Abstract
The TGFβ1/Smad pathway plays a critical role in cholestasis and liver fibrosis. Previous studies show that TGFβ1, TNFα, and insulin inhibit cholesterol 7α-hydroxylase (CYP7A1) gene transcription and bile acid synthesis in human hepatocytes. In this study, we investigated insulin, TGFβ1, and TNFα regulation of rat Cyp7a1 gene transcription. In contrast to inhibition of human CYP7A1 gene transcription, TGFβ1 stimulates rat Cyp7a1 reporter activity. Smad3, FoxO1, and HNF4α synergistically stimulated rat Cyp7a1 gene transcription. Mutations of the Smad3, FoxO1, or HNF4α binding site attenuated the rat Cyp7a1 promoter activity. Furthermore, TNFα and cJun attenuated TGFβ1 stimulation of rat Cyp7a1. Insulin or adenovirus-mediated expression of constitutively active AKT1 inhibited FoxO1 and Smad3 synergy. In streptozotocin-induced diabetic rats, Cyp7a1 mRNA expression levels were induced and insulin attenuated CYP7A1 mRNA levels. Chromatin immunoprecipitation assay showed that FoxO1 binding to Cyp7a1 chromatin was increased in diabetic rat livers and insulin reduced FoxO1 binding. These results suggest a mechanistic basis for induction of Cyp7a1 activity and bile acid synthesis in cholestatic rats and in diabetic rats. The crosstalk of insulin, TGFβ and TNFα signaling pathways may regulate bile acid synthesis and lipid homeostasis in diabetes, fatty liver disease, and liver fibrosis.
Keywords: CYP7A1, bile acid synthesis, liver fibrosis, nuclear receptors, Smad, FoxO1, TGFβ, cholestasis
Bile acids are synthesized from cholesterol in the liver, stored in the gallbladder, and secreted into the intestine to facilitate the absorption of dietary lipids and fat-soluble vitamins and disposal of toxic metabolites, drugs, and xenobiotics (1, 2). Bile acids are quantitatively reabsorbed in the intestine and transported back to the liver via portal circulation. Hepatic bile acid synthesis is tightly controlled by virtue of high cytotoxicity of bile acids. During cholestatic liver injury, hepatic nonparenchymal cells including Kupffer cells (hepatic macrophages) and hepatic stellate cells (HSC) release proinflammatory cytokines (IL-1β, TNFα) and growth factors (hepatic growth factor, TGFβ), respectively, to hepatocytes. These cytokines have been shown to inhibit bile acid synthesis through transcriptional repression of the gene encoding cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis in the liver (3–5). Inhibition of bile acid synthesis and stimulation of bile secretion are adaptive responses to protect hepatocytes from injury. Paradoxically, Cyp7a1 activity and mRNA expression are stimulated in bile duct-ligated (BDL) rat and mouse livers (6–8).
TGFβ1 is increased in the livers of BDL rats and mice, and in patients with liver fibrosis (7, 9). TGFβ1 is a potent profibrogenic factor secreted by activated HSCs to stimulate HSC transdifferentiation and proliferation, extracellular matrix expression, hepatocyte apoptosis, and liver fibrosis (10, 11). Liver fibrosis is associated with nonalcoholic fatty liver disease and diabetes (12, 13). Transcription factors Smad2 and Smad3 are major down stream mediators of TGFβ1 signaling (11, 14). TGFβ1 binds to and activates cell surface TGFβ receptor type I (TβRI) and type II (TβRII) that possess intracellular serine/threonine kinase activity. TGFβ1-activated TβRI then recruits and phosphorylates Smad2 and Smad3 at a conserved C-terminal SSXS motif. Phosphorylation of Smads by TβRI increases Smad protein nuclear localization and its transactivating activity. Smad3 binds GTCT and AGAC sequences in its target genes. In contrast, Smad2 does not bind directly to DNA because the amino acids that are responsible for DNA binding are displaced in Smad2.
Recently, several insulin-regulated genes, FoxO1, FoxO3a and FoxO4, have been identified as interacting partners of Smad3 (15). Smad3 and FoxO1 bind to the cyclin-dependent kinase inhibitor p21 gene promoter and synergistically stimulate its transcription in neuroepithelial and glioblastoma cell proliferation. A subsequent study has identified many TGFβ1 target genes that are synchronously regulated by FoxO-Smad in response to diverse cellular activities including cell cycle control, inflammation, and stress in human epithelial cells (16). FoxO1 plays a critical role in mediating insulin signaling (17). Protein kinase B (AKT/PKB) phosphorylation of FoxO1 results in FoxO1 nuclear exclusion and inhibition of FoxO1-regulated genes. It is thought that FoxO1 converges two antagonistic pathways, PI3K/AKT and TGFβ1/Smad (18).
In this study, we investigated Smad3 regulation of the rat Cyp7a1 gene, and its possible implications in bile acid homeostasis during liver fibrosis and diabetes. Our results show that TGFβ1-activated Smad3 stimulates the rat Cyp7a1 gene. Smad3 directly binds to the rat Cyp7a1 promoter and acts in synergy with FoxO1 and HNF4α to induce rat Cyp7a1 promoter activity. Smad3 and FoxO1 may crosstalk and synchronize insulin inhibition and TGFβ1 activation of Cyp7a1 in diabetes and liver fibrosis. On the other hand, TNFα signal antagonizes TGFβ1 activation of CYP7A1 gene transcription. These three cell-signaling pathways may crosstalk in response to extracellular signals to regulate bile acid synthesis and maintain lipid homeostasis.
