Abstract
The purpose of this review is to highlight recent advances toward the refinement of a three-dimensional structure for lipid-bound apolipoprotein A-I (apoA-I) on recombinant HDL. Recently, X-ray crystallography has yielded a new structure for full-length, lipid-free apoA-I. Although this approach has not yet been successful in solving the three-dimensional structure of lipid-bound apoA-I, analysis of the X-ray structures has been of immense help in the interpretation of structural data obtained from other methods that yield structural information. Recent studies emphasize the use of mass spectrometry to unambiguously identify cross-linked peptides or to quantify solvent accessibility using hydrogen-deuterium exchange. The combination of mass spectrometry, molecular modeling, molecular dynamic analysis, and small-angle X-ray diffraction has provided additional structural information on apoA-I folding that complements previous approaches.
Keywords: cholesterol, mass spectrometry, hydrogen-deuterium exchange, molecular dynamic modeling, chemical cross-linking, apolipoprotein A-I
To understand the mechanism of HDL apolipoprotein A-I (apoA-I) protection against the development of coronary heart disease, recent investigations have focused on apoA-I's dual functions in promoting lipid efflux (1–4) and its role in modulating immune cell activation (5–10). Recent attention has now focused on elucidating the three-dimensional conformation of apoA-I, the most abundant protein constituent within the HDL particle, in response to its acquisition of phospholipid and cholesterol to form nascent lipoprotein particles.
ApoA-I is a 28 kDa protein synthesized by the liver and small intestine. It plays a key role in the formation, metabolism, and catabolism of HDL, a plasma lipoprotein whose levels are highly linked to protection against the development of coronary artery disease, even in patients with very low LDL cholesterol levels (11). Formation of plasma HDL particles depends entirely on the initial lipidation of apoA-I by ABCA1 (12–14). Dimers of lipid-free or lipid-poor apoA-I acquire limited amounts of phospholipid and cholesterol from the membrane-bound ABCA1 transporter to form one of several classes of nascent HDL particles (15). Other classes of nascent HDL particles formed from this interaction contain more than two molecules of apoA-I per particle, with increased amounts of phospholipid and cholesterol (16–18).
A second lipidation step essential for plasma HDL maturation is the activation of the enzyme LCAT by lipid-bound apoA-I. Activation of this enzyme results in the synthesis of cholesteryl esters on nascent HDL (19), providing a hydrophobic core and transforming the small, lipid-poor particle to a spherical lipid-rich HDL. Studies of human deficiencies have shown that if either ABCA1 or LCAT are inactive, plasma concentrations of HDL apoA-I are very low, owing to the rapid removal of the small lipid-poor HDL apoA-I from circulation (20, 21). Thus, both of these two major apoA-I lipidation steps are absolutely essential for the formation of mature spherical plasma HDL and, to some extent, the functionality of the HDLs themselves (22–24).
ApoA-I has a primary structure of 243 amino acids, with secondary structural motifs similar to those of other apolipoproteins. The overall lipid-free solution structure has been described as “molten globular,” (25) which suggests a native-like secondary structure and a tertiary structure that is less organized. In the molten globular state, the α-helical regions usually remain intact, with β-sheets and connecting regions disappearing or becoming less ordered. For these proteins, the hydrophobic regions face inward and the hydrophilic faces interact with the aqueous environment. The α-helix is the primary secondary structure motif within lipid-free apoA-I. Early studies showed that the first 43 residues of the N-terminal globular domain are relatively disordered but does contain a G* amphipathic α-helix from residues 8–33 (26). The amino acids that comprise approximately 80% of the protein, residues 44–243, are composed of a series of 10 tandem, repeating units with high amino acid conservation within repeats. Eight of these repeats contain 22 amino acids, whereas two contain 11 amino acids. Helix-breaking amino acids such as proline (27) separate seven out of the ten repeats. Several investigators have suggested that the frequency of these “breaks” may impart flexibility to apoA-I, allowing it to bend around the periphery of the spherical HDL particle (28, 29).
The description of apoA-I as molten globular is not necessarily at odds with reports claiming that lipid-free apoA-I contains two distinct unfolding domains, an N-terminal (1–187) and a C-terminal (188–243), but suggests that the amino acid side chains have not achieved the close packing associated with the native state of the protein. Recent studies by Brouillette et al. (30) have suggested that the N-terminal region of apoA-I may have a more compact structure than that originally proposed and implied that the well-established propensity of apoA-I to oligomerize (31) might explain how earlier studies achieved a molten globular structure. Overall, the emerging view suggests that molten globules represent a third thermodynamic state that allows the protein to assume many different functions requiring interaction with membrane lipids.
A unique feature of apoA-I is the asymmetric distribution of charged amino acids along the face of each 22 and 11 amino-acid α-helical repeat, designated the type A amphipathic α-helix (32), that was first described by Segrest et al. (33) in 1974. Each of the ten helical repeats in apoA-I has one face enriched in hydrophobic amino acids and the opposing face enriched in negatively charged amino acids. The area between the hydrophilic and hydrophobic faces of the helix is enriched in positively charged amino acids, usually lysine and arginine. It is generally accepted that the hydrophobic face of each amphipathic helix interacts with the phospholipid acyl chains in HDL to enhance particle solvation. Whereas the negatively charged hydrophilic face of each helix supports interaction with the aqueous phase, the function of the highly conserved positively charged amino acids remains speculative. It has been suggested that in HDL, the positively charged residues interact with the negatively charged phosphate of the phospholipid head group (the “snorkel” hypothesis) (33, 34). MacRaild, Howlett, and Gooley (35) have provided NMR evidence that is consistent with the snorkel hypothesis for apoC-II, an apoprotein containing amphipathic helices, interacting with dodecylphosphocholine micelles.
LIPID-FREE CONFORMATION OF APOA-I
Studies employing analytical ultracentrifugation suggest that in solution, lipid-free apoA-I is in equilibrium between two conformations: a loosely folded helical bundle and an elongated helical structure, called an “elongated helical hairpin” (36). The “elongated hairpin” model is consistent with the total α-helix content that was estimated by circular dichroism, as well as with the X-ray analysis of similar molecules and other physical biochemical parameters (36–39). The first X-ray crystal structure of a lipid-free apoA-I was obtained at 3.6 Å resolution for a mutant form of the protein, Δ43 apoA-I, which lacks the N-terminal G* amino acid region. The analysis showed a saddle or horseshoe structure with the apoA-I molecules oriented antiparallel to one another (28). Two years later, the same group reported a second crystal form of lipid-free Δ43 apoA-I. Although the structure was not solved, the space group suggested that this second crystal form could accommodate a four-helix bundle (40), as originally suggested by Rogers et al. (36) and then later supported by fluorescence resonance energy transfer (FRET) studies (30) and also demonstrated for truncated lipid-free apoE (41). Almost 10 years later, Ajees et al. (42) reported an X-ray structure at 2.4 Å for full-length lipid-free apoA-I showing that the principal motif for amino acids 1 to 187, the N-terminal domain, was indeed a four-helix bundle. A study conducted by Brouillette et al. (30) also showed that the N-terminal domain was a compact structure and that the remaining C-terminal residues were part of a separate domain, which is important for lipid binding (43, 44). Ren et al. (45) and Davidson et al. (46), using a variety of techniques, provided further evidence that the N-terminus of lipid-free apoA-I exists in a compact bundle. However, these studies did not rule out the possibility that the compact bundle was in equilibrium with an “elongated helical hairpin,” because the separation between donor and acceptor probes greatly exceeded the distance for efficient FRET analysis in the N-(iodoacetyl)′-(5-sulfo-1-naphthyl)ethylenediamine (AEDANS)-tryptophan system (30).
