1 even in the presence of oxygen. To our surprise, sixteen of the twenty-three equivalent mutations in hamster Scap had no effect on Scap function, and only one (V439G) exhibited reduced Insig binding and constitutive activation of SREBP. The four remaining previously uncharacterized mutations (F296R, L431R, F436R, and L440P) show reduced Insig binding yet favor an ER-retained conformation and are unable to facilitate activation of SREBP, even in the absence of sterols. Our results identify 20 new functionally important residues in Scp1 and suggest that fundamental differences exist between the SSDs of fission yeast and hamster Scap despite their conserved function as regulators of lipid homeostasis. Understanding the underlying reasons for these differences will enhance our knowledge of SSD function.
EXPERIMENTAL PROCEDURES
Materials
We obtained yeast extract from BD Biosciences; Edinburgh minimal medium (EMM) and amino acids from Q-Biogene; oligonucleotides from Integrated DNA Technologies; HRP-conjugated, affinity-purified donkey anti-rabbit and anti-mouse IgG from Jackson ImmunoResearch; cholesterol (C6760) from Steraloids; hydroxypropyl-β-cyclodextrin (HPCD) from Cyclodextrin Technologies Development; peptide N-glycosidase F from New England Biolabs; mevalonate, compactin, trypsin (type I from bovine pancreas), soybean trypsin inhibitor, protease inhibitors (leupeptin, pepstatin A, aprotinin, and PMSF), lipoprotein-deficient serum (LPDS), and oleate from Sigma; fatty-acid free BSA from SeraCare Life Sciences; digitonin and N-acetyl-leucyl-leucyl-norleucinal (ALLN) from Calbiochem.
Antibodies
We obtained anti-Myc 9E10 IgG from Santa Cruz, anti-Myc polyclonal IgG from Upstate, and anti-HSV monoclonal IgG from Novagen. Anti-Sre1 polyclonal IgG generated against the cytosolic N-terminus of fission yeast Sre1 was described previously (14). Anti-Scap R139 polyclonal IgG and 9D5 monoclonal IgG generated against the luminal loop between transmembranes seven and eight of hamster Scap were kind gifts from Drs. Michael Brown and Joseph Goldstein (UT-Southwestern) and were described previously (17).
Monoclonal antibodies to 6xHis-Scp1 (amino acids 446–544) were made using recombinant protein that was purified from Escherichia coli by nickel-affinity chromatography (Qiagen). BALB/c mice were immunized with this protein antigen and screened for immunoreactivity to the antigen by ELISA and Western blotting. Spleen cells from immunopositive mice were removed and fused with SP2/0 myeloma cells to make monoclonal antibodies. Positive clones were identified by ELISA screening using the immunizing antigen. Following dilution cloning, antibody specificity of the final clones was tested by immunoblotting against S. pombe extracts from cells overexpressing Scp1. Antibody isotype was determined using Mouse Isotyping Kit (Boehringer Mannheim): 8G4C11 (IgG1κ), 1G1D6 (IgG2aκ), and 7B4A3 (IgG1κ). Final antibodies were purified from either tissue culture supernatant or ascites fluid using protein-G Sepharose (Pharmacia) as previously described (18).
Yeast strains and cell culture
Yeast strains in all experiments were grown in log phase at 30°C in rich medium or EMM plus supplements. Yeast transformations were performed as previously described (14). S. pombe strain scp1Δ::ura4+ ofd1Δ::kanMX (h−) was derived from the wild-type haploid KGY425 (h−, his-D1, leu1-32, ura4-D18, ade6-M210) using homologous recombination and standard genetic techniques (19, 20).
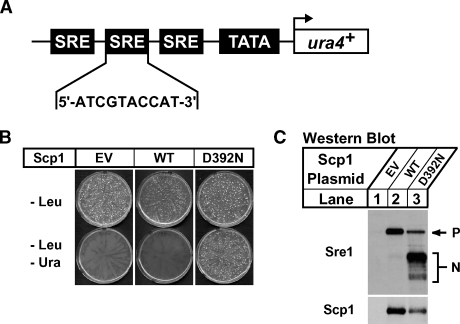
The ura4D-18::3xSRE-ura4+-KanMx scp1Δ::kanMX Sre1 reporter strain used for Scp1 mutant selection harbors an integrated ura4+ gene driven by a minimal promoter containing three tandem copies of the fission yeast Tf2-1 Sre1-regulatory element (SRE) with sequence 5′-ATCGTACCAT-3′ (21). To create this strain, SRE oligomers were generated from complementary oligonucleotides containing a single SRE sequence flanked by restriction sites NheI and SpeI. Oligos were phosphorylated, annealed, ligated, and digested with NheI and SpeI according to the manufacturer's instructions (New England Biolabs). The resulting directional SRE oligomers were resolved on a 20% polyacrylamide gel and eluted with the crush-and-soak method following standard molecular biology protocols (22). Oligomers containing three tandem SRE repeats were ligated into the NheI and SpeI restriction sites of the previously described lacZ reporter plasmid pES10 (15). A minimal promoter containing three SRE sequences and the TATA and transcriptional start site from the fission yeast nmt1+ gene was PCR amplified from the resulting plasmid and inserted into pFA6A-3HA-KanMX6 upstream of ura4+ that was also inserted into the plasmid using PacI and AscI restriction sites (19). The entire reporter cassette was PCR amplified and integrated at the ura4D-18 locus of the wild-type haploid KGY461 (h+, his-D1, leu1-32, ura4-D18, ade6-M210) by homologous recombination. This strain was subsequently mated to previously described scp1Δ::kanMX (14) to create the 3xSRE-ura4+ scp1Δ Sre1 reporter strain (h−, his-D1, leu1-32, ade6-M210, ura4-D18::3xSRE-ura4+-kanMX, scpΔ::kanMX).
Plasmids
pTK-HSV-SREBP-2, encoding herpes simplex virus (HSV)-tagged human SREBP-2 driven by the thymidine kinase (TK) promoter (23), pCMV-Insig-1-Myc, encoding human Insig-1, followed by six tandem copies of the c-Myc tag driven by the cytomegalovirus (CMV) promoter (5), pCMV-Scap, encoding hamster Scap driven by the CMV promoter (17), and pCMV-Scap D443N, encoding hamster Scap D443N driven by the CMV promoter (12) were all kind gifts from Drs. Michael Brown and Joseph Goldstein (UT-Southwestern) and were previously described. pCMV-Scap D428A and all twenty-three pCMV-Scap mutant plasmids listed in Table 1 were generated by mutation of the appropriate codon in pCMV-Scap using QuikChange II XL PCR mutagenesis (Stratagene).
TABLE 1.