MATERIALS AND METHODS
Cell culture
The human hepatoblastoma cell line, HepG2, was purchased from American Type Culture Collection (Manassas, VA). The cells were cultured in Dulbecco's modified Eagle medium and F-12 (Sigma, St. Louis, MO) supplemented with 100U/ml penicillin G/streptomycin sulfate (Mediatech, Herndon, VA) and 10% (v/v) heat inactivated fetal bovine serum (Irvine Scientific, Santa Ana, CA).
Reporters and expression plasmids
Human and rat CYP7A1/Luc reporters were constructed as previously described (19). Expression plasmids pFLAG-CMV2-Smad2, pFLAG-CMV2-Smad3, and pFLAG-CMV2-Smad4 were kindly provided by Dr. David Jones (University of Utah, Salt Lake City, UT). The construction of the expression plasmid for HNF4α (pCMV-HNF4α) was previously described (20). The β-galactosidase expression plasmid (pCMV-β-gal), and the mammalian expression vector pcDNA3.1 were obtained from Clonetech (Palo Alto, CA). The reporter vector pGL2-Basic was purchased from Promega (Madison, WI). Expression plasmid for pCMV-rHNF3β (FoxA2) was kindly provided by Robert H. Costa (University of Illinois at Chicago, IL). pCMV5-FoxO1 was kindly provided by Dr. D. Accili (Columbia University, NY). cJun expression plasmid was originally provided by Dr. Inder Verma (The Salk Institute, San Diego, CA) and subcloned into pcDNA3.
Transient transfection assay
Luciferase reporters and expression plasmids were transfected into HepG2 cells using Lipofectamine 2000 reagent (Life Technologies, Inc., Gaithersbrug, MD) following manufacturer's instructions. Luciferase reporter activities were assayed and expressed as relative luciferase units divided by β-galactosidase activity as described previously. Assays were performed in triplicates and expressed as mean ± SD.
Recombinant adenovirus
Recombinant Adenovirus Ad-EGFP was obtained from Dr. Li Wang (Univ. of Utah, Salt Lake City, UT), Ad-Akt1 expressing myristoylated and constitutively active mutant form of Akt1 was purchased from Vector Biolabs (Philadelphia, PA). Recombinant adenoviruses were amplified in HEK293A cells and purified with Adeno-X Virus mini purification kit (BD Biosciences, San Jose, CA). Virus titer was determined by Adeno-X rapid titer kit (BD Biosciences). Expression of Akt1 in HepG2 cells were confirmed by Western Blot analysis using anti-Akt1 antibody (Cell Signaling Technology, Danvers, MA).
Electrophoretic mobility shift assay
Smad3 was synthesized in vitro using the transcription/translation system (Promega, WI) programmed with the Smad3 expression plasmid according to the manufacturer instruction. Synthetic oligonucleotides of complementary strands were labeled with (α-32P) dCTP and incubated with in vitro translated proteins (5 μl) as described previously. Gels were dried, autoradiographed by PhosphorImaging. The probes (Smad binding site underlined) used in the experiments were: Tinman 4 RLL: gatcGGAGTCTAGACCGGC; rat Cyp7a1 Smad3: gatcAAAGTCTAGTCAGACCCAC.
Site-directed mutagenesis
A PCR-based Quick-Change Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used for mutation of the reporter construct. Mutations of the Smad3 binding site were introduced into complementary oligonucleotides, which were used as PCR primers. Thermal cycling was performed with the primers and wild-type reporter constructs as templates according to the manufacturer's instruction. The sequence of the mutation was confirmed by DNA sequencing. One of the two complementary PCR primers used was: -34 GCACATATAAAGTtTAtTCAaACCCACTGTTTCG+1 (lower cases indicate mutations in Smad 3 site, which is underlined).
RNA isolation and quantitative real-time PCR
Rat livers were kindly provided by Dr. Meszaros (NEOUCOM, Rootstown, OH). Briefly, 8-week-old male Sprague-Dawley rats were randomly divided into two groups. Rats were given either vehicle or a single intraperitoneal. injection of streptozotocin (STZ) (60 mg/Kg body weight). One week after STZ injection, hyperglycemia was confirmed in diabetic group by determining blood glucose level of above 300 mg/dl. Some of these diabetic rats were then given daily insulin injection (0.5 mg/kg) subcutaneously for 6 weeks. The average blood glucose concentrations (mg/dl) in these three treatment groups were: control: 100 ± 2.05; STZ: 402.6 ± 54.6; STZ + insulin: 285.8 ± 58.0. Rats were then sacrificed. Liver tissues were homogenized and RNA isolation, reverse transcription reactions and real-time PCR were performed as described previously (20). All primers/probe used were TaqMan Gene Expression Assays (Applied Biosystems). Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2 -ΔΔCt.
Chromatin immunoprecipitation assay
Rat livers (∼1 g/rat, n = 5) from the same group were pooled and homogenized. Cell nuclei were purified and cross-linked in formaldehyde. Chromatin immunoprecipitation (ChIP) assays were performed with ChIP assay kit (Upstate Biotech, Charlottesville, VA) following manufacturer's instruction. Antibodies used were: Anti-HNF4α (Santa Cruz biotechnology, Santa Cruz, CA); Anti-Smad3 (Santa Cruz Biotechnology); Anti-FoxO1 (Cell Signaling Technology, Danvers, MA). PCR primers F: -579 GCTGTGGCTTCCTGGTAGATG and R: +2 CCGAAACAGTGGGTCTGACTAG were used to amplify a 581 bp rat Cyp7a1 promoter region that contains FoxO1, HNF4α, and Smad3 binding sites.