Chemical cross-linking/mass spectrometry (CCL/MS) combined with homology modeling has provided additional information on the structure of lipid-free apoA-I (47). Like the X-ray analysis by Ajees et al. (42), this study also suggested that lipid-free apoA-I contains a four-helix bundle as its principal motif. However, the in-solution conformation obtained from CCL/MS appears to be more flexible and less organized than the crystal structure, but agrees with studies suggesting that most of the organized, α-helical character was located in the first four repeats (36, 48). The greatest differences between the solution structure proposed by Silva et al. (47) and the X-ray crystal structure of Ajees et al. (42) were located in the N- and C-terminal regions, roughly amino acids 1–24 and 181–243. Recent electron paramagnetic resonance spectroscopy studies of lipid-free apoA-I support the idea that the conformation in the N-terminal region (49, 50) differs from that in the crystal structure as well as that deduced from CCL/MS. An in-depth discussion by Davidson and Thompson (50) suggests that the crystal structure of full-length apoA-I does not appear to accurately accommodate many physical biochemical measurements, like α-helical content (51, 52), which has been accommodated in earlier models of lipid-free apoA-I (36). Brouillette et al. (30) used FRET to study the spatial relationships between tryptophan at residue position 50 with AEDANS labels attached to cysteines engineered at positions 83 and 173. The proposed structure was more compact than that reported from the X-ray crystallography of apoA-I, and the calculated separations appear shorter than those in the structure reported by Ajees et al. (42), as well as those deduced from chemical cross-linking by Silva et al (47).
To summarize, several analyses of full-length lipid-free apoA-I suggest that there are distinct structure domains: the N-terminal region comprising amino acids 1 through 187 and the C-terminal region including amino acids 188 through 243. However, differences in lipid-free apoA-I crystal structures clearly imply that apoA-I is a highly dynamic and flexible molecule (25) whose conformation may be altered through processes like crystallization. As the X-ray analysis of Δ43 apoA-I rekindled interest in the structure of lipid-bound apoA-I, the crystal structure of native full-length lipid-free apoA-I and the Brouillette et al. model have provided a sound framework for future studies.
LIPID-BOUND CONFORMATION OF APOA-I: HELIX REGISTRY OF TWO APOA-I MONOMERS ON RECOMBINANT HDL
Previous investigations have raised some interesting and at times contradictory concepts relating to the functionality of the various domains within apoA-I. Studies have shown that removal of the N-terminal region causes the protein to fold in alternative conformation(s) that affects its overall stability (53), whereas other studies show that removal of this region alters the protein's ability to bind lipid (54). Even simple mutations in the N-terminal region, such as G26R, have been shown to cause decreased lipid binding capability (55). The C-terminal domain (helices 8 through 10) appears to promote “self-association” of apoA-I with itself (56–58) as well as to lipid (44, 53). Although helices 1 and 10 have the greatest lipid binding affinity (59), their removal still leaves a protein that readily associates with phospholipid (60).
Because the lipid-bound form of apoA-I possesses so many unique and important biological functions, many groups have focused on solving its conformation using recombinant HDL (rHDL) particles reconstituted with POPC. Some of the first reports published in support of the “horseshoe-shaped” structure implied by the Δ43 apoA-I crystal structure (42) coined the term “belt” model to describe the two antiparallel monomers of apoA-I wrapped around a bilayer of phospholipids (61, 62), with the hydrophobic face of the amphipathic α-helix in close contact with the fatty acid acyl chains (63). Other studies using rHDL and employing FRET also confirmed the Δ43 apoA-I crystal structure by showing that repeat 5 (121–142) of one molecule of apoA-I lies adjacent to repeat 5 of the second monomer, repeat 5′ (64–66). Both FRET and fluorescence lifetime measurements (67) showed that two molecules of apoA-I bound to either rHDL or spherical HDL could exist in a variable helix-to-helix registry centered on helix 5, as opposed to a fixed helix registry implied by the X-ray crystal structure of Δ43 apoA-I (28).
Salt bridges formed between two adjacent apoA-I molecules are thought to contribute to significant stabilization of the lipid-bound conformation (59, 61), although conclusive experimental evidence proving this concept remains elusive. The possibility that the individual monomers undergo large shifts in their registry was excluded by fluorescence lifetime measurements (67), but smaller shifts reported in this study reinforce the concept of lipid-bound apoA-I as a dynamic protein, i.e., helix 5 in one apoA-I molecule can interact with both helix 4 or helix 5 in the second molecule of apoA-I (67). The dynamic nature of apoA-I was also shown using nitroxide spin labels. In this important study, helices 5, 6, and 7 (residues 121–186) were shown to exhibit significant differences in their exposure to lipid and were thus highly dynamic (68). The authors concluded that a central “hinge” domain responds to particle diameter or lipid content by folding or unfolding, in agreement with later studies demonstrating the flexibility within this domain (69).
REFINEMENT OF THE LIPID-BOUND APOA-I MODEL: ROLE OF THE N- AND C-TERMINAL ENDS
Recently a battery of molecular tools have been used to refine the conformation of apoA-I on rHDL, including CCL/MS, spin labeling, and hydrogen-deuterium exchange (H/DX). In 2003, Davidson and Hilliard were the first to report the use of CCL/MS to describe the lipid-bound conformation of apoA-I on 96 Å POPC rHDL. They suggested that their results were more consistent with a “double belt,” or a “helical hairpin,” rather than the “Z-belt” conformation (70). In 2005, a subsequent CCL/MS study by Silva et al. (71) employing MS/MS sequencing refined their earlier results and allowed them to definitely exclude the Z-belt conformation and hairpin (72). They also concluded that apoA-I had similar “belt” configurations on both 96 Å and 80 Å diameter POPC rHDL and could exist in two major helix registries, a helix 5-5 and a helix 5-2 registry (72). Concurrently in 2005, Bhat et al. (73) described the lipid-bound conformation of apoA-I on 96 Å diameter POPC rHDL using CCL/MS. Only cross-linked peptides whose identity was verified using MS/MS were used to develop the proposed structures. The conclusions from Bhat et al. (73) were generally consistent with those of Silva et al. (71); however, there were important differences between the two studies. Bhat et al. (73) discovered that two distinct cross-linked dimers of apoA-I could be separated by their differential migration on 12% SDS-PAGE. They then demonstrated that these two dimers corresponded to distinct intermolecular cross-links involving either the N-terminus or the central region of the protein (73). With this information, and the structural constraints imposed by intra- and intermolecular cross-links, N- and C-terminal folds were hypothesized to form and interact with each other, forming a “buckle” (73, 74). The “belt-buckle” model is illustrated in Fig. 1, middle panel. Continued investigation by Bhat et al. (74) showed that 80 Å diameter POPC rHDL also assumed a conformation similar to that found for the 96 Å diameter rHDL, except that the N- and C-terminal ends appeared to be further folded back onto the “belt,” suggesting that both belt and hairpin conformations may contribute to the lipid-bound conformation of apoA-I (64, 65). It has been hypothesized that the N- and C-terminal folds function enable the protein to organize and solubilze different amounts of phospholipid as well as to increase the number of apoA-I monomers or other apoproteins as an HDL particle is metabolized in plasma.