Scp1 mutations identified in selection
| Scp1 Residue | Mutations (# Times)b | Scap Mutation Tested | SREBPc Activation |
|---|---|---|---|
| F245 | R(11), A(1), E(1), S(1) | F296R | − |
| Y247 | P(5), C(1), G(1), H(1), R(1) | Y298P | WT |
| Y249 | N(3), P(3), R(3), I(1), l(1), S(1), V(1) | Y300N | WT |
| R259a | H(4), C(2) | K310H | WT |
| A260a | V(4), T(4), D(1) | S311V | WT |
| K261 | I(4), H(2), Q(2), T(2), V(2), C(1), S(1) | K312I | WT |
| G263 | l(4), P(3), A(2), R(2), D(1), E(1), I(1) | G314L | WT |
| L264 | P(10), R(4) | L315P | WT |
| F286 | l(9), I(3) | F337L | WT |
| V326 | R(7), Y(1) | V377R | WT |
| F369 | R(12), G(1), V(1) | F420R | WT |
| L380 | R(16) | L431R | − |
| F384 | V(2), C(1), G(1), I(1), l(1), S(1), T(1) | F435V | WT |
| F385 | R(9), P(6) | F436R | − |
| V388 | G(4), S(3), l(2), D(1), E(1) | V439G | + |
| L389 | P(6), Δ389(1) | L440P | − |
| S390 | l(4), K(2), Y(2), R(1), W(1) | S441L | WT |
| D392 | E(9), H(2), N(1), R(1) | D443E | WT |
| I393 | P(8) | I444P | WT |
| R394 | P(6), D(4), E(1), N(1), S(1), T(1) | R445P | WT |
| R395 | l(2), A(2), G(2), I(2), C(1), E(1), N(1), S(1) | R446L | WT |
| E397 | P(10) | E448P | − |
| L398 | W(5), P(3), R(1) | L449W | − |
Residues identified as second-site mutations in original selection.
Mutations in bold were remade and confirmed as Sre1-activating mutations.
Class abbreviations for SREBP-2 activation: none (−), wild-type (WT), constitutive (+).
pCaMV-Scp1, encoding fission yeast scp1+ driven by the cauliflower mosaic virus (CaMV) promoter, was created in two steps. First, scp1+ cDNA was reverse transcribed from S. pombe poly(A)+ RNA and inserted into pSL1180 (GE Healthcare). scp1+ was subsequently removed from pSL1180 and inserted downstream of the CaMV promoter in pSLF101, which carries a leu2+ auxotrophic marker (24). pCaMV-Scp1 D392N and all twenty-three pCaMV-Scp1 mutant plasmids listed in Table 1 were generated by mutation of the appropriate codon in pCaMV-Scp1 using Quikchange II XL PCR mutagenesis.
Scp1 mutant library construction
Fifty individual pCaMV-Scp1 mutant plasmid libraries representing all 64 codons at each of the identical 50 amino acids in the SSDs of fission yeast Scp1 and hamster Scap (Fig. 1) were generated using the QuikChange multi site-directed mutagenesis kit (Stratagene). To generate each library, a site-directed mutagenesis reaction was performed using pCaMV-Scp1 as a template and an oligo degenerate at the codon of interest. The reaction was subsequently digested with DpnI to remove parental template and transformed into XL-10 Gold Ultracompetent DH5α according to the manufacturer's instructions. For library amplification, the transformation was used to inoculate a 25 ml overnight bacterial culture grown in Luria-Bertani (LB) medium + 100 μg/ml ampicillin. Plasmid DNA was purified from the overnight culture using a Qiagen midiprep kit. One-tenth of the library transformation was plated onto LB + 100 μg/ml ampicillin agar for library size estimation and sequencing of individual library clones. This process was repeated for each of the 50 identical amino acids in the SSDs of Scp1 and Scap.
Fig. 1.
Membrane topology and amino acid comparison of hamster sterol-regulatory element binding protein (SREBP) cleavage-activating protein (Scap) and fission yeast Scp1. Amino acids 1–452 of hamster Scap are shown. Transmembrane segments 2–6 comprise the sterol sensing domain (SSD). The 50 identical amino acids in the SSDs of hamster Scap and fission yeast Scp1 are shown as black and gray filled circles. Residues identified in selection as harboring Sre1-activating Scp1 mutations are black filled circles. Hamster Scap SREBP-activating mutations are shown in white boxes. Hamster Scap SREBP-inhibiting mutations are shown in black boxes. * Previously characterized mutations. # Residues at which Sre1-activating second-site mutations were identified in Scp1.
Each of the 50 libraries constructed were designed to represent all possible codons at a given conserved amino acid in the SSD of Scp1. We utilized the multi site-directed mutagenesis kit from Stratagene for library production to minimize codon bias in the mutagenesis reaction. Library sizes ranged from 4,000–7,040 clones, with a mean of 5,090. Assuming no bias in library production, an average-sized library provided ∼80-fold coverage of all possible codons. To test for bias, we sequenced 16 bacterial clones from the pCaMV-Scp1 D392X (X = any amino acid) library. Out of 16 clones, 10 were wild type (WT), 5 were unique single base pair changes, and 1 was a triple base pair change from the original codon. The high amount of WT codons may be due to incomplete DpnI digestion of parental template. Nevertheless, this small sampling and our results below suggest that each library probably represents all 64 possible codons at a given amino acid.
Scp1 mutant selection
Ten micrograms of each pCaMV-Scp1 mutant plasmid library, pSLF101, pCaMV-Scp1, or pCaMV-Scp1 D392N, was transformed into 1 × 108 logarithmically growing 3xSRE-ura4+ scp1Δ cells. Of each transformation, 1/16 and 1/8 was plated on minimal medium lacking leucine or lacking leucine and uracil, respectively, and cells were incubated for 5 days at 30°C. Growth on minimal medium lacking leucine selected for leu2+-containing pCaMV-Scp1 plasmids and allowed a count of total transformants. On average, ∼12,000 transformants were obtained, representing ∼2-fold coverage of an average Scp1 plasmid library. Eight to sixteen individual colonies from each transformation that produced growth on minimal medium lacking leucine and uracil were picked and restreaked for single colonies two times on the same medium. Restreaking was necessary to isolate a single Scp1 mutation in a transformant, because multiple plasmids were taken up during transformation. Individual plasmid clones were sequenced across the codon of interest after colony PCR amplification using oligos specific to the Scp1 SSD.
