Abstract
Pre-existing neutralizing antibody provides the first line of defense against pathogens in general. For influenza virus, annual vaccinations are given to maintain protective levels of antibody against the currently circulating strains. Here we report that after booster vaccination there was a rapid and robust influenza-specific IgG+ antibody-secreting plasma cell (ASC) response that peaked at approximately day 7 and accounted for up to 6% of peripheral blood B cells. These ASCs could be distinguished from influenza-specific IgG+ memory B cells that peaked 14 to 21 days after vaccination and averaged 1% of all B cells. Importantly, as much as 80% of ASCs purified at the peak of the response were influenza specific. This ASC response was characterized by a highly restricted B cell receptor (BCR) repertoire that in some donors were dominated by only a few B cell clones. This pauci-clonal response, however, showed extensive intraclonal diversification from accumulated somatic mutations. We used the immunoglobulin variable regions isolated from sorted single ASCs to produce over fifty human monoclonal antibodies (mAbs) that bound to the three influenza vaccine strains with high affinity. This strategy demonstrates that we can generate multiple high affinity mAbs from humans within a month after vaccination. The panel of influenza virus specific human mAbs allowed us to address the issue of original antigenic sin (OAS) - the phenomenon where the induced antibody shows higher affinity to a previously encountered influenza virus strain compared to the virus strain present in the vaccine1. However, we found that the vast majority of the influenza virus specific mAbs showed the highest affinity for the current vaccine strain. Thus, OAS does not seem to be a common occurrence in normal healthy adults receiving influenza vaccination.
Influenza causes 36,000 deaths annually in the United States alone and the influenza pandemic of 1918 caused an estimated 50 million deaths worldwide2. Outbreaks of avian influenza infections in human populations that caused substantially higher mortality rates foresee the possibility of another deadly pandemic3. The challenge of influenza has long been to design vaccines that induce long lasting immunity against a pathogen that rapidly alters its appearance to the immune system by mutating (antigenic drift) and exchanging (antigenic shift) its components. Antibodies play a key role in protection against influenza infection4-7. However, the underlying B cell response leading to the rapid production of ASCs that secrete antibodies is only beginning to be understood8-12. Critically, we do not yet know if B cell memory can provide sufficient protection early in the response to counteract variant strains of influenza or if rather the response is dominated by antibodies previously generated against divergent viruses in an OAS fashion. Finally, of profound clinical significance is the possibility that the early ASC response observed after immunization can be exploited to rapidly generate therapeutic or diagnostic mAbs to emerging influenza virus strains, or in fact to any immunizing antigen.