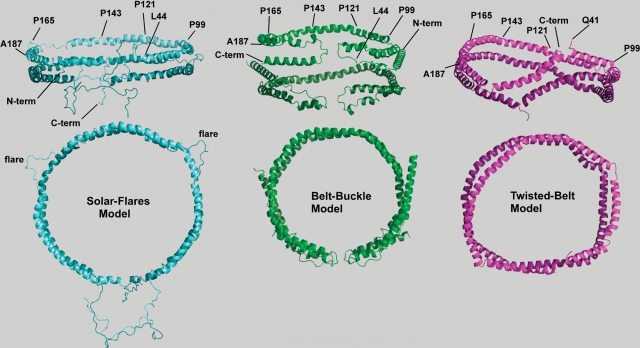
Fig. 1.
Three-dimensional models for two apoA-I molecules bound to ∼96 Å diameter POPC rHDL. Two views of lipidated apoA-I on recombinant HDL with the upper views illustrating an “on edge” view of the parallel strands of apoA-I for each model, whereas the second or lower view represents the top-down view. Phospholipid molecules have been left out of the figures for better visualization of protein conformation. Diameters, measured from the center of the helical strands, are given for each of the three-dimensional figures. Left: “Solar-flares” model showing the conformation for two apoA-I molecules bound to ∼104 Å diameter rHDL containing one hundred molecules of POPC to one full-length apoA-I molecule (75). The “solar-flares” name comes from the regions of apoA-I that are more solvent accessible. They are shown as flaring out into solution and correspond to amino acids 159–178. This model shows the N-terminal end extending into solvent away from the lipid, and the hydrophobic C-terminal helix 10 interacts with the 1–43 amino acid region also not associated with lipid (75). Coordinates were downloaded from the Protein Model Data Base, code PM0074956. Middle: “Belt-buckle” model showing the conformation for two apoA-I molecules bound to a 96 Å diameter rHDL containing seventy-five POPC molecules to one full-length apoA-I molecule (74). The N- and C-terminal ends are also closely associated, but in this model, fold back onto the central region of the apoA-I belt. In this way, the globular N-terminal domain of each apoA-I molecule folds or loops back onto itself, maximizing interaction with the C-terminal highly hydrophobic domain of apoA-I, functioning to adjust the “belt” in order to accommodate different amounts of phospholipids and cholesterol. Right: “Twisted-belt” model derived from molecular dynamic simulations of two apoA-I molecules bound to a 100 Å by 83 Å ellipsoidal rHDL containing eighty molecules of POPC to one truncated Δ40 apoA-I molecule (92). There is considerable agreement among these three three-dimensional models of lipid-bound apoA-I on rHDL. All models suggest that the central region of lipid-bound apoA-I assumes an antiparallel double belt with amphipathic helix 5 on each apoA-I strand situated adjacent to the same region of the second molecule of apoA-I. Differences between the models appear to involve the interaction of the N- and C-terminal regions of the two monomers.
Wu et al. (75) used MS in which H/DX was employed to study the conformation of lipid-free and lipid-bound apoA-I on ∼104 Å POPC rHDL. They measured deuterium incorporation on 48 pairs of peptic peptides across the entire sequence of apoA-I to determine the solvent accessibility of different regions. The “double belt” model (61), along with H/DX data, was used as the basis for computational modeling of apoA-I on rHDL. Additional constraints included particle size and composition, as well as selected information from CCL/MS studies (70, 73). Calculations involving over 2,500 energy-minimization steps gave the “solar flares” model shown in Fig. 1, left panel. This model was in agreement with other models for apoA-I bound to POPC rHDL, because it was based on a “belt” of two apoA-I molecules in which helix 5 in one molecule was in registry with helix 5 of a second apoA-I molecule. However, in contrast to the “belt-buckle” model, the “solar flares” model predicts that the N-termini of two apoA-I molecules form a symmetric, globular domain, with amino acids 14–18 of each of the two molecules in close association. In this model, the C-termini of both apoA-I molecules interact with each other, and the hydrophobic, distal C-terminal residues of helix 10, amino acids 236–243, are exposed to the solvent. As reported by Wu et al. (75), refinements to the “solar-flares” model using secondary structural CCL/MS constraints from either the “hairpin model” (71) or the “N-terminal hairpin” model (a.k.a. “belt-buckle” model) (73) did not improve the quality of the model. This model has been the subject of additional scrutiny using dynamic simulations (76).
Interestingly, the “solar flares” model derives its name from the amino acids, residues 159–178 (portions of helices 6 and 7), that protrude from the edge of the disc. These loops are the proposed LCAT binding site because this region displayed reduced deuterium exchange when LCAT was present (75). Studies employing a variety of techniques have suggested that the region corresponding to helices 6 and 7 is involved in binding LCAT and its activation (20, 77, 78). A unique aspect of helices 5, 6, and 7 relates to the position-specific conservation of charged amino acids (79, 80) that have been suggested to either interact directly with LCAT or to participate in intrahelical salt bridges that form the local conformation(s) necessary for interaction and activation of LCAT. The concept of an “intramolecular hairpin” for a central region within lipid-bound apoA-I was actually suggested to lie within helices 5, 6, and 7 (amino acids 130–174) prior to the report by Wu et al. (68). Later, Martin et al. (37) reported a loop region located between amino acids 133 and 146 (portions of helices 5 and 6) using spin coupling between nitroxide spin probes to assess positional relationships. The conclusions of Martin et al. (37) were supported by FRET analysis using a series of apoA-I variants with cysteines engineered into the loop region. In contrast to the structural requirements demonstrated for LCAT activation associated with the central region of apoA-I involving residues 159–178, lipidation of apoA-I by ABCA1 requires interaction with the C-terminal end of apoA-I, residues 220–231 (51, 52), allowing more-complex interactions between apoA-I and ABCA1 in forming nascent HDL (81, 82).