Statistical Analysis
Real-time PCR results of rat liver mRNA expression are expressed as mean ± SEM. Reporter assays were performed in triplicates and expressed as mean ± SD. Student's t-test was used for statistical analysis. A P value of < 0.05 was considered to be statistically significant difference between treated and untreated control.
RESULTS
Smad3 stimulates rat Cyp7a1 reporter activity
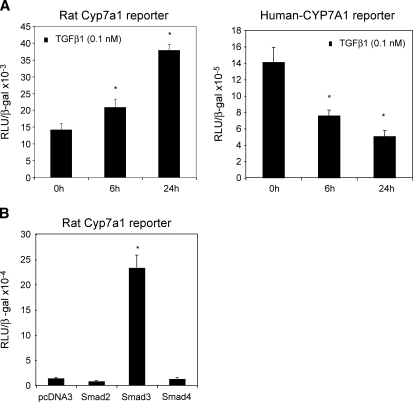
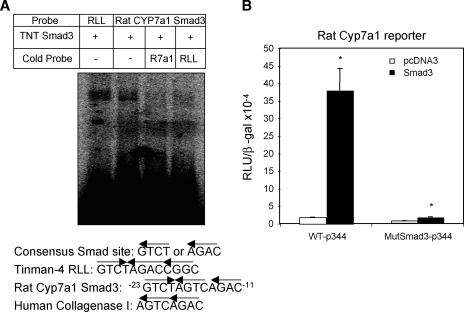
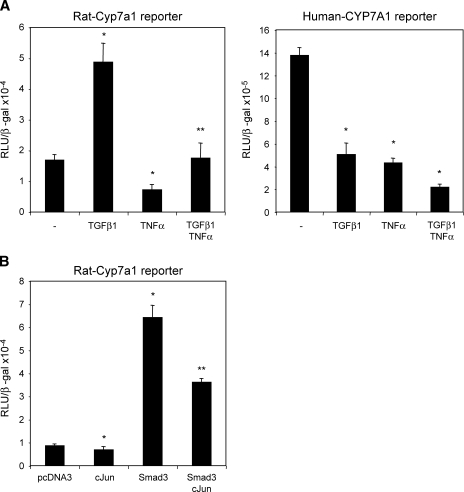
TGFβ1 level is increased in cholestatic liver injury. To understand the mechanism of previously reported induction of CYP7A1 activity and bile acid synthesis in cholestatic rat model (6, 7), we studied the effect of TGFβ1 on rat Cyp7a1 reporter activity. TGFβ1 time-dependently stimulated rat Cyp7a1 promoter activity (Fig. 1A, left panel). In contrast, TGFβ1 strongly inhibited human CYP7A1 reporter activity under the same experimental condition (Fig. 1A, right panel) (5). We then studied the effect of cotransfection of Smad2, Smad3, and Smad 4 on rat CYP7a1 reporter activity. Fig. 1B shows that only Smad3 strongly stimulated the rat Cyp7a1 promoter/reporter activity by approximately 16-fold. Smad3 stimulated rat Cyp7a1 reporter activity in a dose-dependent manner (data not shown). An analysis of the rat Cyp7a1 proximal promoter sequence identified a potential Smad3 binding sequence, -18 AGTCAGAC−11, which is identical to the Smad3 binding site in the human collagenase-I promoter (Fig. 2A) (21). This sequence is not conserved in the human or mouse CYP7A1 promoter. Electrophoretic mobility shift assay shows that in vitro synthesized Smad3 binds to this sequence, and the binding is competed out by unlabeled Smad3 probe or a consensus Smad3 probe from the Tinman-4 gene (Fig. 2A). The Smad3 binding activity to DNA is very weak as reported previously (22). Thus, to further confirm the functional role of this Smad3 binding site, we mutated this Smad3 binding site in the rat Cyp7a1 reporter p-344 and performed reporter assays. Fig. 2B shows that the mutant Cyp7a1 promoter/reporter had much lower basal activity, which was not stimulated by Smad3. These results suggest that Smad3 binds to the rat CYP7A1 promoter and is required for basal transcription of the rat Cyp7a1 gene. These results provide a molecular basis for reported induction of Cyp7a1 activity in BDL rats (6, 7).
Fig. 1.
TGFβ1 and Smad3 stimulate rat Cyp7A1 reporter activity. A: Rat Cyp7A1 reporter construct p3644 (0.2 μg, left panel) or human CYP7A1 reporter construct ph-1887 (0.2 μg, right panel) was transfected into HepG2 cells. Twenty-four h after transfection, cells were treated with 0.1 nM TGFβ1 for 6 or 24 h and harvested for luciferase activity assays. * Statistically significant difference (P < 0.05), TGFβ1-treated vs. nontreated control. B: Rat Cyp7A1 reporter construct p3644 (0.2 μg) was cotransfected with 0.1 μg expression plasmids Smad2, Smad3, or Smad4 into HepG2 cells. Cells were harvested for reporter assays 48 h after transfection. All luciferase activities were normalized to β-gal activity and expressed as relative Luciferase unit (RLU)/β-gal activity. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), Smads vs. pcDNA3 empty vector.
Fig. 2.