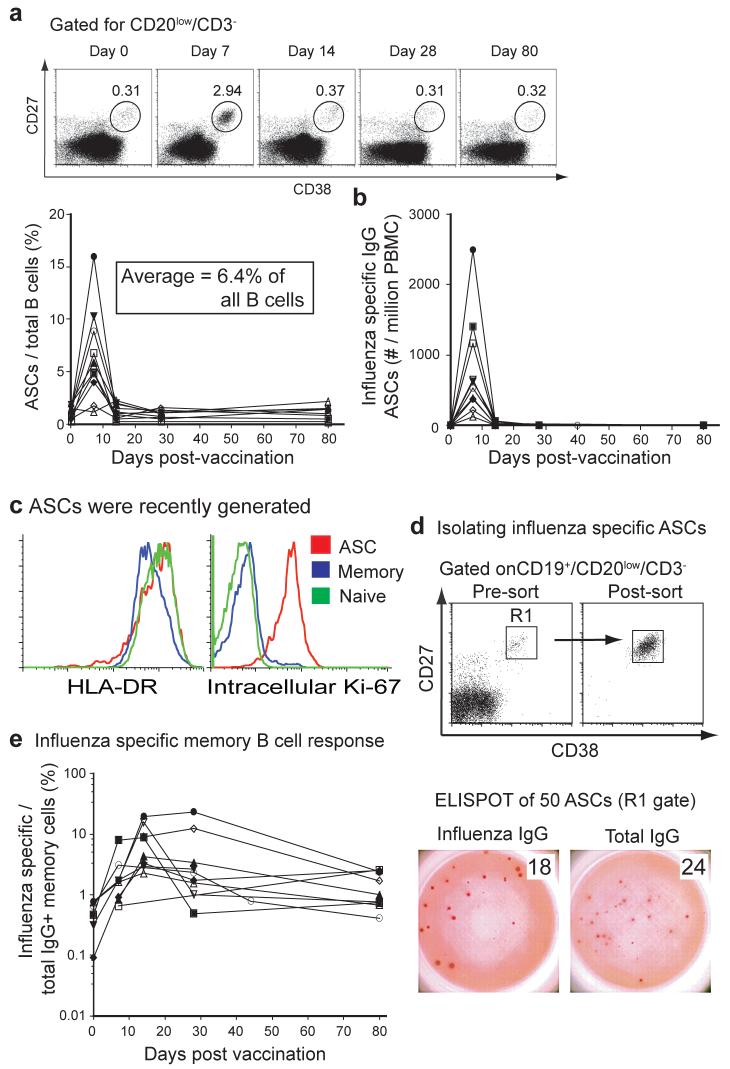
In order to determine the dynamics and magnitude of the human anti-influenza response we analyzed the frequency of ASCs and memory B cells in a time-course following vaccination. The ASC response was quite transient, peaking at approximately day seven and returning to barely detectable levels by day 14 after vaccination (Fig. 1a and 1b). The frequency of influenza-specific ASCs averaged 6.4% (or ∼2,500 ASCs per ml of blood) at day 7, and accounted for up to 16% of all B cells (range for ten donors: 1.1-16%, Fig. 1b). Also, most of these ASCs were generated during the vaccination response as they were almost entirely Ki-67 positive, indicating recent proliferation, and most expressed homogenously high levels of HLA-DR13 (Fig. 1c). Importantly, analysis of IgG secreting ASCs isolated by cell sorting at day 7 post-immunization demonstrated that the vast majority were influenza vaccine specific (ranging from 20-85% and averaging 70%, Fig. 1d). The ASCs were mainly IgG positive, with minor components of IgA and IgM positive cells (data not shown), suggesting an origin from the memory B cell compartment. The memory B cell response was also quantified14. Increasing from low levels prior to vaccination, influenza-specific memory B cells peaked a week after the ASC response at 14 to 28 days after vaccination and averaged 8.2% of the IgG+ memory B cells or ∼1% of all B cells (Fig. 1e). We conclude that influenza vaccination results in a massive burst of IgG+ ASCs that are predominantly influenza-reactive and peak at approximately day 7 post-immunization.
Figure 1. Analysis of the B cell response induced by influenza vaccination.
a, PBMCs collected from 10 donors were assayed for influenza specific IgG secreting ASCs by ELISPOT assay at days 0, 7, 14, 28 and 80 days after vaccination. Each sample was measured in duplicate, averaged and plotted as ASC/106 PBMCs over time post-vaccination. b, ASCs were measured in blood by flow cytometric analysis. Shown is the frequency of the ASC gate (CD3-/CD20-/low/CD19+/CD27hi/CD38hi) for a representative donor and a summary for all ten donors normalized to total CD20+/CD19+ B cell numbers. c, HLA-DR and Intracellular expression of Ki-67 by ASCs compared to naive or memory B cells. d, The majority of ASCs at day 7 after influenza vaccination are influenza specific. Influenza and total IgG-specific ELISPOT assays from several donors were similar. e, Percentage of influenza specific memory cells per total IgG positive memory cells after mitogen stimulation as measured by ELISPOT at days 0, 7, 14, 28 and 80 days post vaccination as previously described14.