METHIONINE OXIDATION ALTERS N- AND C- TERMINI OF LIPID-FREE AND LIPID-BOUND APOA-I
Differences in the degree of N- and C-terminal solvent exposure predicted by the different models can be analyzed with respect to results from limited proteolysis of apoA-I bound to rHDL. If parts of apoA-I have considerable solvent exposure, then these regions should be highly susceptible to proteolysis. Lins et al. (83), employing trypsin, proteinase K, and pronase treatment, reported limited proteolysis of the N-terminus at amino acids 43, 45, 47, or 48 and 65 for 1,2-dimyristoyl-sn-glycerol-3-phosphocholine (DMPC)-apoA-I rHDL complexes. In the same year, Dalton and Swaney (84) reported that apoA-I on POPC rHDL was subject to proteolysis at amino acids around 208, 103, and 92–94 when treated with trypsin, V8, and elastase. Two other studies on lipid-free and lipid-bound apoA-I on POPC rHDL using trypsin or chymotrypsin reported that a predominant cleavage site was located at Y192 (85, 86). However, another study employing chymotrypsin and V8 reported that the principal cut site, at Y192, was found mainly in lipid-free apoA-I, whereas only the methionine-oxidized lipid-bound apoA-I showed any significant cleavage at Y192, as well as at numerous other sites (87). In this study, the peptides formed in greatest yield from unoxidized lipidated apoA-I were from proteolysis at E205, L42, and E34. Oxidation of two methionines substantially altered the proteolytic profile, with the major cut site disappearing and new sites appearing (87). These findings suggest that oxidation of two of the three methionines within apoA-I causes a dramatic change in the conformation of the lipid-bound protein, with the N-terminal region becoming much more solvent exposed than in its native, unoxidized form (87). Recently Brock et al. (88) reported that the methionine sulfoxide content of apoA-I was more abundant in the HDL of patients with type-1 diabetes, a disease for which cardiovascular events are a more significant complication, than in control patients.
DYNAMIC SIMULATIONS
Molecular dynamics simulations of apoA-I bound to rHDL have recently provided valuable new insight into how the conformation of apoA-I is influenced ratio of phospholipid to apoprotein within the particle. These studies directly address the question of how lipid-poor apoA-I structure adapts to increased phospholipid content, e.g., as apoA-I is lipidated by ABCA1 (89–93). Using a mutant form of apoA-I lacking the N-terminal amino acids 1–40, Catte et al. (92) reported an extensive in silico investigation of the relationship between the lipid-to-apoA-I molar ratio and apoA-I conformation. These simulations showed that at low lipid-to-protein ratios, e.g., at POPC-to-apoA-I molar ratios of 50:1 and 25:1, the conformation assumed by both the lipid bilayer and protein was a “twisted belt.” The “twist” gradually relaxed as the molar ratio of POPC to apoA-I increased. At an 80 to 1 molar ratio of POPC to apoA-I, the conformation of apoA-I was nearly a “double belt” as shown in Fig. 1, right structure. The concept of a “twisted bilayer” has also been proposed and supported by studies of lipid-bound apoE through the use of small-angle X-ray scattering (SAXS) and limited proteolysis combined with mass spectrometry (94). A double belt conformation was also reported in an in silico study by Shih et al. (93) using 1,2-dipalmitoyl-sn-glycerol-3-phosphocholine (DPPC) and apoA-I lacking N-terminal amino acids 1–54 or 1–65 at a ratio of DPPC to apoA-I of 80:1. They substantiated their findings by preparing rHDL with DPPC (or DMPC) and truncated apoA-I lacking residues 1–54. Experimental SAXS measurements were similar to the theoretically calculated values and gave diameters in the range of 102 Å to 103 Å (93).
Recently, the “solar flare” HDL model was subjected to a rigorous test by Shih, Sligar, and Schulten (76) using molecular dynamics simulations. After a 10 ns all-atom simulation, both “flares” rapidly collapsed onto the main body of the HDL particle, and the N-terminal globular region was unstable, as evidenced by excessive mobility. Using a coarse-grained simulation without the first 1–43 residues gave a structure with a 45 Å gap between the N and C-termini of the protein strands. In response, the lipid in the gap formed a micelle-like structure to shield the hydrophobic lipid tails from the solvent (76).
SPHERICAL HDL
The majority of plasma HDL is derived from two major steps involving the lipidation of apoA-I. First, ABCAI adds phospholipid and cholesterol, producing small, lipid apoA-I-containing particles. In plasma, these particles are modified by LCAT to generate cholesteryl ester-rich particles that are larger and more spherical. Most of the effort to define lipidated apoA-I structure has been directed at studying rHDL that can be reproducibly prepared in homogenous size and composition. Circulating HDLs are spherical and, as Borhani et al. (28) have suggested, apoA-I floats on the surface monolayer of the particle, with the hydrophobic faces of each helix penetrating past the polar head groups to interact with the fatty acid acyl chains (34, 95). Jonas et al. (96) reported that the α-helix content, tryptophan fluorescence, and antibody binding of apoA-I on 93 Å POPC spheres were similar to those of 96 Å POPC discs and suggested that the secondary structure of apoA-I was similar on both discs and spheres. FRET measurements of apoA-I-labeled rHDL spheres prepared by the action of LCAT on apoA-I-labeled POPC rHDL discs showed that the labeled helical repeats were farther apart in the rHDL spheres than in the rHDL discs (67). In plasma, mature HDLs carry three to four apoA-I molecules, and the LCAT-transformed rHDL spheres studied by Li et al. (67) were shown to carry three molecules of apoA-I. The increased number of apoA-I molecules carried by each rHDL showed that the labeled residues were farther apart, but assignment of specific distance relationships between the labels was not possible (67). Recently, the spherical particle composed of two apoA-Is, 56 POPC and 16 cholesteryl oleate core, has been the subject of theoretical analysis. These studies suggest a shape that resembles a prolate ellipsoid (91), not unlike the smaller HDL particles, but also indicate that the cholesterol moiety of the cholesteryl oleate core is in contact with apoA-I and may contribute to the stabilization and regulation of the structure and function of the particle.