Mammalian cell culture
Cells were maintained as a monolayer at 37°C in 5% CO2. SRD-13A cells, a Scap-deficient cell line derived from γ-irradiation of Chinese hamster ovary (CHO) cells, were a kind gift of Drs. Michael Brown and Joseph Goldstein (UT-Southwestern) and were previously described (25). The cells were maintained in medium A (a 1:1 mixture of Ham's F12 medium and DMEM containing 5% FCS, 5 μg/ml cholesterol, 20 μM BSA-conjugated oleate, 1 mM mevalonate, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate). Specific experimental growth conditions utilizing medium B (a 1:1 mixture of Ham's F12 and DMEM containing 5% LPDS, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate) are described in the figure legends. Transient transfections were adjusted to a total of 6 μg of DNA per 10 cm dish with pcDNA3 (Invitrogen) and carried out using FuGENE 6 reagent according to the manufacturer's instructions (Roche).
Protein preparation and Western blot analysis
Whole-cell lysate protein preparation, protein quantification, and Western blot analysis for S. pombe experiments were described previously (14). To isolate fission yeast membrane protein, 8 × 107 cells were resuspended in 50 μl B88 buffer (20 mM Hepes, pH 7.2, 150 mM KOAc, 5 mM MgOAc, and 250 mM sorbitol) plus protease inhibitors and lysed with glass beads for 10 min in a Vortex Genie cell disrupter at 4°C. Five-hundred microliters of B88 plus protease inhibitors was added to the sample, and lysate was centrifuged at 1,000 g for 5 min at 4°C. The supernatant of this spin was centrifuged at 20,000 g for 10 min at 4°C, and the membrane pellet was resuspended in 100 μl of SDS-lysis buffer (10 mM Tris-HCl, pH 6.8, 100 mM NaCl, 1% SDS, 1 mM EDTA, and 1 mM EGTA) plus protease inhibitors. Prior to SDS-PAGE, membrane samples were diluted with an equal volume of urea-lysis buffer (10 mM Tris-HCl, pH 6.8, 2% SDS, 6 M urea) and heated at 37°C for 30 min to minimize aggregation of Scp1 protein.
Membrane and nuclear protein extracts for mammalian Western blot analysis were prepared from a transfected 10 cm dish of SRD-13A as previously described, except that the initial lysis buffer A contained 50 mM Hepes instead of 10 mM (26).
Coimmunoprecipitation
For Insig-1 and Scap coimmunoprecipitation, membrane extracts from 1/2 of a 10 cm dish of transfected SRD-13A cells isolated as described above were solubilized in 1 ml of digitonin lysis buffer [6 mM Na2HPO4, 4 mM NaH2PO4, 1% (w/v) digitonin, 150 mM NaCl, 4 mM EDTA, 50 mM NaF, and 0.3 mM Na3VO4] containing protease inhibitors for 1 h at 4°C. The lysate was cleared by centrifugation at 100,000 g for 10 min, and soluble proteins were immunoprecipitated by addition of 2 μg anti-Myc 9E10 IgG and 25 μl protein A-agarose (Repligen) followed by incubation at 4°C overnight. Beads were collected by centrifugation and washed three times with 1 ml digitonin lysis buffer. After the final wash, beads were resuspended in 100 μl SDS-lysis buffer plus protease inhibitors and were analyzed by Western blot analysis.
Scap trypsin cleavage assay
Membrane protein isolated as described above from 3/4 of a 10 cm dish of transfected SRD-13A cells was subjected to trypsin digestion, PNGase F treatment, and Western blot analysis using 10% Tris-Tricine gels as previously described (27). Scanning of protein for quantification was performed using a Versadoc imaging system (Biorad).
RESULTS
Plasmid-based site-directed mutagenesis selection for Sre1-activating Scp1 mutants
Figure 1 shows the amino acid sequence and membrane topology of the hamster Scap SSD (6). This region of Scap is important for sterol sensing, because mutations Y298C, L315F, and D443N render Scap insensitive to sterols and Insig and cause constitutive activation of SREBP (12). These three residues are among the 50 amino acids of Scap that are identical in the SSD of fission yeast Scp1 (Fig. 1, black and gray filled circles). As in mammals, mutations Y247C, L264F, and D392N in Scp1 render Scp1 insensitive to sterols and cause constitutive activation of Sre1, which suggests that fission yeast Scp1 can serve as a functional model of the mammalian Scap SSD (16). To identify other amino acids important for the function of the SSD in Scap, we utilized fission yeast genetics to evaluate the role of the other 47 identical residues in the SSD of Scp1 and Scap. To this end, we generated a fission yeast Sre1 reporter strain that grows on minimal medium lacking uracil supplementation only when Sre1 is activated. This strain, designated 3xSRE-ura4+ scp1Δ, lacks Scp1 and contains an integrated reporter construct that consists of three tandem SREs from the fission yeast Tf2-1 long terminal repeat fused to a minimal promoter driving expression of ura4+ (Fig. 2A) (21). When Sre1 is inactive, ura4+ is expressed at minimal levels, and cells do not grow on medium lacking uracil. When Sre1 is active, cleaved Sre1 binds to the SREs, activates expression of ura4+, and allows growth in the absence of uracil.
Fig. 2.
Genetic selection design for identifying Sre1-activating mutations in Scp1. A: Diagram of the integrated 3xSRE reporter construct in the Sre1 reporter strain, 3xSRE-ura4+ scp1Δ. Three tandem Sre1-regulatory element (SRE) sequences fused to a minimal promoter drive expression of ura4+, allowing growth of fission yeast on medium lacking uracil only under conditions of Sre1 activation. SRE sequence is shown. B: 3xSRE-ura4+ scp1Δ yeast cells were transformed with 10 μg of empty vector pSLF101, pCaMV-Scp1, or pCaMV-Scp1 D392N plasmid DNA, and plated on minimal medium lacking leucine or leucine and uracil, respectively. Plates were incubated for 5 days at 30°C. C: scp1Δ ofd1Δ yeast cells containing either empty vector (EV), pCaMV-Scp1 (WT), or pCaMV-Scp1 D392N plasmids were grown in minimal medium lacking leucine for 16 h. Total cell extracts (40 μg) and urea-solubilized membranes (40 μg) were subjected to Western blot analysis with anti-Sre1 IgG or a mixture of anti-Scp1 IgG monoclonal antibodies 8G4C11, 1G1D6, and 7B4A3, respectively. P and N denote the precursor and nuclear forms of Sre1, respectively.