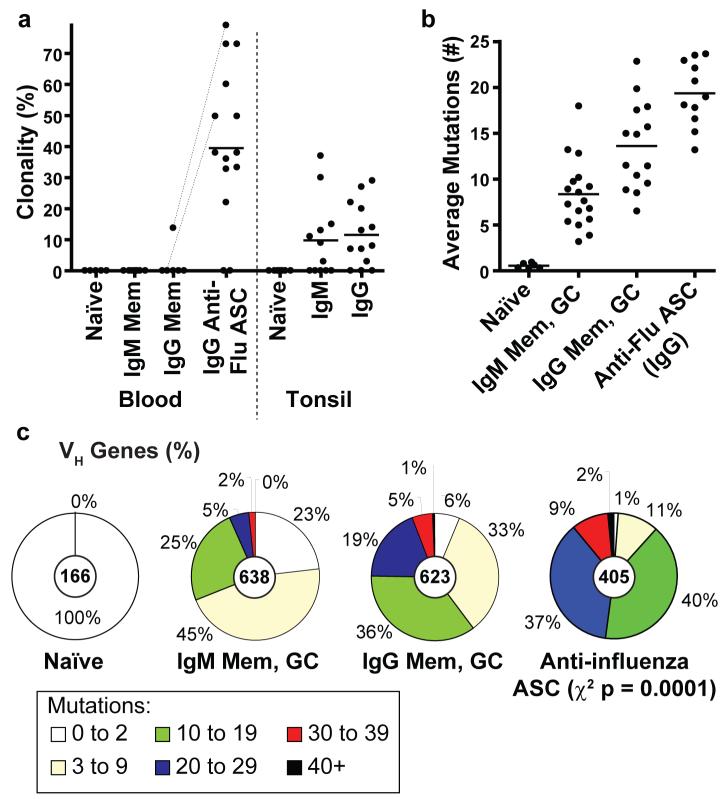
The rapid accumulation of ASCs suggests that the response could be highly clonal in nature, limiting the early influenza response. Some clonal activation of ASCs occurs after tetanus vaccination12. We therefore analyzed the immunoglobulin repertoire breadth (i.e., the variable genes and junctional diversity) of the influenza specific ASCs. Influenza vaccination caused a surprisingly pauci-clonal response, with some donors being dominated by the progeny of only a few expanded B cell clones (Supp. Fig. 1a and Fig. 2a). Clonal expansions accounted for 43% of the ASC variable regions from the 14 immunized donors including three donors with over 70% clonality (Fig. 2a). In stark contrast, based on VH regions sequenced from our laboratory in a comparable fashion15-17, naïve and memory B cells (IgM or IgG) isolated from blood were rarely or never clonal, while for tonsillar B cells only 10% of IgM and 12% of IgG GC and memory cells were clonally related.
Figure 2. The ASC response after influenza vaccination is pauci-clonal and highly diversified by somatic hypermutation.
a, Comparison of the mean proportion (line) of all clonal variable region sequences from day 7 ASCs of 14 donors (points), including: the bulk RNA of 104 to 105 ASCs from 10 donors and verification by single cell RT-PCR for 4 donors (average 37 sequences per donor). The ASCs were the most clonally-related population (t-test p ≤ 0.0003). Dotted lines indicate donors from which memory and ASCs were analyzed simultaneously. Other B cell populations were from historical data analyzed in a similar fashion from our laboratory15-17 (see Methods, Supplemental data at Nature online) b, Each point is the average frequency of somatic mutations per sequence from each donor (n values within Methods). On average the anti-influenza ASCs had accumulated more mutations than either the IgG (t-test p =0.003) or IgM (p = <0.0001) memory and GC populations. c, Indicated is the proportion of all variable genes from each B cell population with the number of somatic mutations denoted in the legend (n-values are at the center of each pie chart).
Immunoglobulin variable gene somatic hypermutation allows for the generation of high affinity antibodies18,19. Surprisingly, the influenza specific ASCs had accumulated more somatic mutations than any normal population of B cells. Considering the various donors (Fig. 2b), the ASCs averaged 19.4 ± 3.5 VH gene mutations, which is greater than that of germinal center or memory B cells that average 13.6 ± 4.8 mutations for IgG or 8.4 ± 3.8 mutations for IgM. A surprising 11% (41/405) of the ASC VH gene segments have more than 30 of 300 (or ∼10%) of the total nucleotides altered (Fig. 2c). A preference for CDR replacement mutations suggests that the ASCs were functionally selected (Sup. Table 1). The observations herein suggest that the origin of the anti-influenza ASCs is predominantly memory B cells that probably accumulated new mutations on this and on previous rounds of activation.
It is not known how often the ASCs that are induced by vaccination produce high affinity antibodies against influenza. Immunoglobulin variable region genes from ASCs can be used to express specific antibodies20. We therefore used the variable gene transcripts of isolated single ASCs to express recombinant monoclonal antibodies in the human 293 cell line (Sup. Fig. 1b and methods). From day 7 post vaccination ASCs of five donors, 71% (61/86) of the antibodies bound with high affinity to either native antigens of the influenza vaccine strains (53/86 or 61%), or to components of the vaccine only (8/86 or 9%) (Fig. 3, Table 1 and Sup. Fig. 2). We suspect that the epitopes found only in the vaccine are exposed on the fixed virions or are from added preservatives. In comparison, none of the 86 mAbs generated from naïve B cells15 (Fig. 3b) and only one of 54 antibodies from random IgG memory B cells bound to the influenza vaccine strains with appreciable affinity (data not shown). The antibodies produced from the influenza specific ASCs bound to any of the three vaccine components with similar frequency (Table 1 and Sup. Fig. 2). Analysis of viral antigen specificity by immunoprecipitation and Western blot (Sup. Fig. 3) found that 60% of the influenza-reactive antibodies bound to HA, of which half were hemagglutination-inhibiting (Fig. 3c and Table 1). Twelve percent of the antibodies bound to neuramininidase (NA) or to other minor components of the vaccine likely residual to the purification of HA and NA during vaccine production. Ten percent of the antibodies did not precipitate native antigens and bound only to epitopes on denatured viral proteins detectable by Western blot. Importantly, each of three representative HAI+ antibodies against influenza-A (anti-H3N2) and one against influenza-B from the day 7 ASCs (Table 1, bold) were found to neutralize viral infection of MDCK cells in vitro (each neutralized virus at <1ug/ml antibody, Sup. Fig. 4). In conclusion, after influenza vaccination early ASCs produce functional antibodies that bind with high affinity and likely provide early protection.
