Abstract
Phospholipid biosynthesis is a vital facet of bacterial physiology that begins with the synthesis of the fatty acids by a soluble type II fatty acid synthase. The bacterial glycerol-phosphate acyltransferases utilize the completed fatty acid chains to form the first membrane phospholipid and thus play a critical role in the regulation of membrane biogenesis. The first bacterial acyltransferase described was PlsB, a glycerol-phosphate acyltransferase. PlsB is a key regulatory point that coordinates membrane phospholipid formation with cell growth and macromolecular synthesis. Phosphatidic acid is then produced by PlsC, a 1-acylglycerol-phosphate acyltransferase. These two acyltransferases use thioesters of either CoA or acyl carrier protein (ACP) as the acyl donors and have homologs that perform the same reactions in higher organisms. However, the most prevalent glycerol-phosphate acyltransferase in the bacterial world is PlsY, which uses a recently discovered acyl-phosphate fatty acid intermediate as an acyl donor. This unique activated fatty acid is formed from the acyl-ACP end products of the fatty acid biosynthetic pathway by PlsX, an acyl-ACP:phosphate transacylase.
Keywords: acyl carrier protein, type II fatty acid synthase, acylphosphate
Bacterial phospholipid synthesis is a vital facet of bacterial physiology, and the phospholipid head group structures found in the bacterial world come in a truly bewildering variety (1). Phosphatidic acid is a universal intermediate in the biosynthesis of these membrane glycerophospholipids in eubacteria, and this review focuses on the two acyltransferase steps that are common reactions in all glycerophospholipid biosynthesis in bacteria, the glycerol-phosphate and 1-acylglycerol-phosphate (LPA) acyltransferases. These enzymes sit at the interface between the soluble type II fatty acid biosynthetic pathway and the creation of a phospholipid molecule that drives membrane expansion. This pivotal position makes the glycerol-phosphate acyltransferases key regulators of both fatty acid and phospholipid synthesis and has spurred considerable research into the function, selectivity, and regulation of the acyltransferase systems. This review will cover the two acyltransferase systems involved in bacterial glycerophospholipid synthesis, the origin and utilization of acyl donors by these pathways, and their roles in regulating membrane biogenesis.
ACYL DONORS IN ACYLTRANSFERASE REACTIONS
The most important acyl donor in bacterial glycerolipid synthesis is acyl-acyl carrier protein (ACP). ACP is a 9 kDa protein that is the acyl group carrier in type II fatty acid synthesis and shuttles the intermediates attached to the sulfhydryl group at the terminus of its 4′-phosphopantetheine prosthetic group between the pathway enzymes (2, 3). These acyl donors are the end products of the bacterial dissociated type II fatty acid synthesis pathway, and in most bacteria, type II fatty acid synthesis is the sole source of fatty acids for membrane phospholipid synthesis. An acyl-ACP intermediate has two possible fates. It can reenter the fatty acid elongation cycle and be extended by two carbons by the action of the elongation-condensing enzymes of fatty acid synthesis, or the acyl-ACP can be used by the acyltransferase system. The fate of a particular acyl-ACP chain length is determined by the competition between the elongation-condensing enzyme and the PlsB glycerol-phosphate acyltransferase for this intermediate based on their opposing substrate specificities. The 16–18 carbon acyl-ACPs are poorer substrates for the elongation-condensing enzymes than their shorter chain precursors (4), whereas 16 and 18 carbon acyl-ACPs are the preferred substrates for the acyltransferases (5), thus accounting for the preponderance of 16–18 carbon fatty acids in bacterial membrane phospholipids. The importance of the relative activities of the competing enzymes, as well as their substrate specificities, are revealed by in vivo experiments in Escherichia coli showing that the inactivation of PlsB results in the accumulation of abnormally long-chain acyl-ACPs due to the continued elongation by the condensing enzymes of fatty acid synthesis (6), whereas overproduction of the elongation-condensing enzyme FabB leads to a higher proportion of 18 carbon fatty acids in the membrane (7). Long-chain acyl-ACP is not commercially available but is efficiently synthesized from ACP using the acyl-ACP synthetase reaction (8). The idea that there are physical interactions between the enzymes of type II fatty acid synthesis enzymes, between the membrane-associated acyltransferases, or between the type II enzymes and the acyltransferases is intriguing, but to date there is no experimental support for this hypothesis.
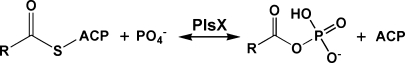
A surprising recent finding was that the most widely distributed bacterial glycerol-phosphate acyltransferase system (PlsY) uses a novel acyl donor, acyl-phosphate (acyl-PO4), produced by the PlsX enzyme (Fig. 1). Acyl-PO4 is a mixed anhydride of phosphoric acid and a fatty acid that was first synthesized in 1945 by Lehninger (9), who prepared these fatty acid derivatives to determine whether they had a role in fatty acid metabolism in mammalian cell extracts. Acyl-PO4 exhibits a higher degree of instability in water than their thioester counterparts (9). However, long-chain acyl-PO4 has a half-life of 12 h at pH 7.4 and 37°C (compared with 3 h for acetyl-PO4) (9) and is clearly sufficiently stable to play its role as an ephemeral metabolic intermediate. PlsX generates the acyl-PO4 intermediates from the acyl-ACP end products of fatty acid synthesis in a reaction analogous to phosphotransacetylase (10). The plsX gene was first recognized as a second site mutation required for the glycerol-phosphate auxotrophic phenotype of plsB mutants (11). These data suggested that plsX either encoded a second acyltransferase or was involved in glycerol-phosphate metabolism. However, only the Km defective acyltransferase activity was detected in membranes prepared from cells possessing a wild-type plsX and a mutant plsB gene, suggesting that PlsX is not an alternative glycerol-phosphate acyltransferase. The plsX gene is most often found associated with genes encoding enzymes of bacterial fatty acid synthesis, reflecting the connection between PlsX and fatty acid biosynthesis. For example, in E. coli, a fab gene cluster consists of plsX-fabH-fabD-fabG-acpP-fabF (12). The PlsX reaction is readily reversible. PlsX is a soluble protein and its crystal structure is known (Protein Data Bank accession numbers 1vi1 and 1u7n), although there is no information available on the role of specific residues in substrate binding or catalysis. However, the PlsX protein appears to associate with the Bacillus subtilis cell membrane in vivo (13), although the molecular determinants of PlsX-membrane interactions have not been explored. Bioinformatic database searches do not identify any PlsX homologs in mammalian systems.