CONCLUSIONS
In conclusion, there is considerable agreement among the various models of apoA-I on POPC rHDL presented in this review and shown in Fig. 1. Taken together, these models predict that the central region of lipid-bound apoA-I assumes an antiparallel “double belt” with a helix 5 to 5 registry, i.e., central amphipathic helices on each apoA-I strand are situated adjacent to one another. The results of Wu et al. (75) and Martin et al. (37) suggest that the two apoA-I chains do not wrap around the lipid particle to give a smooth belt, but that there may be additional protein structure important for LCAT activation and reverse cholesterol transport. Both Wu et al. (75) and Bhat et al. (73, 74) suggest that the C-terminal end is somewhat folded back onto the central region or belt. However, the principal debate or difference between the models centers on ascertaining the conformation assumed by the N- and C-terminal ends. Wu et al. (75) suggest that the N-terminal end is relatively disordered, whereas Bhat et al. (73, 74) have suggested that both the N- and C-termini are relatively ordered and interact with the central part of the apoA-I molecule. All of the models suggest that the N- and C-terminal ends come in close contact with one another. For smaller particles with diameters of ∼80 Å, cross-linking suggests that there is increased interaction between the N terminus and the central region of the belt. Because the results from CCL/MS yield only a time-averaged conformation, these measurements suggest that the N-terminal end spends a significant amount of time close to the “belt,” but do not rule out appreciable mobility for the N-terminus that would allow this region to extend into solution and to possibly overlap with its own C-terminus. Because CCL/MS only provides a snapshot of the protein's conformation at the time the covalent bonds are formed, the N- and C-terminal folds may exist transiently, as represented by the “belt-buckle” model shown in Fig. 2, left panel, or interact with the N-termini from the second apoA-I molecule, as shown in Fig. 2, right panel. In both models, the N-terminal ends of the two apoA-I molecules are folded and interact with other parts of the molecule. Functionally, it is hypothesized that these models exist in equilibrium and enable the lipid-bound protein to adjust to increasing amounts of phospholipid and cholesterol from ABCA1, or as the particle becomes spherical, from the action of LCAT. Recent studies suggest that combining experimentally derived structural data with rigorous tests using molecular dynamics simulations may provide the most accurate models of protein conformation. Future studies will most likely concentrate on refining the conformation of the N- and C-terminal ends of apoA-I bound to rHDL and on determining whether the conformation(s) of apoA-I on nascent HDL lipidated by ABCA1 is the same as that reported for apoA-I on reconstituted HDL discs prepared by sodium cholate dialysis.
Fig. 2.
Three-dimensional model for two apoA-I molecules with a “belt-buckle” conformation and a flexible N-terminal end. Left: The protein component of the “belt-buckle” model for a ∼96 Å diameter rHDL particle with two molecules of apoA-I. Right: The same model, but with the N-terminal end of one apoA-I molecule, residues 1–49, folded over onto the N-terminal end of the second apoA-I molecule. Residues 1–49 are colored red for clarity.
Acknowledgments
The authors gratefully acknowledge Drs. Sean Davidson, Stan Hazen, Jere Segrest, Michael Oda, and John Voss for providing the coordinates of their respective molecular models.
Abbreviations
AEDANS, N-(iodoacetyl)′-(5-sulfo-1-naphthyl)ethylenediamine
apoA-I, apolipoprotein A-I
CCL/MS, chemical cross-linking/mass spectrometry
DMPC, 1,2-dimysteroyl-sn-glycerol-3-phosphocholine
DPPC, 1,2-dipalmitoyl-sn-glycerol-3-phosphocholine
FRET, fluorescence resonance energy transfer
H/DX, hydrogen-deuterium exchange
POPC, 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine
rHDL, recombinant HDL
SAXS, small-angle X-ray scattering
Published, JLR Papers in Press, May 30, 2008.
Footnotes
These studies were supported by The National Heart, Lung, and Blood Institute, National Institutes of Health (Grants HL-49373 and HL-64163 to M.G.S-T.) and an International HDL Pfizer Award to M.J.T.
References
- 1.Zannis V. I., A. Chroni, and M. Krieger. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 84 276–294. [DOI] [PubMed] [Google Scholar]
- 2.Yancey P. G., A. E. Bortnick, G. Kellner-Weibel, M. de la Llera-Moya, M. C. Phillips, and G. H. Rothblat. 2003. Importance of different pathways of cellular cholesterol efflux. Arterioscler. Thromb. Vasc. Biol. 23 712–719. [DOI] [PubMed] [Google Scholar]
- 3.Rader D. J. 2006. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 116 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boden W. E. 2000. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: assessing the data from Framingham to the Veterans Affairs High-Density Lipoprotein Intervention Trial. Am. J. Cardiol. 86 19L–22L. [DOI] [PubMed] [Google Scholar]
- 5.Luoma P. V. 1997. Gene activation, apolipoprotein A-I/high density lipoprotein, atherosclerosis prevention and longevity. Pharmacol. Toxicol. 81 57–64. [DOI] [PubMed] [Google Scholar]
- 6.Kim K. D., H. Y. Lim, H. G. Lee, D. Y. Yoon, Y. K. Choe, I. Choi, S. G. Paik, Y. S. Kim, Y. Yang, and J. S. Lim. 2005. Apolipoprotein A-I induces IL-10 and PGE2 production in human monocytes and inhibits dendritic cell differentiation and maturation. Biochem. Biophys. Res. Commun. 338 1126–1136. [DOI] [PubMed] [Google Scholar]
- 7.Norata G. D., P. Marchesi, A. Pirillo, P. Uboldi, G. Chiesa, V. Maina, C. Garlanda, A. Mantovani, and A. L. Catapano. 2008. Long pentraxin 3, a key component of innate immunity, is modulated by high-density lipoproteins in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 28 925–931. [DOI] [PubMed] [Google Scholar]
- 8.Gotsman I., R. Gupta, and A. H. Lichtman. 2007. The influence of the regulatory T lymphocytes on atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27 2493–2495. [DOI] [PubMed] [Google Scholar]
- 9.Hansson G. K., and P. Libby. 2006. The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 6 508–519. [DOI] [PubMed] [Google Scholar]
- 10.Binder C. J., M. K. Chang, P. X. Shaw, Y. I. Miller, K. Hartvigsen, A. Dewan, and J. L. Witztum. 2002. Innate and acquired immunity in atherogenesis. Nat. Med. 8 1218–1226. [DOI] [PubMed] [Google Scholar]