We then tested the ability of the Sre1 reporter strain to differentiate between Sre1 activation levels in cells expressing Scp1 or Sre1-activating Scp1 mutants. We transformed 3xSRE-ura4+ scp1Δ yeast with leu2+ marked empty vector or plasmids expressing scp1+ or scp1-D392N from the constitutive CaMV promoter. Cells were plated on minimal medium lacking leucine or medium lacking leucine and uracil to select for cells that contained a plasmid (-leu) or for cells that contained a plasmid which facilitated Sre1 activation (-leu-ura). Yeast containing any of the three plasmids grew equally well on medium lacking leucine, but only cells that expressed sterol-resistant Scp1 D392N were able to activate the Sre1 reporter and grow on medium lacking leucine and uracil (Fig. 2B). To ensure that growth on medium lacking uracil correlated with increased Sre1 activation, we assayed Sre1 cleavage in yeast expressing scp1+ or scp1-D392N from the CaMV promoter by Western blotting. As in mammals, nuclear Sre1 is rapidly degraded in the presence of oxygen and is difficult to visualize without inhibiting its turnover (28). To enhance detection of cleaved Sre1 in the presence of oxygen, we performed these cleavage assays in cells lacking Ofd1, a protein required for efficient turnover of nuclear Sre1 (29). scp1Δofd1Δ cells containing empty vector or plasmids expressing scp1+ or scp1-D392N from the CaMV promoter were cultured in minimal medium lacking leucine for 16 h in the presence of oxygen. Expression of Scp1 stabilized the Sre1 precursor but did not induce Sre1 activation under these conditions (Fig. 2C, lanes 1, 2). However, consistent with Sre1 reporter activation, Sre1 was constitutively active in cells expressing Scp1 D392N, even in the presence of oxygen (Fig. 2C, lane 3). Both Scp1 and Scp1 D392N were expressed to similar levels. Collectively, these results establish the Sre1 reporter strain as an effective tool for a plasmid-based selection to identify Sre1-activating mutations in Scp1.
To identify new Sre1-activating mutations in the SSD of Scp1, we constructed 50 different Scp1 mutant plasmid libraries using Quikchange multi PCR mutagenesis (Stratagene) as described in Experimental Procedures. One plasmid library was constructed for each of the 50 identical amino acids in the SSDs of Scap and Scp1 (Fig. 1). Each library contained an average of ∼5,000 pCaMV-Scp1 plasmids representing all possible codon sequences at the residue of interest. All libraries were transformed individually into the 3xSRE-ura4+ scp1Δ Sre1 reporter strain, and cells harboring plasmids that expressed Sre1-activating Scp1 mutants were selected for on minimal medium lacking leucine and uracil. As a control, we also transformed 3xSRE-ura4+ scp1Δ cells with empty vector, pCaMV-Scp1, and pCaMV-Scp1 D392N. Similar to results shown in Fig. 2B, cells that contained empty vector or Scp1 did not grow on medium lacking uracil. Library-transformed cells that grew comparably to Scp1 D392N on medium lacking uracil were picked, and Scp1 mutations present on plasmids in these cells were identified by sequencing across the Scp1 mutation site.
Selection identified twenty-three new Sre1-activating mutations in Scp1
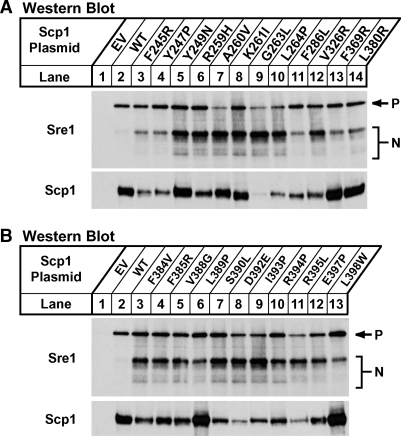
The results of the selection are described in Table 1. By sequencing 8–16 mutants from each library that produced colonies on medium lacking uracil, we identified eighty-nine unique mutations at 23 different amino acids in Scp1 that caused activation of the 3xSRE-ura4+ reporter in the presence of oxygen. Forty-seven of these mutations were identified multiple times. Importantly, eleven mutations were found at Y247, L264, and D392, including mutations Y247C and D392N. Mutations at these residues were previously characterized as Sre1-activating mutations, validating our approach (16). Because the number of mutations was large, we chose to confirm only the most-common change at each of the 23 amino acids identified. Using site-directed mutagenesis, we remade pCaMV-Scp1 plasmids that contained the 23 single-amino-acid substitutions listed in bold in Table 1. To confirm our selection results, 3xSRE-ura4+ scp1Δ cells were transformed with pCaMV-Scp1 or one of the Scp1 mutant plasmids and plated on minimal medium lacking leucine and uracil. Cells that expressed Scp1 did not grow on the selection medium (Fig. 3). However, all cells that expressed one of the twenty-three Scp1 mutant plasmids grew equally well on medium lacking uracil, confirming that these twenty-three Scp1 mutation each caused activation of the 3xSRE-ura4+ reporter (Fig. 3).
Fig. 3.
Growth assay confirmation of twenty-three Sre1-activating Scp1 mutants. 3xSRE-ura4+ scp1Δ yeast cells were transformed with 10 μg of wild-type pCaMV-Scp1 or the indicated mutant pCaMV-Scp1 plasmid DNA and plated on minimal medium lacking leucine or leucine and uracil, respectively. Plates were incubated for 5 days at 30°C.
To determine whether these twenty-three Scp1 mutants caused constitutive Sre1 activation in the presence of oxygen, we assayed Sre1 cleavage by Western blot analysis in yeast that expressed each of the twenty-three Scp1 mutants. scp1Δofd1Δ cells that contained empty vector or plasmids expressing scp1+ or one of the twenty-three scp1 mutants were cultured in minimal medium lacking leucine for 16 h in the presence of oxygen. Similar to results shown in Fig. 2C, expression of Scp1 stabilized the Sre1 precursor compared with vector alone but did not activate Sre1 cleavage (Fig. 4A, B, lanes 1, 2). Importantly, cells that expressed each of the twenty-three Scp1 mutants exhibited constitutive activation of Sre1 even in the presence of oxygen (Fig. 4A, B, lanes 3–14). The expression levels of the Scp1 proteins were variable, but differences in expression did not correlate with Sre1 activation. The combined results of the Sre1 cleavage assays and reporter strain growth assays confirmed that these twenty-three Scp1 mutations were Sre1-activating mutations and identified 20 new functional residues in the SSD of Scp1.
Fig. 4.
All twenty-three Scp1 mutants induce constitutive activation of Sre1. A, B: scp1Δ ofd1Δ yeast cells containing either empty vector, pCaMV-Scp1, or the indicated pCaMV-Scp1 mutant plasmid were grown in minimal medium lacking leucine for 16 h. Total cell extracts (40 μg) and urea-solubilized membranes (40 μg) were subjected to Western blot analysis with anti-Sre1 IgG or a mixture of anti-Scp1 IgG monoclonal antibodies 8G4C11, 1G1D6, and 7B4A3, respectively. P and N denote the precursor and nuclear forms of Sre1, respectively.