Fig. 1.
Structure of acyl-phosphate (acyl-PO4) and its formation by PlsX. Acyl-PO4 is an anhydride between a fatty acid and phosphoric acid that is produced from the long-chain acyl-acyl carrier protein (ACP) end products of bacterial fatty acid synthesis by PlsX, an acyl-ACP:phosphate transacylase.
Some bacterial acyltransferases can use acyl-CoA as an alternative acyl donor in addition to acyl-ACP. In bacteria, acyl-CoAs are derived from exogenous fatty acids that are converted to their acyl-CoA derivatives via an acyl-CoA synthetase (FadD) following their entry into the cell (14). In both Gram-negative (15) and Gram-positive (16) bacteria, the FadD enzymes are associated with an inducible β-oxidation system that allows exogenous fatty acids to be used as a carbon source for growth. Thus, tagging the fatty acid with either CoA or ACP thioesters serves as a molecular marker to distinguish acyl chains destined for degradation from those intended for phospholipid biosynthesis, respectively. Enzymes that transfer long-chain acyl chains between acyl-CoA and acyl-ACP are not present in the bacterial models examined to date, meaning that bacteria may not generally permit exogenous fatty acids to enter the fatty acid biosynthetic pathway and do not have a mechanism to divert newly synthesized products of de novo fatty acid synthesis for degradation. In E. coli, both PlsB and PlsC use acyl-CoA thioesters as readily as acyl-ACPs, and this dual substrate specificity is extrapolated to be the pattern of substrate selectivity in the γ-proteobacteria. The advantage of acyl-CoA utilization by the acyltransferases in these organisms is that exogenous fatty acids can be used for membrane phospholipid formation in lieu of the energy-expensive fatty acid biosynthesis pathway (4, 17). In contrast, the PlsC proteins from Streptococcus pneumoniae (10) and B. subtilis (13) do not accept acyl-CoA as an acyl donor, indicating that Gram-positive bacteria have acyltransferases that cannot utilize acyl-CoA. In the case of S. pneumoniae, this property correlates with the absence of a β-oxidation system and recognizable acyl-CoA synthetases in the genome. On the other hand, B. subtilis has two acyl-CoA synthetases and a β-oxidation pathway (16), but nonetheless, the PlsC from this bacteria does not accept acyl-CoA as substrate (13). Thus, the major route for exogenous fatty acid/acyl-CoA utilization in bacteria is β-oxidation, and the ability of acyltransferases to interchangeably utilize either acyl-ACP or acyl-CoA may potentially be restricted to γ-proteobacteria. More examples of acyl-ACP-specific and dual-specificity acyltransferases need to be definitively characterized in order for refined bioinformatic tools to be developed that can predict the type of acyltransferase based on sequence information alone.
Both acyl-ACP and acyl-CoA have important roles in metabolic regulation in addition to their function as acyltransferase substrates. Long-chain acyl-CoA is a ligand for the FadR transcriptional regulator in E. coli that represses the expression of β-oxidation genes (15) and activates fabA (18) and fabB(19) gene expression in unsaturated fatty acid synthesis. The DesT transcription factor regulates the expression of genes required for oxidative acyl-CoA desaturation in Pseudomonas aeruginosa (20), and its DNA binding is regulated by the composition of the acyl-CoA pool. Unsaturated acyl-CoA binding results in a tighter association of DesT with DNA repressing desaturase transcription, whereas saturated acyl-CoA triggers the release of DesT from DNA and the induction of desaturase expression (21). Acyl-ACP is an allosteric regulator of the initiating steps in fatty acid synthesis, acetyl-CoA carboxylase (22) and β-ketoacyl-ACP synthase III (23). Modulation of the long-chain acyl-ACP concentration underlies the coordinated regulation of fatty acid, phospholipid, and macromolecular biosynthesis by the glycerol-phosphate acyltransferases (see below). These regulatory properties of the acyl-ACP and acyl-CoA donors for phospholipid synthesis in controlling bacterial lipid metabolism suggest that acyl-PO4 may also have a regulatory role in addition to its function as a biosynthetic intermediate. A regulatory function for acyl-PO4 may explain the retention of PlsX in bacteria that possess PlsB, and metabolic labeling experiments point to PlsX as a control point for the coordination of fatty acid synthesis, membrane phospholipid formation, and macromolecular synthesis in B. subtilis (13), a Gram-positive bacterium that lacks PlsB. However, the role, if any, for acyl-PO4 in metabolic regulation remains speculative.
GLYCEROL-3-PHOSPHATE ACYLTRANFERASES
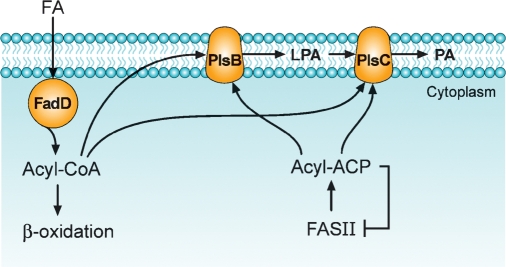
PlsB was the first glycerol-phosphate acyltransferase characterized in bacteria, and it participates in the pathway for phosphatidic acid formation outlined in Fig. 2. A novel mutagenesis strategy allowed the Bell laboratory to isolate glycerol-phosphate auxotrophs that possessed a Km defect in a membrane-associated glycerol-phosphate acyltransferase (24, 25). The availability of the plsB mutants enabled the cloning and extensive characterization of the membrane-bound enzyme that utilizes either acyl-ACP or acyl-CoA thioesters to acylate the 1-position of glycerol-phosphate (26, 27). PlsB is responsible for the selection of fatty acids incorporated into membrane phospholipids and is a key regulatory point in the pathway (4, 5, 28). The selectivity of PlsB for particular acyl chains is responsible for the positional asymmetry in the fatty acid composition of E. coli phospholipids (5), whereas the availability of glycerol-phosphate does not affect the positional distribution of acyl chains (29). In bacteria, there is a marked positional asymmetry in the incorporation of acyl chains into the 1- and 2-positions of glycerol-phosphate. The 1-position is occupied by 16:0 and 18:1 fatty acids, and the 2-position is occupied primarily by 16:1 and 18:1 fatty acids. However, the exclusion of 16:0 from the 2-position is not is not absolute, because fabA mutants that are unable to produce unsaturated fatty acids produce glycerolipids with 16:0 in both positions (30). A detailed biochemical analysis of the positional specificity of PlsB using native acyl-ACP substrates reveals that control is mainly exerted by the exclusion of 16:1 from the 1-position (5).