- 11.Barter P., A. M. Gotto, J. C. LaRosa, J. Maroni, M. Szarek, S. M. Grundy, J. J. Kastelein, V. Bittner, and J. C. Fruchart. 2007. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N. Engl. J. Med. 357 1301–1310. [DOI] [PubMed] [Google Scholar]
- 12.Timmins J. M., J. Y. Lee, E. Boudyguina, K. D. Kluckman, L. R. Brunham, A. Mulya, A. K. Gebre, J. M. Coutinho, P. L. Colvin, T. L. Smith, et al. 2005. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Invest. 115 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oram J. F., and J. W. Heinecke. 2005. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 85 1343–1372. [DOI] [PubMed] [Google Scholar]
- 14.Brunham L. R., R. R. Singaraja, and M. R. Hayden. 2006. Variations on a gene: rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu. Rev. Nutr. 26 105–129. [DOI] [PubMed] [Google Scholar]
- 15.Krimbou L., H. Hajj Hassan, S. Blain, S. Rashid, M. Denis, M. Marcil, and J. Genest. 2005. Biogenesis and speciation of nascent apoA-I-containing particles in various cell lines. J. Lipid Res. 46 1668–1677. [DOI] [PubMed] [Google Scholar]
- 16.Doung P. T., H. L. Collins, M. Nickel, S. Lund-Katz, G. H. Rothblat, and M. C. Phillips. 2006. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J. Lipid Res. 47 832–843. [DOI] [PubMed] [Google Scholar]
- 17.Mulya A., J. Y. Lee, A. K. Gebre, M. J. Thomas, P. L. Colvin, and J. S. Parks. 2007. Minimal lipidation of prebeta HDL by ABCA1 results in reduced ability to interact with ABCA1. Arterioscler. Thromb. Vasc. Biol. 8 1828–1836. [DOI] [PubMed] [Google Scholar]
- 18.Doung P. T., G. L. Weibel, S. Lund-Katz, G. H. Rothblat, and M. C. Phillips. 2008. Characterization and properties of preβ-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtiss L. K., D. T. Valenta, N. J. Hime, and K. A. Rye. 2006. What is so special about apolipoprotein AI in reverse cholesterol transport? Arterioscler. Thromb. Vasc. Biol. 26 12–19. [DOI] [PubMed] [Google Scholar]
- 20.Sorci-Thomas M. G., and M. J. Thomas. 2002. The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc. Med. 12 121–128. [DOI] [PubMed] [Google Scholar]
- 21.von Eckardstein A. 2006. Differential diagnosis of familial high density lipoprotein deficiency syndromes. Atherosclerosis. 186 231–239. [DOI] [PubMed] [Google Scholar]
- 22.Koukos G., A. Chroni, A. Duka, D. Kardassis, and V. I. Zannis. 2007. LCAT can rescue the abnormal phenotype produced by the natural apoA-I mutations (Leu141Arg)Pisa and (Leu159Arg)FIN. Biochemistry. 46 10713–10721. [DOI] [PubMed] [Google Scholar]
- 23.McPherson P. A., I. S. Young, and J. McEneny. 2007. A dual role for lecithin:cholesterol acyltransferase (EC 2.3.1.43) in lipoprotein oxidation. Free Radic. Biol. Med. 43 1484–1493. [DOI] [PubMed] [Google Scholar]
- 24.Asztalos B. F., E. J. Schaefer, K. V. Horvath, S. Yamashita, M. Miller, G. Franceschini, and L. Calabresi. 2007. Role of LCAT in HDL remodeling: investigation of LCAT deficiency states. J. Lipid Res. 48 592–599. [DOI] [PubMed] [Google Scholar]
- 25.Gursky O., and D. Atkinson. 1996. Thermal unfolding of human high-density apolipoprotein A-1: implications for a lipid-free molten globular state. Proc. Natl. Acad. Sci. USA. 93 2991–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segrest J. P., M. K. Jones, H. De Loof, C. G. Brouillette, Y. V. Venkatachalapathi, and G. M. Anantharamaiah. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33 141–166. [PubMed] [Google Scholar]
- 27.Chan L. 1989. The apolipoprotein multigene family: structure, expression, evolution, and molecular genetics. Klin. Wochenschr. 67 225–237. [DOI] [PubMed] [Google Scholar]
- 28.Borhani D. W., D. P. Rogers, J. A. Engler, and C. G. Brouillette. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94 12291–12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouillette C. G., G. M. Anantharamaiah, J. A. Engler, and D. W. Borhani. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531 4–46. [DOI] [PubMed] [Google Scholar]
- 30.Brouillette C. G., W. J. Dong, Z. W. Yang, M. J. Ray, I. I. Protasevich, H. C. Cheung, and J. A. Engler. 2005. Forster resonance energy transfer measurements are consistent with a helical bundle model for lipid-free apolipoprotein A-I. Biochemistry. 44 16413–16425. [DOI] [PubMed] [Google Scholar]
- 31.Yokoyama S., S. Tajima, and A. Yamamoto. 1982. The process of dissolving apolipoprotein A-I in an aqueous buffer. J. Biochem. 91 1267–1272. [DOI] [PubMed] [Google Scholar]
- 32.Segrest J. P., D. W. Garber, C. G. Brouillette, S. C. Harvey, and G. M. Anantharamaiah. 1994. The amphipathic a-helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 45 303–369. [DOI] [PubMed] [Google Scholar]
- 33.Segrest J. P., R. L. Jackson, J. D. Morrisett, and A. M. Gotto. 1974. A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 38 247–258. [DOI] [PubMed] [Google Scholar]
- 34.Mishra V. K., M. N. Palgunachari, J. P. Segrest, and G. M. Anantharamaiah. 1994. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J. Biol. Chem. 269 7185–7191. [PubMed] [Google Scholar]
- 35.MacRaild C. A., G. J. Howlett, and P. R. Gooley. 2004. The structure and interactions of human apolipoprotein C-II in dodecyl phosphocholine. Biochemistry. 43 8084–8093. [DOI] [PubMed] [Google Scholar]
- 36.Rogers D. P., L. M. Roberts, J. Lebowitz, J. A. Engler, and C. G. Brouillette. 1998. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 37 945–955. [DOI] [PubMed] [Google Scholar]
- 37.Martin D. D., M. S. Budamagunta, R. O. Ryan, J. C. Voss, and M. N. Oda. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281 20418–20426. [DOI] [PubMed] [Google Scholar]
- 38.Barbeau D. L., A. Jonas, T. Teng, and A. M. Scanu. 1979. Asymmetry of apolipoprotein A-I in solution as assessed from ultracentrifugal, viscometric, and fluorescence polarization studies. Biochemistry. 18 362–369. [DOI] [PubMed] [Google Scholar]
- 39.Rogers D. P., L. M. Roberts, J. Lebowitz, G. Datta, G. M. Anantharamaiah, J. A. Engler, and C. G. Brouillette. 1998. The lipid-free structure of apolipoprotein A-I: effects of amino-terminal deletions. Biochemistry. 37 11714–11725. [DOI] [PubMed] [Google Scholar]
- 40.Borhani D. W., J. A. Engler, and C. G. Brouillette. 1999. Crystallization of truncated human apolipoprotein A-I in a novel conformation. Acta Crystallogr. D Biol. Crystallogr. 55 1578–1583. [DOI] [PubMed] [Google Scholar]