Smad3 binds to rat Cyp7A1 proximal promoter. A: P32 -labeled probes and in vitro translated Smad3 protein were used in electrophoretic mobility shift assay. A probe containing Smad3 binding site from Tinman-4 gene was used as a positive control. Excess unlabeled probes (50×) were used in competition assays. B: Wild-type or mutant rat Cyp7a1 reporter construct containing mutations in Smad3 binding site were cotransfected with 0.1 μg Smad3 expression plasmid into HepG2 cells. Cells were harvested for reporter activity assays 48 h after transfection. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), Smads vs. pcDNA3 empty vector.
Smad3 and FoxO1 synergistically stimulate rat Cyp7a1 promoter/ reporter activity
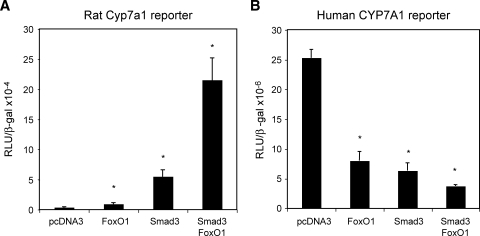
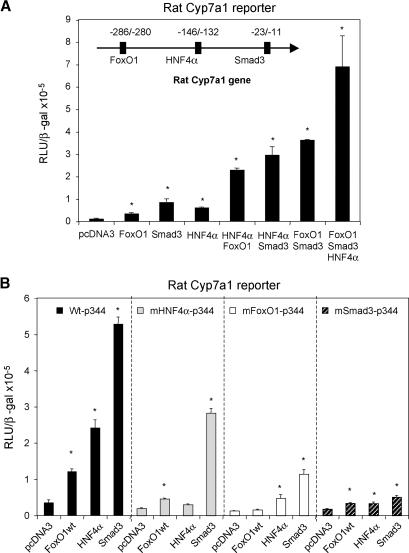
Smad3 is known to interact with other DNA binding factors to exert its gene specific transcriptional regulation (22). We have previously shown that FoxO1 binds to an insulin response element in the rat Cyp7a1 promoter and stimulates rat Cyp7a1 promoter activity (23). Because synergy between Smad3 and FoxO1 in transcriptional regulation has been established in a group of genes that contain both Smad3 and FoxO1 binding sites (15, 16), we therefore tested if Smad3 and FoxO1 could regulate rat Cyp7a1 synergistically. Fig. 3A shows that FoxO1 alone stimulates the rat Cyp7a1 reporter activity by ∼4-fold, while Smad3 alone stimulates the rat Cyp7a1 reporter by ∼15-fold. When both Smad3 and FoxO1 were cotransfected, the rat Cyp7a1 reporter activity was induced by ∼50-fold. These results indicate that Smad3 and FoxO1 may act in synergy to stimulate rat Cyp7a1 gene expression. In contrast, human CYP7A1 promoter/reporter activity was strongly inhibited by cotransfection of Smad3 and FoxO1 (Fig. 3B). As a negative control, Smad3 did not show synergy with FoxA2 (data not shown), which was also shown to bind the insulin response element in the rat Cyp7a1 promoter (23). Because HNF4α is a critical activator of both human and rat Cyp7a1, we next tested if HNF4α had a synergistic effect on rat Cyp7a1 stimulated by Smad3 and FoxO1. Interestingly, reporter assays revealed synergistic stimulation of the rat Cyp7a1 reporter by HNF4α and FoxO1, and HNF4α and Smad3 (Fig. 4A). When three transcriptional factors were cotransfected together, the rat Cyp7a1 reporter was maximally stimulated. We next mutated HNF4α, FoxO1, or Smad3 binding site in the rat Cyp7a1 reporter construct and performed reporter assays to evaluate the relative importance of each of these three transcriptional factors in the rat Cyp7a1 gene expression. As shown in Fig. 4B, Smad3 showed a stronger stimulatory effect on the wild-type rat Cyp7a1 reporter than HNF4α or FoxO1. Interestingly, mutation of the HNF4α binding site not only abolished HNF4α stimulation of the reporter activity, but also attenuated the stimulating effect of FoxO1 and HNF4α. Similarly, mutation of the FoxO1 binding site significantly reduced HNF4α and Smad3 stimulation of the reporter activity. When the Smad3 binding site was mutated, the HNF4α and FoxO1 stimulatory effects were almost abolished. These results indicate that the synergy between FoxO1, Smad3, and HNF4α are critical in rat Cyp7a1 gene expression, and Smad 3 plays a central role in coordinating transcriptional regulation of the rat Cyp7a1 gene. Disruption of such a transcriptional complex by inhibiting any of these transcriptional factors will result in strong repression of rat Cyp7a1 gene expression.
Fig. 3.
Smad3 and FoxO1 synergistically stimulate rat Cyp7A1 reporter activity. Rat Cyp7A1 reporter construct p344 (0.2 μg) (A) or human CYP7A1 reporter construct ph-1887 (0.2 μg) (B) were cotransfected with 0.1 μg expression plasmids Smad3 and/or FoxO1 in HepG2 cells as indicated. Cells were harvested for reporter assays 48 h after transfection. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), FoxO1 and/or Smads vs. pcDNA3.
Fig. 4.
Rat Cyp7A1 is stimulated by Smad3/FoxO1/HNF4α. A: Rat Cyp7A1 reporter p344 (0.2 μg) was cotransfected with Smad3, FoxO1, and/or HNF4α in HepG2 cells as indicated for reporter assays. The FoxO1, HNF4α, and Smad3 binding sites in rat Cyp7a1 gene promoter are illustrated. B: Wild-type or rat Cyp7A1 reporter constructs containing mutations in Smad3, FoxO1, or HNF4α binding site (0.2 μg) were cotransfected with FoxO1, Smad3, or HNF4α. Cells were harvested for reporter assays 48 h after transfection. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), FoxO1, Smads, and/or HNF4α vs. pcDNA3 empty plasmid.