Fig. 2.
The pathway for phosphatidic acid (PA) formation in E. coli (Gram-negative). The acylation of glycerol-phosphate is carried out by PlsB, an integral membrane protein that transfers the acyl chain to the 1-position of glycerol-phosphate. 1-Acylglycerol-phosphate (LPA) is then acylated by the second integral membrane protein, PlsC. The acyl-ACP end products of type II fatty acid biosynthesis are the primary acyl donors in both reactions. Acyl-ACPs are feedback inhibitors of type II fatty acid synthesis (FASII), and regulation at the PlsB step coordinates fatty acid production with membrane phospholipid formation. E. coli has an acyl-CoA synthetase (FadD) that funnels exogenous fatty acids to either β-oxidation or glycerolipid synthesis via their utilization of acyl-CoA as an acyl donor for PlsB and PlsC.
There are four conserved blocks of amino acids in the PlsB class of acyltransferases. Much of what we know about these motifs has been learned from site-directed mutagenesis and kinetic analysis of the mammalian PlsB homolog in the Coleman laboratory and has been reviewed in detail (31). Briefly, motif 1 is an HX4D sequence located at amino acid 306 (E. coli PlsB numbering) that directly participates in catalysis (32, 33). The invariant aspartate sets up a charge relay system with the histidine to facilitate the deprotonation of the hydroxyl group and promote its nucleophilic attack on the acyl donor. Motif 2 (348-GAFFIRRTF) and motif 3 (383-FVEGGRSRTG) are important for binding glycerol-phosphate by interacting with the phosphate group via the arginine residues underlined in the sequences. Motif 4 (417-ITLIPIYI) is also thought to be involved in catalysis based on the inactivation of PlsB when residues in this motif are mutated. However, a structural role for proline at this position cannot be ruled out. Experimentally verifying the correct folding of membrane proteins is a difficult undertaking. Understanding the function of specific residues in the acyltransferases is important because there are several human genetic disorders that involve mutations in acyltransferase genes, such as Barth syndrome (34), rhizomelic chondrodysplasia punctata type 2 (35), and AGPAT-2-related lipodystrophy (36). PlsB has two membrane-spanning domains with the catalytic motifs separated in the C- and N-terminal halves that interact to affect catalysis (37, 38). Although this suggests that the extracellular loop (or luminal loop in mammals) has little role in catalysis, modifications in this loop do result in a loss of activity, suggesting that it has a structural role in enzyme function.
Plants have a variation on the bacterial acyl-ACP-dependent glycerol-phosphate acyltransferase that is a soluble protein localized in the plastid. These properties have allowed the determination of a high-resolution crystal structure of the glycerol-phosphate acyltransferase that informs us concerning the specific functions of the conserved amino acid blocks in this group of proteins (39–41). Motif 1 containing the HX4D motif is oriented with the carboxyl of the aspartate, which is hydrogen bonded to the histidine to leave the nonbonding electron pair on the histidine facing the active site, where the lone electron pair participates in abstracting a proton from the 1-position hydroxyl of glycerol-phosphate to activate this atom for nucleophilic attack on the acyl thioester. This configuration is reminiscent of the active site of serine hydrolases, where the histidine-aspartate pair activates the active site serine. The plant PlsB structure places the 1-position hydroxyl of glycerol-phosphate in the position of the serine hydroxyl of the hydrolases. These data strongly support the key catalytic role for the HX4D motif in acyltransferases. The phosphate of glycerol-phosphate is held in place in the plant acyltransferase structure by four basic residues arranged on two loops separated by a 42 residue spacer between each pair of basic residues. This organization is reminiscent of the 38 residue spacing between the twin arginine residues in PlsB located in motifs 2 and 3, supporting a role for these motifs in forming the phosphate binding pocket. One phosphate binding consensus sequence in plant PlsB is GGRxR, which matches the motif 3 sequence of bacterial PlsB.
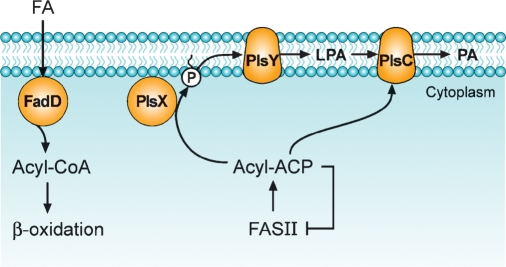
Most bacteria, including important human Gram-positive pathogens such as S. pneumoniae and Staphylococcus aureus, lack a plsB gene, and the pathway for the acylation of glycerol-phosphate proceeds as outlined in Fig. 3. The PlsY protein family (pfam02660/COG0344) encompassing the domain of unknown function, DUF205, is an acyl-PO4-dependent glycerol-phosphate acyltransferase. Most bacteria have only a single plsY gene, but some members of the Bacillus genus have multiple plsY genes. Bacillus anthracis has three plsY homologs in its genome, and it is unclear whether these three genes have the redundant functions or they catalyze acyltransferase reactions with substrates other than glycerol-phosphate.