- 41.Wilson C., M. R. Wardell, K. H. Weisgraber, R. W. Mahley, and D. A. Agard. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 252 1817–1822. [DOI] [PubMed] [Google Scholar]
- 42.Ajees A. A., G. M. Anantharamaiah, V. K. Mishra, M. M. Hussain, and H. M. Murthy. 2006. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. USA. 103 2126–2131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Tanaka M., M. Koyama, P. Dhanasekaran, D. Nguyen, M. Nickel, S. Lund-Katz, H. Saito, and M. C. Phillips. 2008. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 47 2172–2180. [DOI] [PubMed] [Google Scholar]
- 44.Saito H., P. Dhanasekaran, D. Nguyen, P. Holvoet, S. Lund-Katz, and M. C. Phillips. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278 23227–23232. [DOI] [PubMed] [Google Scholar]
- 45.Ren X., L. Zhao, A. Sivashanmugam, Y. Miao, L. Korando, Z. Yang, C. A. Reardon, G. S. Getz, C. G. Brouillette, W. G. Jerome, et al. 2005. Engineering mouse apolipoprotein A-I into a monomeric, active protein useful for structural determination. Biochemistry. 44 14907–14919. [DOI] [PubMed] [Google Scholar]
- 46.Davidson W. S., K. Arnvig-McGuire, A. Kennedy, J. Kosman, T. L. Hazlett, and A. Jonas. 1999. Structural organization of the N-terminal domain of apolipoprotein A-I: studies of tryptophan mutants. Biochemistry. 38 14387–14395. [DOI] [PubMed] [Google Scholar]
- 47.Silva R. A., G. M. Hilliard, J. Fang, S. Macha, and W. S. Davidson. 2005. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 44 2759–2769. [DOI] [PubMed] [Google Scholar]
- 48.Davidson W. S., T. Hazlett, W. W. Mantulin, and A. Jonas. 1996. The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. USA. 93 13605–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagerstedt J. O., M. S. Budamagunta, M. N. Oda, and J. C. Voss. 2007. EPR spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apo-AI in solution. J. Biol. Chem. 282 9143–9149. [DOI] [PubMed] [Google Scholar]
- 50.Davidson W. S., and T. B. Thompson. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282 9143–9149. [DOI] [PubMed] [Google Scholar]
- 51.Leroy A., and A. Jonas. 1994. Native-like structure and self-association behavior of apolipoprotein A-I in a water/n-propanol solution. Biochim. Biophys. Acta. 1212 285–294. [DOI] [PubMed] [Google Scholar]
- 52.Saito H., S. Lund-Katz, and M. C. Phillips. 2004. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 43 350–380. [DOI] [PubMed] [Google Scholar]
- 53.Rogers D. P., C. G. Brouillette, J. A. Engler, S. W. Tendian, L. Roberts, V. K. Mishra, G. M. Anantharamaiah, S. Lund-Katz, M. C. Phillips, and M. J. Ray. 1997. Truncation of the amino terminus of human apolipoprotein A-I substantially alters only the lipid-free conformation. Biochemistry. 36 288–300. [DOI] [PubMed] [Google Scholar]
- 54.Zhu H. L., and D. Atkinson. 2004. Conformation and lipid binding of the N-terminal (1–44) domain of human apolipoprotein A-I. Biochemistry. 43 13156–13164. [DOI] [PubMed] [Google Scholar]
- 55.Lagerstedt J. O., G. Cavigiolio, L. M. Roberts, H. S. Hong, L. W. Jin, P. G. Fitzgerald, M. N. Oda, and J. C. Voss. 2007. Mapping the structural transition in an amyloidogenic apolipoprotein A-I. Biochemistry. 46 9693–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu H. L., and D. Atkinson. 2007. Conformation and lipid binding of a C-terminal (198–243) peptide of human apolipoprotein A-I. Biochemistry. 46 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laccotripe M., S. C. Makrides, A. Jonas, and V. I. Zannis. 1997. The carboxyl-terminal hydrophobic residues of apolipoprotein A-I affect its rate of phospholipid binding and its association with high density lipoprotein. J. Biol. Chem. 272 17511–17522. [DOI] [PubMed] [Google Scholar]
- 58.Oda M. N., T. M. Forte, R. O. Ryan, and J. C. Voss. 2003. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat. Struct. Biol. 10 455–460. [DOI] [PubMed] [Google Scholar]
- 59.Palgunachari M. N., V. K. Mishra, S. Lund-Katz, M. C. Phillips, S. O. Adeyeye, S. Alluri, G. M. Anantharamaiah, and J. P. Segrest. 1996. Only the two end helixes of eight tandem amphipathic helical domains of human apo A-I have significant lipid affinity. Implications for HDL assembly. Arterioscler. Thromb. Vasc. Biol. 16 328–338. [DOI] [PubMed] [Google Scholar]
- 60.Beckstead J. A., B. L. Block, J. K. Bielicki, C. M. Kay, M. N. Oda, and R. O. Ryan. 2005. Combined N- and C-terminal truncation of human apolipoprotein A-I yields a folded, functional central domain. Biochemistry. 44 4591–4599. [DOI] [PubMed] [Google Scholar]
- 61.Segrest J. P., M. K. Jones, A. E. Klon, C. J. Sheldahl, M. Hellinger, H. De Loof, and S. C. Harvey. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274 31755–31758. [DOI] [PubMed] [Google Scholar]
- 62.Koppaka V., L. Silvestro, J. A. Engler, C. G. Brouillette, and P. H. Axelsen. 1999. The structure of human lipoprotein A-I. Evidence for the “belt” model. J. Biol. Chem. 274 14541–14544. [DOI] [PubMed] [Google Scholar]
- 63.Mishra V. K., G. M. Anantharamaiah, J. P. Segrest, M. N. Palgunachari, M. Chaddha, S. W. Sham, and N. R. Krishna. 2006. Association of a model class A (apolipoprotein) amphipathic alpha helical peptide with lipid: high resolution NMR studies of peptide.lipid discoidal complexes. J. Biol. Chem. 281 6511–6519. [DOI] [PubMed] [Google Scholar]
- 64.Tricerri M. A., A. K. Behling Agree, S. A. Sanchez, J. Bronski, and A. Jonas. 2001. Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 40 5065–5074. [DOI] [PubMed] [Google Scholar]
- 65.Li H. H., D. S. Lyles, M. J. Thomas, W. Pan, and M. G. Sorci-Thomas. 2000. Structural determination of lipid-bound apoA-I using fluorescence resonance energy transfer. J. Biol. Chem. 275 37048–37054. [DOI] [PubMed] [Google Scholar]
- 66.Li H. H., M. J. Thomas, W. Pan, E. Alexander, M. Samuel, and M. G. Sorci-Thomas. 2001. Preparation and incorporation of probe-labeled apoA-I for fluorescence resonance energy transfer studies of rHDL. J. Lipid Res. 42 2084–2091. [PubMed] [Google Scholar]
- 67.Li H. H., D. S. Lyles, W. Pan, E. Alexander, M. J. Thomas, and M. G. Sorci-Thomas. 2002. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J. Biol. Chem. 42 39093–39101. [DOI] [PubMed] [Google Scholar]
- 68.Maiorano J. N., R. J. Jandacek, E. M. Horace, and W. S. Davidson. 2004. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 43 11717–11726. [DOI] [PubMed] [Google Scholar]