TNFα counteracts Smad3 stimulation of rat Cyp7a1 reporter activity
Since proinflammatory cytokines (TNFα and IL-1β) are known to antagonize TGFβ1-induced gene expression (24), and TNFα and IL-1β rapidly inhibit CYP7A1 gene expression (3, 25, 26), we studied the effect of TNFα on TGFβ1 regulation of Cyp7a1 reporter activity. TGFβ1 stimulated, whereas TNFα (50 ng/ml) repressed, rat Cyp7a1 reporter activity as expected (Fig. 5A, left panel). Addition of TNFα attenuated TGFβ1 stimulation of rat Cyp7a1 reporter activity. TGFβ1 or TNFα repressed human Cyp7a1 reporter activity as expected (Fig. 5A, right panel). Combination of TNFα and TGFβ1 strongly repressed human CYP7A1 reporter activity. IL-1β had the same effect as TNFα (data not shown). Studies have shown that TNFα activates cJun, which interacts with and inhibits Smad-dependent gene expression (24). Fig. 5B shows that cotransfection of cJun partially repressed Smad3 stimulation of rat Cyp7a1 reporter activity. These results showed that TNFα antagonized TGFβ1 stimulatory effect on rat Cyp7a1 gene transcription, and this effect is at least in part mediated by TNFα induction of c-Jun-Smad3 interaction.
Fig. 5.
TNFα antagonizes TGFβ1 stimulation of rat Cyp7A1 reporter activity. A: Rat Cyp7A1 reporter construct p344 (0.2 μg, left panel) or human CYP7A1 reporter construct ph-1887 (0.2 μg, right panel) was transfected into HepG2 cells. Twenty-four h after transfection, cells were treated with 0.1 nM TGFβ1 or 50 ng/ml TNFα as indicated for 24 h and harvested for luciferase activity assays. * Statistically significant difference (P < 0.05), TGFβ1 and/or TNFα vs. untreated control. ** Statistically significant difference (P < 0.05), TGFβ + TNFα vs. TNFα. B: Rat Cyp7A1 reporter construct p-344 (0.2 μg) was cotransfected with 0.1 μg c-Jun or Smad3 into HepG2 cells. Cells were harvested for reporter assays 48 h after transfection. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), cJun and/or Smad3 vs. pcDNA3 empty plasmid. ** Statistically significant difference (P < 0.05), Smad3 + cJun vs. Smad3.
Insulin blocks FoxO1 and Smad3 stimulation of rat Cyp7a1
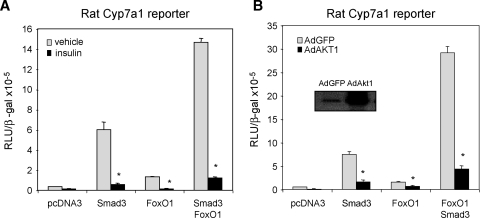
It has been reported for a long time that insulin inhibits Cyp7a1 expression and bile acid synthesis in rat livers (27). However, the underlying mechanism of insulin inhibition of Cyp7a1 activity is still not fully understood. Recent studies show that insulin signaling activates AKT/PKB, which phosphorylates and inhibits FoxO1 activity (17). We therefore tested if insulin inhibition of FoxO1 activity could impair FoxO1/Smad3/HNF4α synergistic stimulation of rat Cyp7a1 and thus result in inhibition of rat Cyp7a1 gene transcription. Reporter assays show that insulin treatment (100 nM) or adenovirus-mediated gene transfer of a constitutively active form of AKT1 inhibited basal rat Cyp7a1 reporter activity and prevented FoxO1 and Smad3 stimulation of the rat Cyp7a1 reporter activity (Fig. 6A and 6B).
Fig. 6.
Insulin inhibits FoxO1 and Smad3 stimulation of rat Cyp7A1 gene expression. A: Rat Cyp7A1 reporter p344 (0.2 μg) was cotransfected with Smad3 and/or FoxO1 into HepG2 cells. Twenty-four h after transfection, cells were either treated with insulin (100nM) for 24 h or harvested for luciferase activity assays. * Statistically significant difference (P < 0.05), insulin-treated vs. vehicle control. B: Rat Cyp7a1 reporter p344, Smad3, and FoxO1 were cotransfected as in A. Cells were infected with adenovirus (MOI = 10) expressing either GFP or constitutively active Akt1 for 48 h. The inset figure indicates increased Akt1 expression by western blot analysis. Cells were harvested for reporter assays 48 h after transfection. Experiments are performed in triplicates and results are expressed as mean ± SD. * Statistically significant difference (P < 0.05), AdAKT1-transfected vs. AdGFP control.
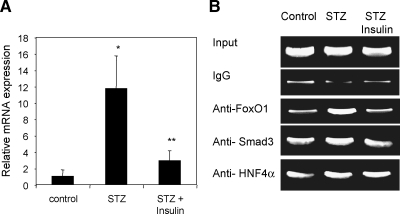
We then assayed Cyp7a1 mRNA expression levels in STZ-induced diabetic rats. Cyp7a1 mRNA expression was strongly induced in the livers of STZ-induced type-I diabetic rats. Daily administration of insulin to the diabetic rats inhibited the Cyp7a1 mRNA expression in rat livers (Fig. 7A). These results are consistent with study reported 25 years ago that bile acid synthesis and cholesterol 7α-hydroxylase activity were induced in diabetic rats (27). ChIP assays showed that FoxO1 binding to the Cyp7a1 promoter was increased in livers of STZ-treated rats, while insulin administration reduced FoxO1 binding to the control level (Fig. 7B). Smad3 or HNF4α binding to Cyp7a1 chromatin was not altered significantly in each group of rats. These results suggest that insulin may inhibit the Cyp7a1 gene in rat livers by inhibiting FoxO1 activity and thus reduces its synergistic activation with Smad3 and HNF4α on Cyp7a1 gene transcription.