Figure 3. High affinity mAbs generated from single influenza specific ASCs.
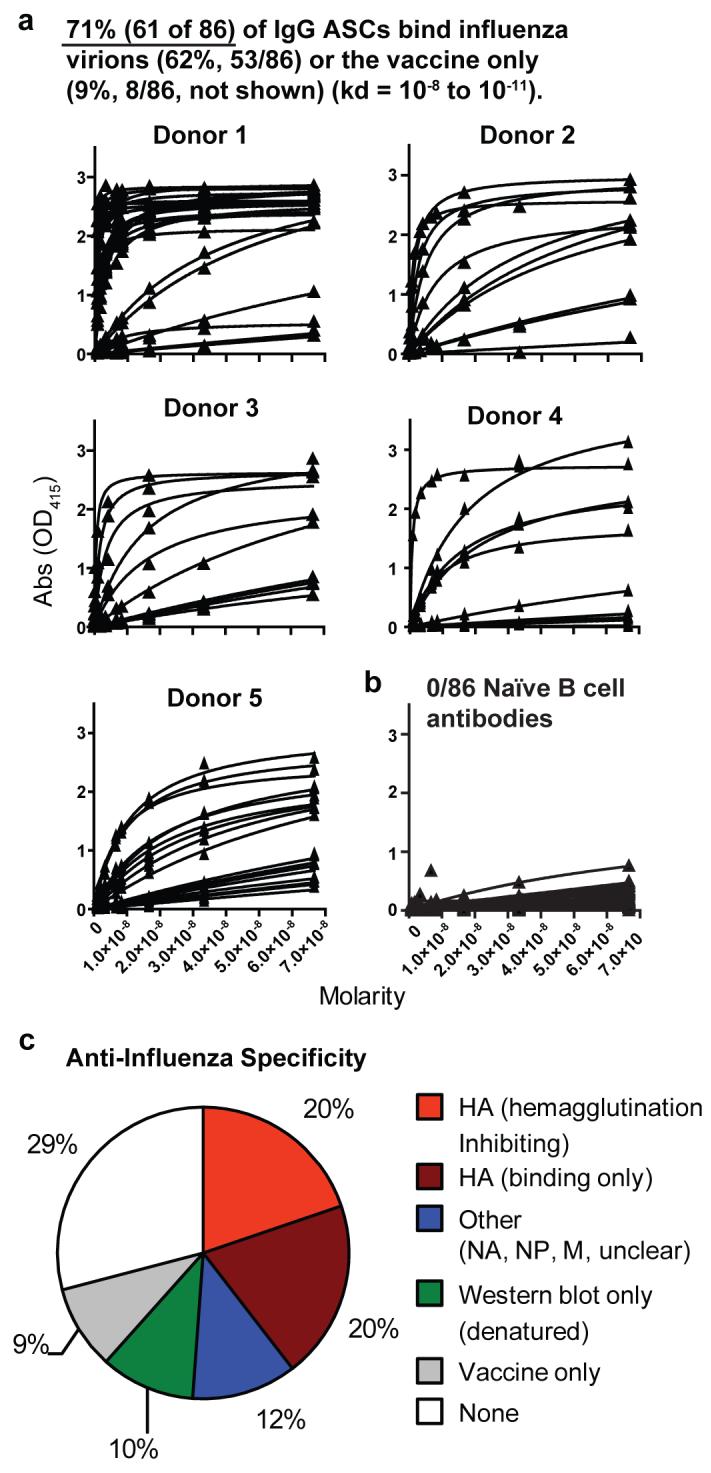
a, Recombinant monoclonal antibodies from day 7 IgG anti-influenza ASCs (Sup. Fig. 1b) bind to a mixture of the three influenza vaccine strain virions with high affinity. In total, 71% of the ASC antibodies bound either native antigens of influenza viruses freshly grown in eggs (62% or 53/86), or to antigens within the vaccine only (9% or 8/86, not shown). Each of the five donors were influenza specific (by donor 34, 13, 11, 15, and 21 antibodies were generated of which 45% to 85% were influenza specific). Individual antibody strain specificities are in Table 1 and Sup. Fig. 2. b, 0/86 naïve B cell antibodies bound influenza. c, Analysis by immunoprecipitation and Western blot (Sup. Fig. 3) identified the specific viral antigens bound. Hemagglutination assays were used to identify those antibodies that were inhibiting (Table 1 and Methods).