Fig. 3.
The pathway for phosphatidic acid (PA) formation in B. subtilis (Gram-positive). The acyl-ACP end products of type II fatty acid synthesis are converted to acyl-PO4 by the soluble protein PlsX. These acyl-ACPs are feedback inhibitors of type II fatty acid synthesis (FASII), and it is thought that regulation at the PlsX step coordinates fatty acid and phospholipid synthesis. Acyl-PO4 then serves as a substrate for PlsY, an integral membrane protein that acylates the 1-position of glycerol-P. The LPA is converted to phosphatidic acid by the acyl-ACP-specific PlsC. B. subtilis has two acyl-CoA synthetases to convert exogenous fatty acids to acyl-CoA and a fatty acid β-oxidation system; however, the acyl-CoA derived from exogenous fatty acid is not used as an acyl donor in glycerolipid synthesis.
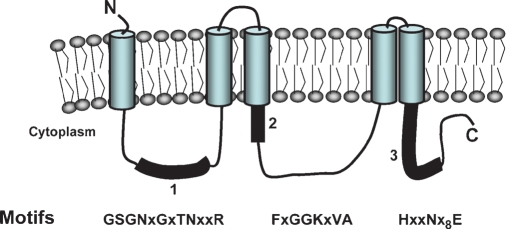
PlsY is an integral membrane protein with five membrane-spanning segments (Fig. 4) (42). There are three short, nonconserved extracellular loops providing minimal exposure of the protein to the extracellular side of the cell membrane, with three to four amino acids from the N terminus and two to four residues in extracellular loop 1 and only one to three residues in loop 2. The more extensive cytoplasmic loops contain the three conserved sequence motifs that are required for catalysis. The site-directed mutants in motif 1 located in the first cytoplasmic loop reveal that it is important for catalysis. Arginine-46 is essential for enzyme activity, and mutation of asparagine-43 impairs activity. An NxxR motif acts as the γ-phosphate binding motif in the Hsp90-ATP complex (43), suggesting a similar phosphate binding role for motif 1. Thus, motif 1 may be critical for the binding of acyl-PO4 based on the assignment of glycerol-phosphate binding to motif 2. Motif 2 in the second cytoplasmic loop is proposed as the glycerol-phosphate binding site based on the glycerol-phosphate Km defects associated with the PlsY[G102A] and PlsY[G103A] mutants. This motif is similar to the ATP binding sites of prototypical kinases that have phosphate binding loops consisting of a glycine-rich sequence followed by a lysine residue (44, 45). The glycines are conserved because the presence of side chains would interfere with substrate binding, consistent with the Km defects observed in the PlsY[G102A] and PlsY[G103A] mutants. Positively charged side chains are important for phosphate binding (46), and motif 2 contains lysine-104 downstream of the glycines. Site-directed mutagenesis demonstrates lysine-104 is essential for activity and is likely a key player in binding the phosphate of glycerol-phosphate. Motif 3 is in the C-terminal cytoplasmic segment and contains a conserved histidine that contributes to catalysis based on the compromised activity of the PlsY[H185A] mutation. The proposed role of histidine-185 in PlsY is the same as the proposed role of the conserved histidine within the HX4D motif found in the PlsB/PlsC group of acyltransferases (see above). Histidine-185 in motif 3 may act as a general base to abstract a proton from the hydroxyl group of glycerol-phosphate to facilitate the nucleophilic attack on the phosphoanhydride bond of acylphosphate. The conserved glutamate in motif 3 certainly has a role in protein folding, because the PlsY[E197A] mutant fails to correctly assemble into the membrane. However, formation of a glutamate-histidine charge relay system that is structurally important and intimately involved in catalysis cannot be ruled out.
Fig. 4.
Topology and active site motifs that define the PlsY acyltransferase family. PlsY is an integral membrane protein with five transmembrane segments. There are three domains exposed to the interior of the cell that contain conserved sequence motifs that are involved in enzyme activity. Motif 1 is proposed to govern acyl-PO4 binding, motif 2 is the site for glycerol-phosphate binding, and motif 3 is thought to be directly involved in catalysis.
There is little known about mechanisms that control the activity of PlsY and its role in regulating fatty acid and membrane lipid synthesis. PlsY from S. pneumoniae is noncompetitively inhibited by long-chain acyl-CoA (42). This regulatory property is understood within the context of bacteria, such as P. aeruginosa, that import fatty acids from the environment and convert them to acyl-CoA derivatives. These acyl-CoAs are not only used for β-oxidation but also are incorporated into membrane phospholipid by PlsB, therefore reducing the demand for acyl moieties derived from fatty acid synthesis (Fig. 1). Acyl-CoA inhibition of PlsY contributes to the utilization of exogenous fatty acids by shutting down the use of acyl moieties from fatty acid biosynthesis (acyl-ACP) when an alternative source of fatty acid (acyl-CoA) is present. However, S. pneumoniae lacks both β-oxidation and a prototypical acyl-CoA synthetase, and the PlsC in this organism does not use acyl-CoA substrates, so the relevance of this potential mechanism of regulation in most Gram-positive bacteria is questionable. The mutational inactivation of PlsY in B. subtilis leads to continued fatty acid synthesis, acyl-PO4 formation, and the accumulation of nonesterified fatty acids via the slow hydrolysis of acyl-PO4 (13). In contrast, inactivation of PlsX leads to the cessation of membrane lipid synthesis without the accumulation of fatty acids or other pathway byproducts in the cell (13). Thus, it appears that PlsX is the key regulatory point where fatty acid and phospholipid synthesis are coordinated in bacteria that lack a plsB gene. Genetic regulation of the PlsX/PlsY/PlsC pathway is puzzling. The global transcriptional regulator FapR in B. subtilis regulates plsX and plsC expression, but not plsY (47). However, in S. pneumoniae, fatty acid synthesis genes and plsC are regulated by the FabT transcriptional repressor, but plsX and plsY are not (48). The importance of these differences in transcriptional regulation in bacteria that rely on the PlsX/PlsY/PlsC pathway as the sole route to membrane phospholipids remains an important avenue to explore.