Fig. 7.
FoxO1 mediates insulin inhibition of Cyp7a1 in rat livers. A: Real-time PCR assays of Cyp7a1 mRNA expression in the livers of rats of control, streptozotocin (STZ), or STZ plus insulin group. Results are expressed as mean ± SEM. Student's t-test was used to compare two groups. A P value < 0.05 was considered to be significant. * Statistically significant difference (P < 0.05, n = 5), STZ-treated vs. vehicle control; ** statistically significant difference (P < 0.05, n = 5), STZ + insulin vs. STZ. B: Rat liver nuclei were used in chromatin immunoprecipitation (ChIP) assay. ChIP assays were performed as described in Materials and Methods. Ten percent lysate was used as “input”. Nonimmuno IgG was used as background control. PCR amplified DNA fragments were analyzed on agarose gel.
DISCUSSION
This study shows that Smad3 is a major activator of rat Cyp7a1 gene transcription. Smad3 binds to a consensus sequence (nt -23 to -11) in the core promoter and may be essential for basal transcription of the rat Cyp7a1 gene. Smad3 interacts with HNF4α and FoxO1, which bind to the previously identified bile acid response element-II (-146 to -132), and an upstream insulin response element (-286 to -280), respectively (3). The observed strong synergy of Smad3, FoxO1, and HNF4α may explain much higher Cyp7a1 activity and rate of bile acid synthesis in rats than other species. The rat Cyp7a1 may be a new member of the “FoxO-Smad synexpression group” recently identified (16). The synchronized regulation of rat Cyp7a1 gene expression by insulin and TGFβ1/Smad pathways may provide a balanced and rapid adaptive response to diverse cellular signals in hepatocytes. The insulin and TGFβ1/Smad signaling pathways may antagonize each other to control the rate of bile acid synthesis during liver regeneration, proliferation, and injury. In the human CYP7A1 gene promoter, Smad3 and FoxO1 sites are not conserved, and Smad3 and FoxO1 interact with HNF4α as a corepressor that inhibits human CYP7A1 and bile acid synthesis as a protection against cholestatic liver injury (5, 23).
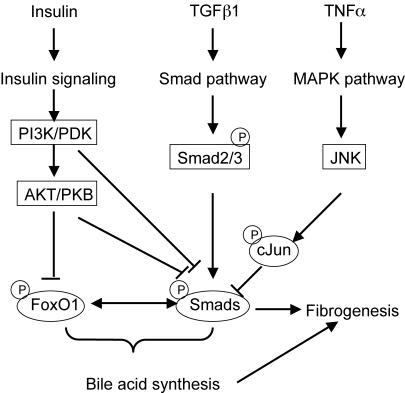
It is well established that FoxO1 activity is inhibited by insulin signaling via a mechanism involving FoxO1 phosphorylation and nuclear exclusion (17). In this study we found that insulin or a constitutively active form of Akt1 inhibited FoxO1 and Smad3 activation of rat Cyp7a1 promoter activity. In addition, livers from STZ-induced type-I diabetic rats showed enhanced FoxO1 binding to Cyp7a1 chromatin and elevated Cyp7a1 mRNA expression, which were inhibited by insulin administration. These results are consistent with previous reports that insulin inhibits cholesterol 7α-hydroxylase activity and mRNA expression in rats (27, 28). Proinflammatory cytokines inhibit CYP7A1 and bile acid synthesis. In this study we show that TNFα signaling also antagonizes TGFβ1 signaling to inhibit rat Cyp7a1 gene transcription. Fig. 8 illustrates a model of signaling crosstalks of the insulin/FoxO1, TGFβ1/Smads and TNFα/cJun pathways in coordinated regulation of bile acid synthesis and fibrogenesis. During liver injury and fibrogenesis, TGFβ1 released from HSC activates Smad3, which stimulates Cyp7a1 gene expression and bile acid synthesis. Insulin signaling activates PI3K/PDK and AKT/PKB, which inhibit FoxO1 and Smad3 interaction, thus antagonizes TGFβ1 signaling regulation of Cyp7a1 gene expression. PI3K/PDK and AKT/PKB have been shown to directly interact with and inactivate Smads (29–32). In acute phase responses, rapid release of TNFα from Kupffer cells may activate MAPK/JNK signaling to attenuate TGFβ1 stimulation of Cyp7a1 gene expression in rats. The crosstalks of these three cell signaling pathways may play critical roles in regulation of bile acid synthesis in liver injury, fibrogenesis, and liver regeneration.
Fig. 8.
A model of insulin/FoxO1, TGFβ1/Smad, and TNFα/cJun signaling pathway crosstalk in coordinated regulation of bile acid synthesis in the cholestatic liver injury, fibrogenesis, and regeneration.