Testing the twenty-three homologous Scp1 mutations in mammalian Scap
Next, we tested whether the twenty-three Sre1-activating mutations in Scp1 also act as SREBP-activating mutations in mammalian Scap. Using site-directed mutagenesis, we made pCMV-Scap plasmids containing each of the twenty-three homologous Sre1-activating mutations listed in Table 1. We tested the ability of Insig-1 to regulate each Scap mutant by monitoring SREBP-2 cleavage. SRD-13A cells, a CHO cell line lacking endogenous Scap (25), were transfected with plasmids that expressed SREBP-2 and WT Scap, D443N Scap, or one of the twenty-three Scap mutants in the absence or presence of Insig-1. Surprisingly, our preliminary screening found that sixteen of the twenty-three mutants functioned like Scap and activated SREBP-2 cleavage in an Insig-1-regulated manner (Table 1). Of the seven remaining Scap mutants, two were located at previously characterized amino acids in the MELADL sequence of Scap required for COPII binding, and they exhibited reduced SREBP-2 activation (6). The five remaining uncharacterized mutations showed altered SREBP-2 activation and were studied further.
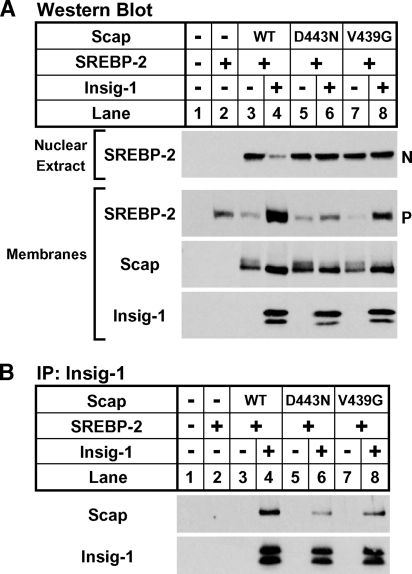
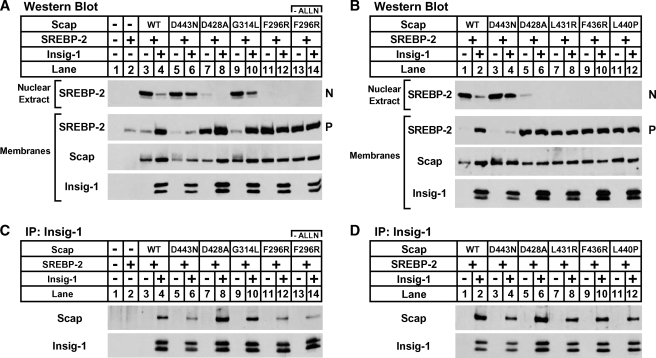
Shown in Fig. 5, SRD-13A cells were transfected with plasmids that expressed SREBP-2 and Scap or the indicated Scap mutant and assayed for activation of SREBP-2 under sterol-depleted conditions in the absence or presence of Insig-1. Cells that expressed SREBP-2 alone had low levels of SREBP-2 precursor in the membrane fraction and no active nuclear SREBP-2 (Fig. 5A, lanes 1, 2). Expression of Scap resulted in SREBP-2 proteolysis and cleaved nuclear SREBP-2 in the absence of sterols, and activation was suppressed by Insig-1 (Fig. 5A, lanes 3, 4). Cells expressing the known sterol and Insig-resistant Scap D443N had equal levels of SREBP-2 activation in the absence or presence of Insig-1 (Fig. 5A, lanes 5, 6). Like Scap D443N, the new Scap mutant, V439G, activated SREBP-2 cleavage equally in the absence or presence of Insig-1 (Fig. 5A, lanes 7, 8). WT and Scap mutant protein levels were similar. Importantly, we also observed high-molecular-weight modifications on Scap that caused the protein to run as a smear in samples that exhibited activated SREBP-2 (Fig. 5A). Unlike Scap, V439G and D443N Scap exhibited high-molecular-weight smearing, even in the presence of Insig-1. These modifications are consistent with previously described Golgi-specific glycosylation resulting from Scap transport to the Golgi apparatus (30). These results suggest that like Scap D443N, Scap V439G facilitates SREBP-2 activation that is resistant to suppression by Insig-1.
Fig. 5.
Hamster Scap mutant V439G mediates insulin-induced gene (Insig)-resistant SREBP activation and is defective in Insig binding. On day 0, SRD-13A cells were set up in medium A at 9 × 105 cells per 10 cm dish. On day 2, cells were transfected in medium B with 4 μg of pTK-HSV-SREBP-2, 0 or 300 ng of pCMV-Insig-1-Myc, and 1 μg of the indicated pCMV-Scap plasmid. Three hours after transfection, cells were refed with medium B containing 50 μM compactin and 50 μM mevalonate and incubated for 18 hours. N-acetyl-leucyl-leucyl-norleucinal (ALLN) (25 μg/ml) was added to each dish 2 hours prior to harvest. Cells were fractionated as described in Experimental Procedures, and membranes were divided into two aliquots and used for both Western blot analysis (A) and immunoprecipitation (B). A: Nuclear extracts (0.3 dish of cells) and membranes (0.25 dish of cells) were subjected to Western blot analysis with anti-herpes simplex virus (HSV) IgG (SREBP-2), anti-Scap IgG 9D5, or anti-Myc IgG 9E10 (Insig-1). N and P denote the nuclear and precursor forms of SREBP-2, respectively. B: Digitonin-solubilized membranes (0.5 dish of cells) were subjected to immunoprecipitation (IP) with anti-Myc IgG 9E10 and protein A-agarose. Bound fractions were subjected to Western blot analysis with polyclonal anti-Myc IgG (Insig-1) or anti-Scap IgG R139.
Scap D443N exhibits sterol and Insig resistance because its binding to Insig is reduced, compared with Scap (12). To determine whether Scap V439G also shows reduced Insig binding, we assayed the interaction of WT, D443N, and V439G Scap with Insig-1 in the same membranes analyzed in Fig. 5A. Insig-1 was immunoprecipitated from digitonin-solubilized membranes and assayed for its interaction with the different Scap proteins by Western blotting. Compared with Scap, both Scap D443N and V439G showed reduced binding to Insig-1 (Fig. 5B). Binding was specific, because no Scap was present in the bound fraction in the absence of Insig-1. These findings support our SREBP cleavage assay results and suggest that V439G is a partially Insig-resistant SREBP-activating mutation in the SSD of Scap.