- 69.Corsico B., J. D. Toledo, and H. A. Garda. 2001. Evidence for a central apolipoprotein A-I domain loosely bound to lipids in discoidal lipoproteins that is capable of penetrating the bilayer of phospholipid vesicles. J. Biol. Chem. 276 16978–16985. [DOI] [PubMed] [Google Scholar]
- 70.Davidson W. S., and G. M. Hilliard. 2003. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass spectrometry study. J. Biol. Chem. 278 27199–27207. [DOI] [PubMed] [Google Scholar]
- 71.Silva R. A., G. M. Hilliard, L. Li, J. P. Segrest, and W. S. Davidson. 2005. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 44 8600–8607. [DOI] [PubMed] [Google Scholar]
- 72.Davidson W. S., and R. A. Silva. 2005. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr. Opin. Lipidol. 16 295–300. [DOI] [PubMed] [Google Scholar]
- 73.Bhat S., M. G. Sorci-Thomas, E. T. Alexander, M. P. Samuel, and M. J. Thomas. 2005. Intermolecular contact between globular N-terminal fold and C-terminal domain of apoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280 33015–33025. [DOI] [PubMed] [Google Scholar]
- 74.Bhat S., M. G. Sorci-Thomas, R. Tuladhar, M. P. Samuel, and M. J. Thomas. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu Z., M. A. Wagner, L. Zheng, J. S. Parks, J. M. Shy III, J. D. Smith, V. Gogonea, and S. L. Hazen. 2008. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction [erratum]. Nat. Struct. Mol. Biol. 15 330, 651. [DOI] [PubMed] [Google Scholar]
- 76.Shih A. Y., S. G. Sligar, and K. Schulten. 2008. Molecular models need to be tested: the case of a solar flares discoidal HDL model. Biophys. J. 94 L87–L89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sorci-Thomas M. G., M. W. Kearns, and J. P. Lee. 1993. Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J. Biol. Chem. 268 21403–21409. [PubMed] [Google Scholar]
- 78.Sorci-Thomas M. G., M. J. Thomas, L. K. Curtiss, and M. Landrum. 2000. Single repeat deletion in apoA-I blocks cholesterol esterification and results in rapid catabolism of D6 and wild-type apoA-I in transgenic mice. J. Biol. Chem. 275 12156–12163. [DOI] [PubMed] [Google Scholar]
- 79.Roosbeek S., B. Vanloo, N. Duverger, H. Caster, J. Breyne, I. De Beun, H. Patel, J. Vandekerckhove, C. Shoulders, M. Rosseneu, et al. 2001. Three arginine residues in apolipoprotein A-I are critical for activation of lecithin:cholesterol acyltransferase. J. Lipid Res. 42 31–40. [PubMed] [Google Scholar]
- 80.Alexander E. T., S. Bhat, M. J. Thomas, R. B. Weinberg, V. R. Cook, M. S. Bharadwaj, and M. G. Sorci-Thomas. 2005. Apolipoprotein A-I helix 6 negatively charged residues attenuate lecithin:cholesterol acyltransferase (LCAT) reactivity. Biochemistry. 44 5409–5419. [DOI] [PubMed] [Google Scholar]
- 81.Chroni A., T. Liu, I. Gorshkova, H. Y. Kan, Y. Uehara, A. Von Eckardstein, and V. I. Zannis. 2003. The central helices of ApoA-I can promote ATP-binding cassette transporter A1 (ABCA1)-mediated lipid efflux. Amino acid residues 220–231 of the wild-type apoA-I are required for lipid efflux in vitro and high density lipoprotein formation in vivo. J. Biol. Chem. 278 6719–6730. [DOI] [PubMed] [Google Scholar]
- 82.Chroni A., G. Koukos, A. Duka, and V. I. Zannis. 2007. The carboxy-terminal region of apoA-I is required for the ABCA1-dependent formation of alpha-HDL but not prebeta-HDL particles in vivo. Biochemistry. 46 5697–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lins L., S. Piron, K. Conrath, B. Vanloo, R. Brasseur, M. Rosseneu, J. Baert, and J. M. Rusysschaert. 1993. Enzymatic hydrolysis of reconstituted dimysteroylphosphatidylcholine-apo A-I complexes. Biochim. Biophys. Acta. 1151 137–142. [DOI] [PubMed] [Google Scholar]
- 84.Dalton M. B., and J. B. Swaney. 1993. Structural and functional domains of apolipoprotein A-I within high density lipoproteins. J. Biol. Chem. 268 19274–19283. [PubMed] [Google Scholar]
- 85.Ji Y., and A. Jonas. 1995. Properties of an N-terminal proteolytic fragment of apolipoprotein A-I in solution and in reconstituted high density lipoproteins. J. Biol. Chem. 270 11290–11297. [DOI] [PubMed] [Google Scholar]
- 86.Calabresi L., G. Tedeschi, C. Treu, S. Ronchi, D. Galbiati, S. Airoldi, C. R. Sirtori, Y. Marcel, and G. Franceschini. 2001. Limited proteolysis of a disulfide-linked apoA-I dimer in reconstituted HDL. J. Lipid Res. 42 935–942. [PubMed] [Google Scholar]
- 87.Roberts L. M., M. J. Ray, T. W. Shih, E. Hayden, M. M. Reader, and C. G. Brouillette. 1997. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 36 7615–7624. [DOI] [PubMed] [Google Scholar]
- 88.Brock J. W., A. J. Jenkins, T. J. Lyons, R. L. Klein, E. Yim, M. Lopes-Virella, R. E. Carter, S. R. Thorpe, and J. W. Baynes. 2008. Increased methionine sulfoxide content of apoA-I in type 1 diabetes. J. Lipid Res. 49 847–855. [DOI] [PubMed] [Google Scholar]
- 89.Shih A. Y., P. L. Freddolino, A. Arkhipov, and K. Schulten. 2007. Assembly of lipoprotein particles revealed by coarse-grained molecular dynamics simulations. J. Struct. Biol. 157 579–592. [DOI] [PubMed] [Google Scholar]
- 90.Shih A. Y., A. Arkhipov, P. L. Freddolino, and K. Schulten. 2006. Coarse grained protein-lipid model with application to lipoprotein particles. J. Phys. Chem. B. 110 3674–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Catte A., J. C. Patterson, D. Bashtovyy, M. K. Jones, F. Gu, L. Li, A. Rampioni, D. Sengupta, T. Vuorela, P. Niemela, et al. 2008. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys. J. 94 2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Catte A., J. C. Patterson, M. K. Jones, W. G. Jerome, D. Bashtovyy, Z. Su, F. Gu, J. Chen, M. P. Aliste, S. C. Harvey, et al. 2006. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys. J. 90 4345–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shih A. Y., A. Arkhipov, P. L. Freddolino, S. G. Sligar, and K. Schulten. 2007. Assembly of lipids and proteins into lipoprotein particles. J. Phys. Chem. B. 111 11095–11104. [DOI] [PubMed] [Google Scholar]
- 94.Peters-Libeu C. A., Y. Newhouse, S. C. Hall, H. E. Witkowska, and K. H. Weisgraber. 2007. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J. Lipid Res. 48 1035–1044. [DOI] [PubMed] [Google Scholar]
- 95.Hristova K., W. C. Wimley, V. K. Mishra, G. M. Anantharamiah, J. P. Segrest, and S. H. White. 1999. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J. Mol. Biol. 290 99–117. [DOI] [PubMed] [Google Scholar]
- 96.Jonas A., J. H. Wald, K. L. Toohill, E. S. Krul, and K. E. Kezdy. 1990. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J. Biol. Chem. 265 22123–22129. [PubMed] [Google Scholar]