Currently, the mechanism of CYP7A1 regulation during obstructive cholestasis is still not fully understood. Previous studies have shown that Cyp7a1 gene expression is paradoxically increased in long-term BDL rat and mouse models (6–8). A recent study reported that Cyp7a1 mRNA expression dramatically decreased 2 days after bile duct ligation in mice (33). Another study reports rapid repression of Cyp7a1 expression after partial hepatectomy and return to normal levels 7 days after partial hepatectomy in mice (34). The early repression of CYP7A1 gene expression could be due to rapid induction of TNFα as an acute response to liver injury. In chronic liver injury, both hepatocytes and nonparenchymal cells secrete various growth factors that mediate hepatocyte proliferation, regeneration, and fibrogenesis. Some of these factors such as TGFβ1 may positively regulate Cyp7a1 expression to increase bile acid synthesis, which activates FXR to stimulate liver regeneration (34). FXR induces a negative nuclear receptor, small heterodimer partner to inhibit Cyp7a1 expression in the liver (35). Recent studies suggest that bile acids activate intestinal FXR to induce fibroblast growth factor 15, which is transported to the liver to inhibit Cyp7a1 expression (8). This recent finding confirmed a hypothesis that bile acids induced an intestinal factor that inhibited hepatic bile acid synthesis, which was based on the observation that hepatic Cyp7a1 gene expression was repressed by intraduodenal, but not intravenous or portal infusion of bile acids in rats (36). Thus, reduced fibroblast growth factor 15 signaling in hepatocytes due to blockage of enterohepatic circulation may also contribute to positive regulation of Cyp7a1 in rodent models of obstructive cholestasis. It has been reported that bile acid synthesis is reduced but CYP7A1 mRNA and enzyme activity are not decreased in a study of a small number of human patients with obstructive cholestasis (37). Cholestasis in human patients could be caused by many different pathological conditions such as bile duct stones, tumors, drugs, genetic diseases, etc. These pathological conditions may alter CYP7A1 expression and bile acid synthesis by different mechanisms. Further studies of cholestatic patients of liver injury are needed to understand the complex interaction of proinflammatory cytokines, growth factors, and nuclear receptors in regulation of CYP7A1 and bile acid synthesis in humans.
In summary, this study identified a distinct regulatory mechanism of rat Cyp7a1 by Smad3 and FoxO1, which may be implicated in the regulation of bile acid synthesis in diabetes and liver fibrosis. This study also suggests that proinflammatory cytokines released from Kupffer cells and TGFβ1 released from HSC cells also crosstalks to regulate bile acid synthesis, fibrogenesis, and liver regeneration to maintain hepatic bile acid homeostasis.
Acknowledgments
T. Li is a recipient of the American Heart Association postdoctoral fellowship from the Great River Affiliate of AHA.
Abbreviations
AKT/PKB, protein kinase B
BDL, bile duct ligation
ChIP, chromatin immunoprecipitation
CYP7A1, cholesterol 7α-hydroxylase
FoxO1, forkhead transcription factor O1
HNF4α, hepatocyte nuclear factor 4α
HSC, hepatic stellate cells
Smad, mothers against decapentaplegic homolog
STZ, streptozotocin
TGFβ1, transforming growth factor β1
TβRI, TGFβ1 receptor type I
TβRII, TGFβ1 receptor type II
TNFα, tumor necrosis factor α
Published, JLR Papers in Press, May 29, 2008.
Footnotes
This study is supported by NIH grants DK58379 and DK44442.
References
- 1.Russell D. W., and K. D. Setchell. 1992. Bile acid biosynthesis. Biochemistry. 31 4737–4749. [DOI] [PubMed] [Google Scholar]
- 2.Chiang J. Y. 1998. Regulation of bile acid synthesis. Front. Biosci. 3 D176–D193. [DOI] [PubMed] [Google Scholar]
- 3.Li T., A. Jahan, and J. Y. Chiang. 2006. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song K. H., E. Ellis, S. Strom, and J. Y. Chiang. 2007. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 46 1993–2002. [DOI] [PubMed] [Google Scholar]
- 5.Li T., and J. Y. Chiang. 2007. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha-hydroxylase gene. Gastroenterology. 133 1660–1669. [DOI] [PubMed] [Google Scholar]
- 6.Dueland S., J. Reichen, G. T. Everson, and R. A. Davis. 1991. Regulation of cholesterol and bile acid homoeostasis in bile-obstructed rats. Biochem. J. 280 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., J. Binz, M. J. Numerick, S. Dennis, G. Luo, B. Desai, K. I. MacKenzie, T. A. Mansfield, S. A. Kliewer, B. Goodwin, et al. 2003. Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J. Clin. Invest. 112 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inagaki T., M. Choi, A. Moschetta, L. Peng, C. L. Cummins, J. G. McDonald, G. Luo, S. A. Jones, B. Goodwin, J. A. Richardson, et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2 217–225. [DOI] [PubMed] [Google Scholar]
- 9.Napoli J., D. Prentice, C. Niinami, G. A. Bishop, P. Desmond, and G. W. McCaughan. 1997. Sequential increases in the intrahepatic expression of epidermal growth factor, basic fibroblast growth factor, and transforming growth factor beta in a bile duct ligated rat model of cirrhosis. Hepatology. 26 624–633. [DOI] [PubMed] [Google Scholar]