Table 1.
Characteristics of anti-Influenza antibodies
| H1N1: A/New Caledonia/20/99 | ||||
|---|---|---|---|---|
| Antibody | Kd(M) | HAI | Antigen | |
| Donor 1 | D1-1 | 2.70E-10 | None | HA |
| D1-2 (×2)* | 7.33E-10 | None | HA | |
| D1-3 (×2)* | 1.55E-09 | None | HA | |
| D1-4 | 2.14E-09 | None | HA | |
| D1-5 | 2.66E-09 | 1 | HA | |
| D1-6 | 3.62E-09 | 1 | HA | |
| Donor 2 | D2-1 | 2.73E-09 | None | 85 kDa band on Western |
| D2-2 | 5.42E-09 | None | 85 kDa band on Western | |
| D2-3 | 5.20E-09 | None | HA | |
| Donor 3 | D3-1 | 1.88E-09 | None | 85 kDa band on Western |
| Donor 4 | D4-1 | 4.00E-08 | None | 85 kDa band on Western |
| D4-2 | 1.65E-08 | None | HA | |
| Donor 5 | D5-1 | 5.01E-11 | None | Multiple bands on Western |
| D5-2 | 1.01E-09 | None | Denatured HA1 on Western | |
| D5-3 | 1.78E-08 | None | NA | |
| D5-10 | 1.78E-08 | None | Multiple bands on Western | |
| H3N2: A/Wisconsin/67/2005(2006/7) or A/California/7/2004 (for Donor 3) | ||||
| Donor 1 | D1-7 | 7.72E-11 | 128 | HA |
| D1-8 | 2.86E-10 | 4 | HA | |
| D1-9 (×4) * | 3.77E-10 | 8 | HA | |
| D1-10 | 4.18E-10 | 4 | HA | |
| D1-11 | 1.57E-09 | None | NP | |
| Donor 2 | D2-4 | 3.62E-10 | 2 | HA |
| D2-5 | 8.29E-09 | None | Unclear | |
| Donor 3 | D3-2 | 3.50E-09 | None | HA |
| D3-3 | 1.56E-08 | None | HA | |
| D3-4 | 4.86E-10 | 32 | HA | |
| Donor 4 | D4-3 | 4.56E-09 | 1 | HA |
| B strains: B/Malaysia/2506/2004 or B/Shanghai/361/2004 (Donors 3 and 6) | ||||
| Donor 1 | D1-12 | 1.93E-10 | None | HA |
| D1-13 | 2.04E-10 | 16 | HA | |
| D1-14 (×2)* | 2.43E-10 | None | HA | |
| D1-15 | 2.47E-10 | 8 | HA | |
| D1-16 | 6.20E-10 | None | HA | |
| D1-17 | 6.33E-10 | None | HA | |
| D1-18 | 4.74E-10 | None | HA | |
| D1-19 | 3.70E-08 | None | M on Western | |
| Donor 2 | D2-6 | 1.71E-09 | None | Unclear |
| D2-7 | 5.77E-08 | None | M on Western | |
| D2-8 | 3.66E-08 | None | HA | |
| Donor 3 | D3-5 | 1.66E-08 | None | Unclear (low affinity) |
| D3-6 | 1.62E-08 | None | Denatured NP Western | |
| Donor 4 | D4-4 | 1.71E-08 | 1 | HA |
| D4-5 | 3.28E-08 | 4 | HA | |
| Donor 5 | D5-5 | 3.78E-08 | None | Unclear (low affinity) |
| D5-6 | 1.11E-08 | None | M on Western | |
| D5-7 | 1.26E-08 | None | Unclear (low affinity) | |
| D5-8 | 4.8E-08 | None | 85 kDa band on Western | |
| Donor 6 | D6-1 | 5.04E-10 | 4 | HA |
Clonal expansions with number of clones indicated; Bold: mAbs tested for viral neutralization
Although most of the ASCs arise only after vaccination (Fig. 1a), twenty-nine percent of the antibodies generated did not detectably bind to the influenza strains or whole vaccine (Fig. 3c). Possible causes include errors introduced by the RT-PCR steps (though PCR errors were rare, Sup. Table 2), targeting of non-viral or denatured components of the vaccine or antigens only evident physiologically, bystander activation of non-specific memory cells8, or displacement of non-specific plasma cells from the bone marrow13. The latter possibility is unlikely as expression of HLA-DR13 and Ki-67 (Fig. 1c) by the ASCs suggests they were newly generated.