The remaining four Scap mutations and one mutation characterized as WT in the initial screening were also tested for their ability to facilitate SREBP-2 cleavage. SRD-13A cells were transfected with plasmids that expressed SREBP-2 and Scap, or the indicated Scap mutant, and assayed for SREBP-2 activation in the absence or presence of Insig-1. Similar to the results shown in Fig. 5A, no SREBP-2 activation was present in cells that expressed SREBP-2 alone in the absence of Scap (Fig. 6A, lanes 1, 2). Cells that expressed Scap activated SREBP-2 cleavage in an Insig-1-dependent manner (Fig. 6A, B, lanes 3, 4 and 1, 2, respectively) and those that expressed Scap D443N exhibited activation of SREBP-2 that was insensitive to the presence of Insig-1 (Fig. 6A, B, lanes 5, 6 and 3, 4, respectively). Scap mutation G314L, which tested WT in our initial screening of all twenty-three mutants, showed Insig-1-regulated SREBP-2 activation similar to Scap (Fig. 6A, lanes 9, 10). Surprisingly, new Scap mutants F296R, L431R, F436R, and L440P all failed to activate SREBP-2, even in the absence of Insig-1 (Fig. 6A, B, lanes 11, 12 and 7–12, respectively). SREBP activation in these mutants resembled that of cells expressing Scap D428A (Fig. 6A, B, lanes 7, 8 and 5, 6, respectively), a previously characterized Scap mutant defective in SREBP-2 activation (13). Despite their failure to activate SREBP-2, all four Scap mutants were expressed at levels similar to WT, D443N, and D428A Scap. Omitting the proteasome inhibitor ALLN that was added to stabilize nuclear SREBP-2 did not affect the expression level of Scap F296R, indicating that equal expression of the Scap mutants was not due to proteasome inhibition (Fig. 6A, lanes 11–14). In addition, all four Scap mutants increased the stability of the SREBP-2 precursor, suggesting that they are sufficiently folded to bind SREBP-2 in the ER membrane. These results show that Scap mutants F296R, L431R, F436R, and L440P are expressed at normal levels but are defective in activation of SREBP-2.
Fig. 6.
Hamster Scap mutants F296R, L431R, F436R, and L440P are defective in SREBP activation and Insig binding. On day 0, SRD-13A cells were set up in medium A at 9 × 105 cells per 10 cm dish. On day 2, cells were transfected in medium B with 4 μg of pTK-HSV-SREBP-2, 0 or 300 ng of pCMV-Insig-1-Myc, and 1 μg of the indicated pCMV-Scap plasmid. Three hours after transfection, cells were refed with medium B containing 50 μM compactin and 50 μM mevalonate and incubated for 18 hours. ALLN (25 μg/ml) was added to each dish 2 hours prior to harvest except where noted. Cells were fractionated as described in Experimental Procedures, and membranes were divided into two aliquots and used for both Western blot analysis (A) and (B) and immunoprecipitation (IP) (C) and (D). A, B: Nuclear extracts (0.3 dish of cells) and membranes (0.25 dish of cells) were subjected to Western blot analysis with anti-HSV IgG (SREBP-2), anti-Scap IgG 9D5, or anti-Myc IgG 9E10 (Insig-1). N and P denote the nuclear and precursor forms of SREBP-2, respectively. C, D: Digitonin-solubilized membranes (0.5 dish of cells) were subjected to immunoprecipitation with anti-Myc IgG 9E10 and protein A-agarose. Bound fractions were subjected to Western blot analysis with polyclonal anti-Myc IgG (Insig-1) or anti-Scap IgG R139.
Previous studies showed that Scap D428A exhibits reduced activation of SREBP-2, because its binding to Insig is increased compared with Scap (13). To test whether Scap mutants F296R, L431R, F436R, and L440P also have increased binding to Insig, we tested binding of these Scap proteins to Insig-1 in the same membranes used in Fig. 6A and B by Insig-1 immunoprecipitation. Consistent with previous results, a greater amount of D428A than WT Scap coimmunoprecipitated with Insig-1 (Fig. 6C, D). Surprisingly, all four new Scap mutants exhibited reduced Insig-1 binding, similar to that of Insig-resistant Scap D443N (Fig. 6C, D). These results show that Scap mutants F296R, L431R, F436R, and L440P have reduced Insig-1 binding compared with Scap.
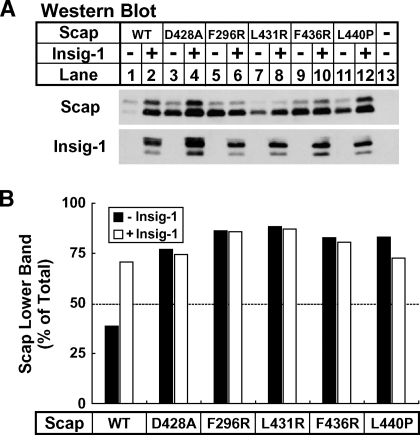
Scap undergoes a conformational change in the presence of sterols and Insig that inhibits its transport to the Golgi apparatus (27). This change in conformation can be monitored by trypsin digestion of intact microsomes and Western blotting against a protected fragment in the luminal loop of Scap between transmembrane segments 7 and 8. This protected fragment is shortened by nine amino acids in the presence of sterols or upon Insig binding due to cleavage of a newly exposed juxtamembranous arginine residue on the cytosolic face of Scap (27). We utilized this assay to probe the conformation of the four Scap mutants that were defective in both Insig-1 binding and SREBP-2 activation. Intact membranes isolated from sterol-depleted SRD-13A cells that expressed Scap or the indicated Scap mutant in the absence or presence of Insig-1 were treated with trypsin and assayed for the protected Scap fragment by Western blotting. In membranes from cells that expressed Scap, the upper ER exit conformation band was predominant in the absence of Insig-1 (Fig. 7A, lane 1). Expression of Insig-1 caused an increase in the amount of ER-retained conformation lower band, and this was the predominant band under these conditions (Fig. 7A, lane 2). Consistent with previous results, Scap D428A exhibited a higher amount of the lower band than upper band, even in the absence of Insig-1, and the ratio of the two bands was not altered by expression of Insig-1 (Fig. 7A, lanes 3, 4) (13). Interestingly, Scap mutants F296R, L431R, F436R, and L440P all resembled Scap D428A and exhibited a predominant amount of lower band in the absence of sterols that was unaffected by the presence of Insig-1 (Fig. 7A, lanes 5–12). Quantification of the Western blot in Fig. 7A showing the fraction of lower band present is shown in Fig. 7B. The amount of lower band increased from 39% to 71% when Insig-1 was added to Scap. D428A and all four Scap mutants had at least 72% lower band, and this was unchanged in the presence of Insig-1. These results are consistent with the failure of these four Scap mutants to activate SREBP-2 and suggest that these mutants assume an ER-retained conformation despite a reduced affinity for Insig-1.
Fig. 7.