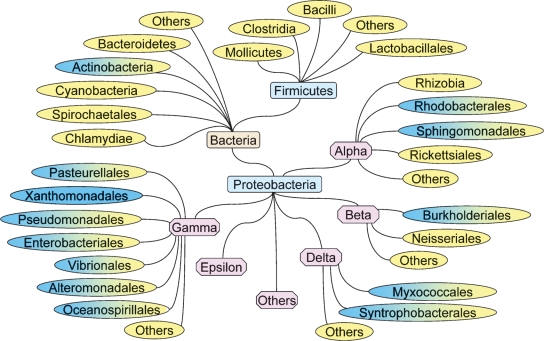
The PlsX/PlsY pathway is widely distributed in bacterial genomes based on the analysis of ∼600 finished and draft bacterial genome sequences deposited in the Integrated Microbial Genomes database (Fig. 5). The only group that did not contain PlsX/PlsY were the Xanthomonadales in the γ-proteobacteria, which have PlsB only (Fig. 5). Archaea and eukarya do not have a recognizable PlsX/Y pathway. The PlsB pathway is found in a subset of the genomes and is restricted primarily to the γ-proteobacteria (Fig. 5). The existence of PlsB in these bacteria provides them with an acyltransferase capable of utilizing acyl-CoA, which allows the incorporation of exogenous fatty acids into membrane phospholipids. The plsB gene may not be essential in many of these bacteria, most of which also contain the PlsX/PlsY system, for example, P. aeruginosa (49). On the other hand, B. subtilis lacks plsB, and both the plsX and plsY genes are essential (50).
Fig. 5.
Phylogenetic distribution of the PlsB/PlsC and PlsX/PlsY/PlsC acyltransferase systems in bacteria. Bacterial groups that contain only the PlsB/PlsC pathway to phosphatidic acid are shown in blue, those that have only the PlsX/PlsY/PlsC pathway are shown in yellow, and those that contain members that have both pathways are shown in both colors.
1-ACYLGLYCEROL-3-PHOSPHATE ACYLTRANSFERASE
A second acyltransferase, PlsC (51, 52), was discovered in E. coli and completes the synthesis of phosphatidic acid by transferring a fatty acid to the 2-position of LPA. PlsC is universally expressed in bacteria. Detailed substrate specificity studies with acyl-ACP substrates have not been reported, but PlsC appears to lack a strict substrate specificity based on the incorporation of saturated and unsaturated fatty acids into the 2-position depending on their abundance. The PlsCs from Gram-positive bacteria, like S. pneumoniae (10) and B. subtilis (13), use only acyl-ACP as the acyl donor. This is in contrast to the E. coli enzyme that is also capable of using acyl-CoA as the acyl donor (51), as do the plant (53) and mammalian (54) homologs. Thus, it appears that Gram-negative bacteria are capable of utilizing acyl-ACP, acyl-PO4, and acyl-CoA to generate membrane glycerophospholipids, whereas Gram-positive bacteria use only acyl-PO4 and acyl-ACP. Most of what can be gleaned about the architecture of PlsC in the membrane and the nature of its key catalytic residues is derived from prediction rather than from direct experiment. PlsC also contains the HX4D motif, indicating that the catalytic mechanism for activating the 2-hydroxyl of glycerol-phosphate is similar to the activation of the 1-position by PlsB. PlsC also contains a consensus sequence corresponding to motif 2 and motif 3 of PlsB containing the critical arginines proposed to anchor the phosphate of LPA. A definitive experimental description of the membrane topology of PlsC coupled with site-directed mutagenesis and enzymatic characterization of these mutants will be required to verify the predictions based on bioinformatic comparisons.
The basic PlsC structure has been adopted by bacteria to perform acyltransferase reactions that do not involve LPA. Some bacteria have multiple PlsC homologs, and their non-PlsC reactions are not always immediately obvious. Rhodobacter capsulatus (55), Pseudomonas fluorescens (56), and Neisseria meningitides (57) have two PlsC homologs that complement the E. coli plsC(Ts) growth phenotype, which suggests that they may all be LPA acyltransferases. However, in R. capsulatus, one of the complementing PlsC homologs was identified as OlsA, a protein that is involved in the final acylation step in the biosynthesis of ornithine lipids (55). All olsA genes are plsC homologs, but the OlsA proteins of Sinorhizobium meliloti (58) and P. fluorescens (56) do not have detectable LPA acyltransferase activity, illustrating that complementation in itself is not a reliable method to definitively determine function. Investigators must check the biochemical properties of the proteins and analyze mutant cells for an underlying biochemical phenotype, such as the accumulation of LPA, which indicates that a complementing gene may encode something other than a bone fide PlsC. Why some bacteria would have multiple LPA acyltransferases is puzzling. In eukaryotes, there are many of these enzymes with differing physiological functions and substrate specificities (31). It has been suggested that enzymes with different substrate specificities are required to produce lipids with defined fatty acid composition; however, extending this argument to bacteria that usually have only three to four major fatty acids may not be valid.
REGULATORY ROLES FOR THE ACYLTRANFERASES
The long-chain acyl-ACP end products of fatty acid biosynthesis have emerged as the key regulators of membrane glycerolipid biosynthesis (3, 59). The importance of long-chain acyl-ACP regulation was realized through the demonstration that acyl-ACP hydrolysis by thioesterases leads to deregulated fatty acid biosynthesis and to the release of fatty acids into the medium (60–62). There are three enzymes that are regulated by acyl-ACP: acetyl-CoA carboxylase (ACC), the initiating condensing enzyme β-ketoacyl-ACP synthase III (FabH), and a rate-controlling step in fatty acid elongation, enoyl-ACP reductase (FabI). The ACC reaction is inhibited by long-chain acyl-ACP-lowering ACC activity, thereby limiting the supply of malonate groups for chain initiation and elongation (22). FabH is also potently inhibited by mature acyl-ACP chain lengths, leading to a decrease in the initiation of new acyl chains to limit the total number of fatty acids produced (23, 63). FabI, which produces acyl-ACP, is inhibited by acyl-ACP via product inhibition (63). Because FabI has a determinant role in completing rounds of fatty acid elongation (64), reduced FabI activity slows the rate of fatty acid elongation.