The long held theory of OAS suggests that new influenza variants will evade surveillance when memory B cells reactive to previous viral strains dominate the response1. In order to consider the impact of OAS directly, we compared the relative affinity to either the current B strain virus (B/Malaysia/2506/2004) or to the two previous ones (B/Shanghai/361/2002 or B/Hong Kong/33/2001, Fig. 4a and b). In the 2006/7 season, antibodies were analyzed from five donors that had also been vaccinated in the 2005/6 season and one in 1991 so that reactive memory cells should be readily available for an OAS response. Importantly, each of the 19 anti-B strain antibodies bound to the new B strain with equal, and in most cases with greater affinity than the previous vaccine strains (Fig. 4c and Sup. Fig. 2). This adaptation occurred despite the only 10% or less difference of the HA sequence of the 2006/7 B strain from those used in previous vaccines. Although previous exposure to B/Malaysia/2506/2004 cannot be entirely ruled out, there was no history of exposure and pre-vaccination serum titers of antibody against B/Malaysia/2506/2004 were not above background levels (data not shown). Thus we conclude that even for the earliest detectable influenza-specific B cells after vaccination, the ASCs, OAS does not limit reactivity to newly introduced influenza strains.
Figure 4. Specificity for the newly introduced influenza B strain in the vaccine suggests a minimal impact of OAS.
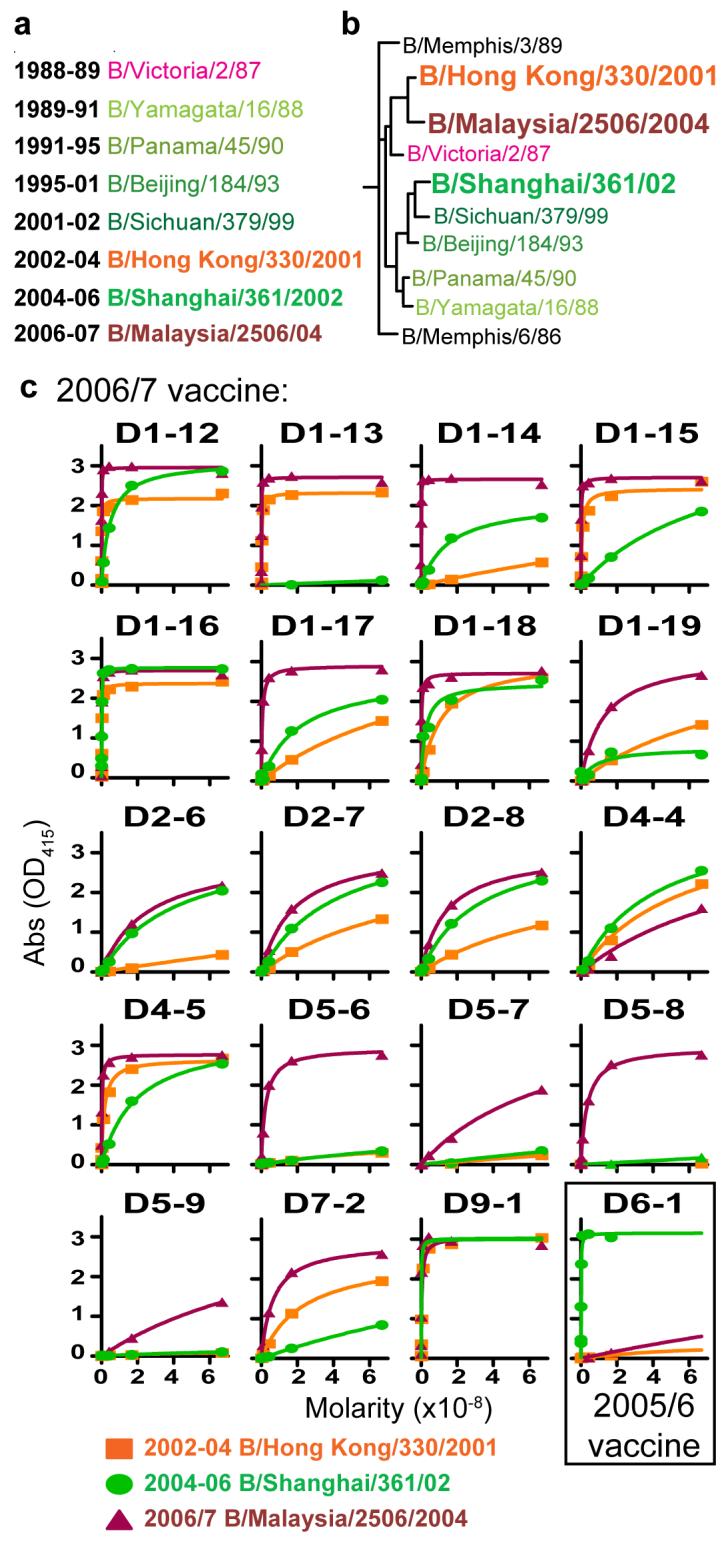
a, Influenza B strains used for the vaccine since 1989. Throughout the Figure, strain names are color coded for the Yamagata lineage (green) and the Victoria lineage (orange/red). b, Phylogenetic tree illustrating the similarity of recent influenza B strains and the years in which each strain was included in the vaccine. The three vaccine strains tested herein (larger font) included B/Malaysia/2506/2004 (2006/7 season) that is most similar to the 2002-2004 strain (B/Hong Kong/33/2001). Conversely, the 2005/6 vaccine strain, B/Shanghai/361/2002, is more divergent. c, All anti-B strain antibodies reacted with equal or greater affinity to the current year’s vaccines when tested by ELISA.
In conclusion, we show that after influenza vaccination we can isolate an almost entirely antigen specific population of ASCs that comprise about 5% of all blood-borne B cells. Further, the findings herein help to resolve a major obstacle longstanding in the field of medicine21: the rapid production of fully human monoclonal antibodies. Antibody or serum therapy has been demonstrated to effectively treat a plethora of diseases, but is not widely used because sometimes fatal anaphylactic