Hamster Scap mutants F296R, L431R, F436R, and L440P assume an endoplasmic reticulum-retained conformation even in the absence of Insig. On day 0, SRD-13A cells were set up in medium A at 9 × 105 cells per 10 cm dish. On day 2, cells were transfected in medium B with 0 or 300 ng of pCMV-Insig-1-Myc and 2 μg of the indicated pCMV-Scap plasmid. Three hours after transfection, cells were refed with fresh medium B and incubated for 16 hours. Cells were then washed with PBS and refed medium B containing 1% hydroxypropyl-β-cyclodextrin, 50 μM compactin, and 50 μM mevalonate. After incubation for 1 h, cells were fractionated, and membrane proteins (0.75 dish of cells) were subjected to trypsin digestion for 30 min at 30°C as described in Experimental Procedures. A: Trypsin-digested proteins (0.37 dish of cells) were subjected to SDS-PAGE using 10% Tris-Tricine gels and Western blot analysis with anti-Scap IgG 9D5. Nontrypsin-treated membrane proteins (0.15 dish of cells) were subjected to Western blot analysis with anti-Myc IgG 9E10 (Insig-1). B: Relative intensities of trypsin-digested proteins from A were quantified using a Versadoc imaging system.
DISCUSSION
Our previous studies characterized a fission yeast SREBP pathway that functions as a sterol-dependent oxygen-sensing system and suggested that fission yeast Scp1 is a functional model for mammalian Scap (14). Like hamster Scap, fission yeast Scp1 regulates the activation of Sre1 in response to changes in cellular sterols. In addition, conserved mutations Y247C (Y298C), L264F (L315F), and D392N (D443N) in the SSDs of Scp1 and Scap render both proteins insensitive to sterols and cause constitutive activation of SREBP (12, 16). Here, we utilized a combination of yeast genetics and site-directed mutagenesis to identify additional residues important for the function of the Scp1 and Scap SSDs. No codon bias was observed in the mutant library production, as discussed in Experimental Procedures. Thus, this approach allowed us to sample every possible amino acid substitution at 50 different conserved amino acids in the SSD of Scp1. By sequencing only a small fraction of the positive transformants, we identified eighty-nine unique mutations at 23 different residues in the SSD of Scp1 that activated the 3xSRE-ura4+ reporter (Table 1). We retested the most-common mutation at each of the 23 amino acids and found that all twenty-three caused constitutive activation of Sre1 even in the presence of oxygen (Table 1 and Fig. 4). Three of these mutations were located at known functional residues Y247, L264, and D392 but represented new substitutions at these amino acids. The other twenty mutations were at new locations in the SSD of Scp1 and predominantly clustered around the three previously known Sre1-activating mutation sites in transmembrane segments 2, 3, and 6 (Fig. 1). Our results indicate that these regions are critical for Scp1 function and support previous data that suggest that Scp1 is required for sterol sensing and regulated Sre1 activation in fission yeast.
Because we only sampled amino acids in Scp1 that were identical in hamster Scap, we were able to test all twenty-three homologous Sre1-activating mutations for their ability to regulate mammalian SREBP activation. Surprisingly, sixteen of the mutations had no detectable effect on the function of Scap, including Y298P, L315P, and D443E, which were located at sites of previously described sterol-resistant mutations (Table 1) (12). Two mutations, E448P and L449W, showed reduced activation of SREBP in the absence of Insig. These residues are located in the MELADL sequence of Scap and are required for incorporation into COPII transport vesicles (6, 7). Thus, it is not surprising that these mutations partially inhibited SREBP activation. However, the homologous mutations in fission yeast Scp1 caused Sre1 activation, which suggests that this region of Scp1 may not serve the same function as MELADL in Scap.
Of the other five Scap mutations tested, only one exhibited an Insig-resistant SREBP-activating phenotype similar to Scap D443N (Fig. 5). This mutation, V439G, is positioned near the end of the 6th transmembrane segment, close to D443, and may affect Scap function in a fashion similar to D443N. The remaining four Scap mutations, F296R, L431R, F436R, and L440P, were defective in activating SREBP-2 cleavage, even in the absence of sterols (Fig. 6). Consistent with this finding, these Scap mutants constitutively assumed an ER-retained conformation, as assayed by trypsin digestion (Fig. 7). These mutations resembled a recently characterized Insig-sensitive mutation in Scap (D428A) that binds Insig better than Scap, exhibits reduced SREBP activation, and favors an ER-retained conformation (13). However, unlike D428A, these new mutations showed reduced Insig-1 binding, compared with Scap (Fig. 6). One explanation is that these amino acid substitutions in Scap are too disruptive and cause retention of Scap by an unknown quality control mechanism. However, these mutant proteins stabilize SREBP-2 precursor and produce a protected fragment of the correct size after trypsin digestion, suggesting that each Scap mutant is folded correctly. Future experiments are required to address the function of these residues in the SSD of Scap and to understand why mutations at these amino acids in fission yeast Scp1 cause constitutive Sre1 activation, but fail to activate mammalian SREBP.
Prior to this study, we hypothesized that Scp1-dependent sterol regulation of Sre1 occurred through a mechanism similar to that seen in mammals. However, in light of our current results, it is clear that significant differences exist between Scp1 and Scap and in how these proteins mediate sterol-regulated activation of Sre1 and SREBP. We identified twenty-three mutations in Scp1 that activated cleavage of Sre1, yet only one of these mutations caused a similar phenotype in mammals. The reasons for this are unclear. However, the sterol signal for SREBP activation differs between fission yeast (4-methyl sterols) and mammals (cholesterol and 25-hydroxycholesterol), and regulation of Sre1 activation does not require the Insig homolog Ins1 (9, 11). These differences may alter the effects of different amino acid substitutions in the SSDs of Scap and Scp1. Alternatively, the differences we observed may suggest that Scp1-dependent sterol regulation of Sre1 occurs through mechanisms other than regulated ER retention. Sequence homologs of the Site-1 and Site-2 SREBP-activating proteases have not been identified in fission yeast, and the mechanism and subcellular location of Sre1 proteolysis are unknown.
The fact that many different amino acid substitutions at a single residue caused constitutive Sre1 activation is consistent with the mutations acting as loss-of-function, rather than gain-of-function, mutations. Despite this, the Scp1 mutations actually led to increased Sre1 cleavage. Interestingly, a large number of the eighty-nine Scp1 mutations replaced a hydrophobic residue in a transmembrane segment with proline or arginine, amino acids not usually conducive to transmembrane segment formation (Table 1). In addition, the twenty-three Scp1 mutants we identified showed wide variation in expression level (Fig. 4 and data not shown), suggesting that a number of the mutant proteins were unstable. This variation in expression was not observed in mammalian cells, where all Scap mutants showed expression levels similar to those in Scap (Figs. 5, 6 and data not shown). These findings are consistent with a model in which Scp1 loss of function, or perhaps misfolding, leads to activation of Sre1. Further experiments are required to fully understand both the mechanism of Scp1-dependent sterol-regulated Sre1 activation in fission yeast and how these twenty-three mutations alter Scp1 activity. This new collection of Sre1-activating Scp1 mutants will serve as a useful tool for addressing these questions.