- 10.Friedman S. L. 2000. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 275 2247–2250. [DOI] [PubMed] [Google Scholar]
- 11.Gressner A. M., and R. Weiskirchen. 2006. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J. Cell. Mol. Med. 10 76–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angulo P. 2002. Nonalcoholic fatty liver disease. N. Engl. J. Med. 346 1221–1231. [DOI] [PubMed] [Google Scholar]
- 13.Friedman S. L. 2003. Liver fibrosis–from bench to bedside. J. Hepatol. 38 (Suppl 1): S38–S53. [DOI] [PubMed] [Google Scholar]
- 14.Heldin C. H., K. Miyazono, and P. ten Dijke. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 390 465–471. [DOI] [PubMed] [Google Scholar]
- 15.Seoane J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 117 211–223. [DOI] [PubMed] [Google Scholar]
- 16.Gomis R. R., C. Alarcon, W. He, Q. Wang, J. Seoane, A. Lash, and J. Massague. 2006. A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA. 103 12747–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakae J., T. Kitamura, D. L. Silver, and D. Accili. 2001. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arden K. C. 2004. FoxO: linking new signaling pathways. Mol. Cell. 14 416–418. [DOI] [PubMed] [Google Scholar]
- 19.Crestani M., D. Stroup, and J. Y. Chiang. 1995. Hormonal regulation of the cholesterol 7 alpha-hydroxylase gene (CYP7). J. Lipid Res. 36 2419–2432. [PubMed] [Google Scholar]
- 20.Li T., and J. Y. Chiang. 2005. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288 G74–G84. [DOI] [PubMed] [Google Scholar]
- 21.Qing J., Y. Zhang, and R. Derynck. 2000. Structural and functional characterization of the transforming growth factor-beta -induced Smad3/c-Jun transcriptional cooperativity. J. Biol. Chem. 275 38802–38812. [DOI] [PubMed] [Google Scholar]
- 22.Attisano, L., and S. Tuen Lee-Hoeflich. 2001. The Smads. Genome Biol 2. RE:view. S3010.1–3010.8. [DOI] [PMC free article] [PubMed]
- 23.Li T., X. Kong, E. Owsley, E. Ellis, S. Strom, and J. Y. Chiang. 2006. Insulin regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes: roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281 28745–28754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verrecchia F., and A. Mauviel. 2004. TGF-beta and TNF-alpha: antagonistic cytokines controlling type I collagen gene expression. Cell. Signal. 16 873–880. [DOI] [PubMed] [Google Scholar]
- 25.Feingold K. R., D. K. Spady, A. S. Pollock, A. H. Moser, and C. Grunfeld. 1996. Endotoxin, TNF, and IL-1 decrease cholesterol 7 alpha-hydroxylase mRNA levels and activity. J. Lipid Res. 37 223–228. [PubMed] [Google Scholar]
- 26.Miyake J. H., S. L. Wang, and R. A. Davis. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J. Biol. Chem. 275 21805–21808. [DOI] [PubMed] [Google Scholar]
- 27.Subbiah M. T., and R. L. Yunker. 1984. Cholesterol 7 alpha-hydroxylase of rat liver: an insulin sensitive enzyme. Biochem. Biophys. Res. Commun. 124 896–902. [DOI] [PubMed] [Google Scholar]
- 28.Twisk J., M. F. Hoekman, E. M. Lehmann, P. Meijer, W. H. Mager, and H. M. Princen. 1995. Insulin suppresses bile acid synthesis in cultured rat hepatocytes by down-regulation of cholesterol 7 alpha-hydroxylase and sterol 27-hydroxylase gene transcription. Hepatology. 21 501–510. [PubMed] [Google Scholar]
- 29.Song K., H. Wang, T. L. Krebs, and D. Danielpour. 2006. Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. EMBO J. 25 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remy I., A. Montmarquette, and S. W. Michnick. 2004. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6 358–365. [DOI] [PubMed] [Google Scholar]
- 31.Conery A. R., Y. Cao, E. A. Thompson, C. M. Townsend, Jr., T. C. Ko, and K. Luo. 2004. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat. Cell Biol. 6 366–372. [DOI] [PubMed] [Google Scholar]
- 32.Seong H. A., H. Jung, K. T. Kim, and H. Ha. 2007. 3-Phosphoinositide-dependent PDK1 negatively regulates transforming growth factor-beta-induced signaling in a kinase-dependent manner through physical interaction with Smad proteins. J. Biol. Chem. 282 12272–12289. [DOI] [PubMed] [Google Scholar]
- 33.Park Y. J., M. Qatanani, S. S. Chua, J. L. Larey, S. A. Johnson, M. Watanabe, D. D. Moore, and Y. K. Lee. 2008. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 47 1578–1586. [DOI] [PubMed] [Google Scholar]
- 34.Huang W., K. Ma, J. Zhang, M. Qatanani, J. Cuvillier, J. Liu, B. Dong, X. Huang, and D. D. Moore. 2006. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 312 233–236. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6 517–526. [DOI] [PubMed] [Google Scholar]
- 36.Pandak W. M., D. M. Heuman, P. B. Hylemon, J. Y. Chiang, and Z. R. Vlahcevic. 1995. Failure of intravenous infusion of taurocholate to down-regulate cholesterol 7 alpha-hydroxylase in rats with biliary fistulas. Gastroenterology. 108 533–544. [DOI] [PubMed] [Google Scholar]
- 37.Bertolotti M., L. Carulli, M. Concari, P. Martella, P. Loria, E. Tagliafico, S. Ferrari, M. Del Puppo, B. Amati, E. De Fabiani, et al. 2001. Suppression of bile acid synthesis, but not of hepatic cholesterol 7alpha-hydroxylase expression, by obstructive cholestasis in humans. Hepatology. 34 234–242. [DOI] [PubMed] [Google Scholar]