The cellular acyl-ACP levels are controlled by regulation at the glycerol-phosphate acyltransferase step (Fig. 2). The blockade of PlsB activity using conditional mutants leads to the accumulation of long-chain acyl-ACP and to the cessation of fatty acid biosynthesis (28). One key physiological regulator of PlsB activity is ppGpp, a global regulator of gene expression and stable RNA synthesis in bacteria (65). Elevating the ppGpp levels in vivo inhibits PlsB activity, which in turn triggers an increase in acyl-ACP and a reduction in fatty acid synthesis (28). Thus, PlsB functions as a sensor of the status of protein synthesis to coordinate membrane phospholipid formation with macromolecular biosynthesis and cell growth. The regulatory loop also works in the other direction. The inhibition of fatty acid synthesis triggers an increase in ppGpp (66), which informs macromolecular biosynthetic systems about the status of membrane biogenesis. The mechanism of ppGpp regulation by fatty acid biosynthesis is not completely clear, but the direct interaction between ACP and ppGpp synthase II (SpoT) (67, 68) suggests the acyl-ACP pool may be involved in this process also.
The control of acyl-ACP levels in the PlsX/PlsY pathway occurs at the PlsX step. The inactivation of PlsY in B. subtilis leads to the continued production of acyl-ACPs, their conversion to acyl-PO4, and their subsequent hydrolysis to fatty acid (13). This biochemical result shows that, unlike PlsB, the PlsY acyltransferase is not positioned in the PlsX/PlsY/PlsC pathway to coordinate fatty acid and membrane phospholipid synthesis. In contrast, PlsX inactivation leads to the coordinated cessation of both fatty acid and phospholipid synthesis in B. subtilis without the accumulation of a lipid intermediate (13). This result is interpreted to reflect the accumulation of long-chain acyl-ACP in the absence of PlsX and the feedback inhibition of fatty acid synthesis at the acetyl-CoA carboxylase and FabH steps, as described for the PlsB regulatory circuit. However, these conclusions are extrapolated from a single study, and more research is needed to identify whether there are any intracellular ligands that regulate PlsX activity. Gram-positive bacteria also produce ppGpp in response to protein synthesis arrest, but it is not known whether this alarmone has a role in regulating PlsX activity.
There is no known regulatory role for the PlsC acyltransferases in bacterial physiology. The inactivation of PlsC leads to the accumulation of LPA (13, 51), which is dephosphorylated to monoacylglycerol and subsequently hydrolyzed to fatty acid (13). Thus, there appears to be no role for PlsC beyond its essential function to acylate all of the LPA that is produced by either PlsB or PlsY.
PERSPECTIVES
The focus of future research on bacterial acyltransferase systems will be the PlsX/PlsY pathway. Little is known about the biochemistry and regulation of this newly discovered system, yet it is apparent from this work that these two enzymes play a key role in regulating fatty acid and phospholipid synthesis in the majority of bacteria. Although the crystal structure of PlsX is known, almost nothing is known about the catalytic mechanism and regulation of this enzyme. Understanding this enzyme is particularly relevant in light of its proposed role in the coordination of fatty acid and phospholipid synthesis in Gram-positive bacteria (13). Some bacteria, like B. anthracis, have multiple plsY homologs in their genomes. These PlsY proteins are sufficiently distinct in sequence to suggest that they may represent acyl-PO4-dependent acyltransferases with alternative acyl acceptors. The discovery of the essential role of PlsX/PlsY in the Firmicutes also has potential medical relevance. This group of bacteria contains the major human Gram-positive pathogens and uses the PlsX/PlsY pathway exclusively for glycerolipid synthesis. These essential enzymes are unique to bacteria and thus represent two new attractive targets for the development of antibacterial agents to combat the growing problem of pathogens resistant to current antibiotics. Type II fatty acid synthesis is a validated target for antibacterial drug discovery, and the compounds identified to date are effective against multidrug-resistant pathogens. These important findings have been reviewed recently in detail (59, 69), and the success of the natural products that target membrane lipid biosynthesis at the fatty acid step suggests that compounds targeting the unique bacterial acyltransferases also hold promise in antibacterial drug discovery.
Published, JLR Papers in Press, March 27, 2008.
Footnotes
This work was supported by National Institutes of Health Grant GM-34496, Cancer Center Support Grant CA21765, and the American Lebanese Syrian Associated Charities.
References
- 1.Cronan J. E., Jr. 2003. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57 203–224. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y-M., H. Marrakchi, S. W. White, and C. O. Rock. 2003. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J. Lipid Res. 44 1–10. [DOI] [PubMed] [Google Scholar]
- 3.Rock C. O., and S. Jackowski. 2002. Forty years of fatty acid biosynthesis. Biochem. Biophys. Res. Commun. 292 1155–1166. [DOI] [PubMed] [Google Scholar]
- 4.Cronan, J. E., Jr., and C. O. Rock. 1996. Biosynthesis of membrane lipids. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. F. C. Neidhardt, R. Curtis, C. A. Gross, et al., editors. American Society for Microbiology, Washington, DC. 612–636.