responses and serum sickness are common. These obstacles can only be overcome by using fully human mAbs. The findings herein demonstrate that we can now generate human monoclonal antibodies from the antigen-specific ASCs directly, and within only weeks of vaccination (Supp. Fig. 1C). With a modern resurgence of interest in monoclonal antibody therapy we anticipate that antibodies produced from post-vaccination ASCs will generate substantial advances for the treatment of infectious diseases.
Conventional wisdom holds that the level of pre-formed antibody is the main correlate of protection against influenza virus. However, our results showing the rapidity of the antibody response after vaccination and the high affinity of the antibodies produced strongly suggests that the recall response could also play a role in protective immunity. This antibody would, of course, not prevent initial infection but could play a crucial role in preventing the spread of virus and bringing about faster resolution of the infection. This notion is supported by our finding that OAS was not a significant aspect of the memory response as the antibodies produced were highly specific to the immunizing antigen.
Summary of Methods
Detailed Methods are found in the supplementary data. Healthy volunteers received either the Fluzone 2005-06 (Aventis Pasteur Inc) or Fluvirin 2006-07 (Chiron) influenza vaccine formulations. This project has IRB approvals from both Emory and OMRF. Enumeration of IgG or influenza-specific B cells were previously described, including: ELISPOT22 and polyclonal activation of memory B cells14,22. HAI titers, inhibiting antibody concentrations, and viral neutralization were determined by standard procedures23. mAbs were tested for HAI at 30 μg/ml and five 2-fold serial dilutions. The ASCs were identified herein as CD3-/CD20-/low/CD19+/CD27hi/CD38hi cells. Isolation of other B cell types and methods for RT-PCR analysis of the variable genes were done as previously described15,17. An average of 35 variable gene sequences was analyzed from each donor (Supplemental methods). The leader and constant regions that are rarely targeted by somatic mutation were primed for RT and PCR to avoid biases between populations for the variable gene repertoire and clonality determinations. The single cell RT-PCR methods and the procedures for production of recombinant mAbs were as previously described15,24. Monoclonal antibodies were screened against fresh influenza virions grown in chicken eggs. Antibody affinities (Kd) were calculated by nonlinear regression analysis (GraphPad Prism software) of influenza ELISA curves plotted from 8 dilutions of antibody ranging from 10 ug/ml to 0.125 ug/ml.