In addition to the implications our results have for Scp1 and Scap, our findings may provide insight into the function of other mammalian SSD-containing proteins, such as HMG-CoA reductase (HMGR), Niemann-Pick type C1, and Patched (31). Several of the Scp1 residues at which Sre1-activating mutations were identified in this study are conserved in the SSDs of these three proteins. This possibility is especially intriguing for HMGR, an SSD-containing protein whose degradation is regulated by lanosterol, a 4-methyl sterol intermediate that also mediates Scp1-dependent Sre1 activation in fission yeast (16, 32). Future studies are required to investigate these interesting possibilities.
Acknowledgments
From UT-Southwestern Medical Center at Dallas, the authors thank Wayne Lai and Ling-Chu Hung in the Department of Pathology for generating monoclonal antibodies to Scp1, and Michael Brown and Joseph Goldstein for their sharing of reagents. The authors thank members of the Espenshade lab for experimental advice and for reviewing the manuscript.
Abbreviations
ALLN, N-acetyl-leucyl-leucyl-norleucinal
CaMV, cauliflower mosaic virus
CHO, Chinese hamster ovary
CMV, cytomegalovirus
EMM, Edinburgh minimal medium
ER, endoplasmic reticulum
HPCD, hydroxypropyl-β-cyclodextrin
Insig, insulin-induced gene
HSV, herpes simplex virus
LPDS, lipoprotein-deficient serum
Scap, SREBP cleavage-activating protein
SRE, Sre1-regulatory element
SREBP, sterol-regulatory element binding protein
SSD, sterol-sensing domain
TK, thymidine kinase
Published, JLR Papers in Press, May 23, 2008.
Footnotes
This work was supported by the National Institutes of Health, Grant HL-077588. A.L.H. is a recipient of the American Heart Association Predoctoral Fellowship 0515394U. P.J.E. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
References
- 1.Goldstein J. L., R. A. DeBose-Boyd, and M. S. Brown. 2006. Protein sensors for membrane sterols. Cell. 124 35–46. [DOI] [PubMed] [Google Scholar]
- 2.Espenshade P. J. 2006. SREBPs: sterol-regulated transcription factors. J. Cell Sci. 119 973–976. [DOI] [PubMed] [Google Scholar]
- 3.Horton J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua X. X., A. Nohturfft, J. L. Goldstein, and M. S. Brown. 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 87 415–426. [DOI] [PubMed] [Google Scholar]
- 5.Yang T., P. J. Espenshade, M. E. Wright, D. Yabe, Y. Gong, R. Aebersold, J. L. Goldstein, and M. S. Brown. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110 489–500. [DOI] [PubMed] [Google Scholar]
- 6.Sun L. P., L. Li, J. L. Goldstein, and M. S. Brown. 2005. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280 26483–26490. [DOI] [PubMed] [Google Scholar]
- 7.Sun L. P., J. Seemann, J. L. Goldstein, and M. S. Brown. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc. Natl. Acad. Sci. USA. 104 6519–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawson R. B. 2003. The SREBP pathway—insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4 631–640. [DOI] [PubMed] [Google Scholar]
- 9.Espenshade P. J., and A. L. Hughes. 2007. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41 401–427. [DOI] [PubMed] [Google Scholar]
- 10.Radhakrishnan A., L. P. Sun, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2004. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell. 15 259–268. [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan A., Y. Ikeda, H. J. Kwon, M. S. Brown, and J. L. Goldstein. 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA. 104 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yabe D., Z. P. Xia, C. M. Adams, and R. B. Rawson. 2002. Three mutations in sterol-sensing domain of SCAP block interaction with Insig and render SREBP cleavage insensitive to sterols. Proc. Natl. Acad. Sci. USA. 99 16672–16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feramisco J. D., A. Radhakrishnan, Y. Ikeda, J. Reitz, M. S. Brown, and J. L. Goldstein. 2005. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl. Acad. Sci. USA. 102 3242–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes A. L., B. L. Todd, and P. J. Espenshade. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 120 831–842. [DOI] [PubMed] [Google Scholar]
- 15.Todd B. L., E. V. Stewart, J. S. Burg, A. L. Hughes, and P. J. Espenshade. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes A. L., C. S. Lee, C. M. Bien, and P. J. Espenshade. 2007. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J. Biol. Chem. 282 24388–24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai J., A. Nohturfft, D. Cheng, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272 20213–20221. [DOI] [PubMed] [Google Scholar]
- 18.Herz J., R. C. Kowal, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1990. Low density lipoprotein receptor-related protein mediates endocytosis of monoclonal antibodies in cultured-cells and rabbit liver. J. Biol. Chem. 265 21355–21362. [PubMed] [Google Scholar]
- 19.Bahler J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14 943–951. [DOI] [PubMed] [Google Scholar]
- 20.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Sehgal A., C. Y. Lee, and P. J. Espenshade. 2007. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS. Genet. 3 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular Cloning: A Laboratory Manual. 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Hua X. X., J. Sakai, M. S. Brown, and J. L. Goldstein. 1996. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271 10379–10384. [DOI] [PubMed] [Google Scholar]
- 24.Forsburg S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21 2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawson R. B., R. DeBose-Boyd, J. L. Goldstein, and M. S. Brown. 1999. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274 28549–28556. [DOI] [PubMed] [Google Scholar]
- 26.Nohturfft A., D. Yabe, J. L. Goldstein, M. S. Brown, and P. J. Espenshade. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102 315–323. [DOI] [PubMed] [Google Scholar]
- 27.Brown A. J., L. P. Sun, J. D. Feramisco, M. S. Brown, and J. L. Goldstein. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 10 237–245. [DOI] [PubMed] [Google Scholar]
- 28.Wang X. D., R. Sato, M. S. Brown, X. X. Hua, and J. L. Goldstein. 1994. Srebp-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 77 53–62. [DOI] [PubMed] [Google Scholar]
- 29.Hughes B. T., and P. J. Espenshade. 2008. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 27 1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nohturfft A., R. A. DeBose-Boyd, S. Scheek, J. L. Goldstein, and M. S. Brown. 1999. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. USA. 96 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewen C. J., and T. P. Levine. 2002. Cholesterol homeostasis: not until the SCAP lady INSIGs. Curr. Biol. 12 R779–R781. [DOI] [PubMed] [Google Scholar]
- 32.Song B. L., N. B. Javitt, and R. A. DeBose-Boyd. 2005. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1 179–189. [DOI] [PubMed] [Google Scholar]