- 5.Rock C. O., S. E. Goelz, and J. E. Cronan, Jr. 1981. Phospholipid synthesis in Escherichia coli. Characteristics of fatty acid transfer from acyl-acyl carrier protein to sn-glycerol-3-phosphate. J. Biol. Chem. 256 736–742. [PubMed] [Google Scholar]
- 6.Cronan J. E., Jr., L. J. Weisberg, and R. G. Allen. 1975. Regulation of membrane lipid synthesis in Escherichia coli. Accumulation of free fatty acids of abnormal length during inhibition of phospholipid synthesis. J. Biol. Chem. 250 5835–5840. [PubMed] [Google Scholar]
- 7.Garwin J. L., A. L. Klages, and J. E. Cronan, Jr. 1980. β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem. 255 3263–3265. [PubMed] [Google Scholar]
- 8.Rock C. O., and J. L. Garwin. 1979. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J. Biol. Chem. 254 7123–7128. [PubMed] [Google Scholar]
- 9.Lehninger A. L. 1945. Synthesis and properties of the acyl phosphates of some higher fatty acids. J. Biol. Chem. 162 333–342. [PubMed] [Google Scholar]
- 10.Lu Y-J., Y-M. Zhang, K. D. Grimes, J. Qi, R. E. Lee, and C. O. Rock. 2006. Acyl-phosphates initiate membrane phospholipid synthesis in Gram-positive pathogens. Mol. Cell. 23 765–772. [DOI] [PubMed] [Google Scholar]
- 11.Larson T. J., D. N. Ludtke, and R. M. Bell. 1984. sn-Glycerol-3-phosphate auxotrophy of plsB strains of Escherichia coli: evidence that a second mutation, plsX, is required. J. Bacteriol. 160 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh W., and T. J. Larson. 1992. Physical location of genes in the rne(ams)-rpmF-plsX-fab region of the Escherichia coli K-12 chromosome. J. Bacteriol. 174 7873–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paoletti L., Y-J. Lu, G. E. Schujman, D. de Mendoza, and C. O. Rock. 2007. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J. Bacteriol. 189 5816–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black P. N., and C. C. DiRusso. 2003. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol. Mol. Biol. Rev. 67 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiRusso C. C., P. N. Black, and J. D. Weimar. 1999. Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog. Lipid Res. 38 129–197. [DOI] [PubMed] [Google Scholar]
- 16.Matsuoka H., K. Hirooka, and Y. Fujita. 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 282 5180–5194. [DOI] [PubMed] [Google Scholar]
- 17.Polacco M. L., and J. E. Cronan, Jr. 1977. Mechanism of the apparent regulation of Escherichia coli unsaturated fatty acid synthesis by exogenous oleic acid. J. Biol. Chem. 252 5488–5490. [PubMed] [Google Scholar]
- 18.Cronan J. E., Jr., and S. Subrahmanyam. 1998. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29 937–943. [DOI] [PubMed] [Google Scholar]
- 19.Campbell J. W., and J. E. Cronan, Jr. 2001. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 183 5982–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu K., K-H. Choi, H. P. Schweizer, C. O. Rock, and Y-M. Zhang. 2006. Two aerobic pathways for the formation of unsaturated fatty acids in Pseudomonas aeruginosa. Mol. Microbiol. 60 260–273. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y-M., K. Zhu, M. W. Frank, and C. O. Rock. 2007. A Pseudomonas aeruginosa transcription factor that senses fatty acid structure. Mol. Microbiol. 66 622–632. [DOI] [PubMed] [Google Scholar]
- 22.Davis M. S., and J. E. Cronan, Jr. 2001. Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J. Bacteriol. 183 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heath R. J., and C. O. Rock. 1996. Inhibition of β-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271 10996–11000. [DOI] [PubMed] [Google Scholar]
- 24.Bell R. M. 1974. Mutants of Escherichia coli defective in membrane phospholipid synthesis: macromolecular synthesis in an sn-glycerol 3-phosphate acyltransferase Km mutant. J. Bacteriol. 117 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell R. M. 1975. Mutants of Escherichia coli defective in membrane phospholipid synthesis: properties of wild type and Km defective sn-glycerol-3-phosphate acyltransfersae activities. J. Biol. Chem. 250 7147–7152. [PubMed] [Google Scholar]
- 26.Lightner V. A., T. J. Larson, P. Tailleur, G. D. Kantor, C. R. H. Raetz, R. M. Bell, and P. Modrich. 1980. Membrane phospholipid synthesis in Escherichia coli: cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyltransferase. J. Biol. Chem. 255 9413–9420. [PubMed] [Google Scholar]
- 27.Green P. R., A. H. Merrill, Jr., and R. M. Bell. 1981. Membrane phospholipid synthesis in Escherichia coli: purification, reconstitution, and characterization of sn-glycerol-3-phosphate acyltransferase. J. Biol. Chem. 256 11151–11159. [PubMed] [Google Scholar]
- 28.Heath R. J., S. Jackowski, and C. O. Rock. 1994. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB). J. Biol. Chem. 269 26584–26590. [PubMed] [Google Scholar]
- 29.Goelz S. E., and J. E. Cronan, Jr. 1980. The positional distribution of fatty acids in Escherichia coli phospholipids is not regulated by sn-glycerol 3-phosphate levels. J. Bacteriol. 144 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson M. B., and J. E. Cronan, Jr. 1978. An estimate of the minimum amount of fluid lipid required for the growth of Escherichia coli. Biochim. Biophys. Acta. 512 472–479. [DOI] [PubMed] [Google Scholar]
- 31.Coleman R. A., and D. P. Lee. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43 134–176. [DOI] [PubMed] [Google Scholar]
- 32.Heath R. J., and C. O. Rock. 1998. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 180 1425–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewin T. M., P. Wang, and R. A. Coleman. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38 5764–5771. [DOI] [PubMed] [Google Scholar]
- 34.Schlame M., and M. Ren. 2006. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580 5450–5455. [DOI] [PubMed] [Google Scholar]
- 35.Ofman R., E. H. Hettema, E. M. Hogenhout, U. Caruso, A. O. Muijsers, and R. J. Wanders. 1998. Acyl-CoA:dihydroxyacetonephosphate acyltransferase: cloning of the human cDNA and resolution of the molecular basis in rhizomelic chondrodysplasia punctata type 2. Hum. Mol. Genet. 7 847–853. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal A. K., E. Arioglu, A. S. De, N. Akkoc, S. I. Taylor, A. M. Bowcock, R. I. Barnes, and A. Garg. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31 21–23. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Baro M. R., D. A. Granger, and R. A. Coleman. 2001. Mitochondrial glycerol phosphate acyltransferase contains two transmembrane domains with the active site in the N-terminal domain facing the cytosol. J. Biol. Chem. 276 43182–43188. [DOI] [PubMed] [Google Scholar]
- 38.Pellon-Maison M., R. A. Coleman, and M. R. Gonzalez-Baro. 2006. The C-terminal region of mitochondrial glycerol-3-phosphate acyltransferase-1 interacts with the active site region and is required for activity. Arch. Biochem. Biophys. 450 157–166. [DOI] [PubMed] [Google Scholar]
- 39.Tamada T., M. D. Feese, S. R. Ferri, Y. Kato, R. Yajima, T. Toguri, and R. Kuroki. 2004. Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea. Acta Crystallogr. D Biol. Crystallogr. 60 13–21. [DOI] [PubMed] [Google Scholar]
- 40.Slabas A. R., J. T. Kroon, T. P. Scheirer, J. S. Gilroy, M. Hayman, D. W. Rice, A. P. Turnbull, J. B. Rafferty, T. Fawcett, and W. J. Simon. 2002. Squash glycerol-3-phosphate (1)-acyltransferase. Alteration of substrate selectivity and identification of arginine and lysine residues important in catalytic activity. J. Biol. Chem. 277 43918–43923. [DOI] [PubMed] [Google Scholar]
- 41.Turnbull A. P., J. B. Rafferty, S. E. Sedelnikova, A. R. Slabas, T. P. Schierer, J. T. Kroon, J. W. Simon, T. Fawcett, I. Nishida, N. Murata, et al. 2001. Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase. Structure. 9 347–353. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y-J., F. Zhang, K. D. Grimes, R. E. Lee, and C. O. Rock. 2007. Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase. J. Biol. Chem. 282 11339–11346. [DOI] [PubMed] [Google Scholar]
- 43.Soti C., A. Racz, and P. Csermely. 2002. A nucleotide-dependent molecular switch controls ATP binding at the C-terminal domain of Hsp90. N-terminal nucleotide binding unmasks a C-terminal binding pocket. J. Biol. Chem. 277 7066–7075. [DOI] [PubMed] [Google Scholar]
- 44.Saraste M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15 430–434. [DOI] [PubMed] [Google Scholar]
- 45.Scholz B., S. Rechter, J. C. Drach, L. B. Townsend, and E. Bogner. 2003. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res. 31 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riordan J. F. 1979. Arginyl residues and anion binding sites in proteins. Mol. Cell. Biochem. 26 71–92. [DOI] [PubMed] [Google Scholar]
- 47.Schujman G. E., L. Paoletti, A. D. Grossman, and D. de Mendoza. 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell. 4 663–672. [DOI] [PubMed] [Google Scholar]
- 48.Lu Y-J., and C. O. Rock. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59 551–566. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 100 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA. 100 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coleman J. 1990. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J. Biol. Chem. 265 17215–17221. [PubMed] [Google Scholar]
- 52.Coleman J. 1992. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC). Mol. Gen. Genet. 232 295–303. [DOI] [PubMed] [Google Scholar]
- 53.Hanke C., F. P. Wolter, J. Coleman, G. Peterek, and M. Frentzen. 1995. A plant acyltransferase involved in triacylglycerol biosynthesis complements an Escherichia coli sn-1-acylglycerol-3-phosphate acyltransferase mutant. Eur. J. Biochem. 232 806–810. [PubMed] [Google Scholar]
- 54.Ganesh B. B., P. Wang, J. H. Kim, T. M. Black, T. M. Lewin, F. T. Fiedorek, and R. A. Coleman. 1999. Rat sn-glycerol-3-phosphate acyltransferase: molecular cloning and characterization of the cDNA and expressed protein. Biochim. Biophys. Acta. 1439 415–423. [DOI] [PubMed] [Google Scholar]
- 55.Agun-Sunar S., R. Bilaloglu, H. Goldfine, and F. Daldal. 2007. Rhodobacter capsulatus OlsA is a bifunctional enzyme active in both ornithine lipid and phosphatidic acid biosynthesis. J. Bacteriol. 189 8564–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cullinane M., C. Baysse, J. P. Morrissey, and F. O'Gara. 2005. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 151 3071–3080. [DOI] [PubMed] [Google Scholar]
- 57.Shih G. C., C. M. Kahler, J. S. Swartley, M. M. Rahman, J. Coleman, R. W. Carlson, and D. S. Stephens. 1999. Multiple lysophosphatidic acid acyltransferases in Neisseria meningitidis. Mol. Microbiol. 32 942–952. [DOI] [PubMed] [Google Scholar]
- 58.Weissenmayer B., J. L. Gao, I. M. Lopez-Lara, and O. Geiger. 2002. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 45 721–733. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y-M., and C. O. Rock. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6 222–233. [DOI] [PubMed] [Google Scholar]
- 60.Voelker T. A., and H. M. Davies. 1994. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J. Bacteriol. 176 7320–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang P., and J. E. Cronan, Jr. 1994. Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. J. Bacteriol. 176 2814–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho H., and J. E. Cronan, Jr. 1995. Defective export of a periplasmic enzyme disrupts regulation of bacterial fatty acid synthesis. J. Biol. Chem. 270 4216–4219. [DOI] [PubMed] [Google Scholar]
- 63.Heath R. J., and C. O. Rock. 1996. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271 1833–1836. [DOI] [PubMed] [Google Scholar]
- 64.Heath R. J., and C. O. Rock. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J. Biol. Chem. 270 26538–26542. [DOI] [PubMed] [Google Scholar]
- 65.Magnusson L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13 236–242. [DOI] [PubMed] [Google Scholar]
- 66.Seyfzadeh M., J. Keener, and M. Nomura. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 90 11004–11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butland G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, et al. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 433 531–537. [DOI] [PubMed] [Google Scholar]
- 68.Battesti A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62 1048–1063. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y-M., S. W. White, and C. O. Rock. 2006. Inhibiting bacterial fatty acid synthesis. J. Biol. Chem. 281 17541–17544. [DOI] [PubMed] [Google Scholar]