Supplementary Material
Acknowledgements
We thank Adam Popkowski, Hong Wu, Leni Abraham and Beth Begley for technical assistance and Rafael Casellas and John Knight for critical reading of this manuscript. This work was funded in parts by NIH grant #s: HHSN266200500026C (PCW), P20 RR018758 (PCW), NIH/NIAID U19-AI057266-04 (RA), NIH/NIAID HHSN266200700006C Center of Excellence for Influenza Research and Surveillance (RA), NIH/NIAID N01-AI-50025-02 (RA and CL). JW was supported by a postdoctoral fellowship from The Swedish Research Council.
Appendix
Online Methods
Cell and serum isolation
All studies were preapproved by the Emory and OMRF review boards. Healthy volunteers received influenza vaccine formulations (Fluzone, Aventis Pasteur, 2005/6, or Fluvirin, Chiron, 2006/7). Peripheral blood mononuclear cells (PBMC) were isolated using Vacutainer tubes (Becton Dickinson, BD) or lymphoprep gradient (CellGro), washed, and resuspended in supplemented culture media or PBS. Plasma was heat inactivated.
ELISPOT and Memory B cell assay
ELISPOT and memory assays were previously described14,25 and detailed in the supplemental data. Total IgG secreting or influenza specific ASCs were detected using 1/20 diluted influenza vaccine in PBS (as above) or with goat anti-human Ig (Caltag). Dilutions of washed PBMCs incubated in supplemented RPMI for 2 hrs were incubated in ELISPOT plates for 6 hours. After washing the plates, ASC antibody was detected with anti-huIgG-biotin (Caltag) and avidin-D-HRP (Vector Laboratories) and developed with AEC substrate (Sigma) before analysis on an ELISPOT counter (Cellular Technologies Ltd.). Memory cells were detected by incubating PBMCs at 5×105 cells/ml in R-10 supplemented with pokeweed mitogen extract (PWM), phosphothiolated CpG ODN-200626, and Staphylococcus Aureus Cowan (SAC) (Sigma). After culture for 6 days the cells were washed and quantified by ELISPOT assay.
Flow cytometry and cell sorting
Flow cytometry analysis was performed on whole blood following lysis of erythrocytes. Mostly Pharmingen antibodies were used for quantifying ASC or memory cells (Fig. 1) except anti-CD27-APC (ebiosciences) and goat anti-huIgG-FITC (Southern Biotechnologies). For single cell analysis and production of mAbs, antibodies used included anti-CD3-FITC, anti-CD20-FITC, anti-CD38-APC-Cy5.5, anti-CD27-PE, anti-IgG-Alexa-647, and anti-CD19-PE-Alexa 610 from Caltag, plus anti-IgD-biotin and strepavidin-Pe-Cy7 (Pharmingen). ASCs were gated as IgG+IgD-CD19+CD3-CD20lowCD27high CD38high. All other cell types were isolated as previously described15-17. Cytometry data was analyzed using FlowJo software
Single cell RT-PCR and PCR of antibody variable region genes
As detailed in the supplemental data, single B cells were sorted into 96-well PCR plates containing RNase inhibitor (Promega). VH and Vκ genes from each cell were amplified by RT-PCR and nested PCR reactions using cocktails of primers as previously described15,24 and then sequenced. To generate recombinant antibodies, restriction sites were incorporated by PCR with primers to the particular variable and junctional genes. RT-PCR of bulk RNA to analyze V genes was as previously described15,17,29.
Analysis of clonality and somatic mutations of variable region genes
To quantify clonality, variable genes were randomly sequenced from the bulk RNA of ASCs from 10 donors (by donor, n = 22, 47, 49, 12, 16, 19, 36, 25, 34, and 63) and verified by single cell RT-PCR analysis of ASCs from four donors (n = 65, 37, 30, and 50). Naïve, Memory, and germinal center (GC) cell variable gene libraries included the following VH gene n-values: Blood naïve (by donor n = 61, 24, 15, 14, and 24), blood IgM memory (by donor n = 28, 17, 27, 11, 23, 12, 29, and 20), blood IgG memory (by donor n = 23, 18, 18,17, 22, and 21), tonsillar naïve B cells (by donor n = 125, 32, 16, 22, 32, 23, 46, and 81), tonsillar IgM and GC/memory (by donor n = 50, 42, 35, 16, 60, 15, 50, 25, 39, 19, 55, and 58 VH genes), and tonsillar IgG GC/memory (by donor n = 113, 25, 14, 40, 12, 41, 11, 23, 18, 51, 15, 54, and 69). The n-values for analysis of somatic hypermutation included: Anti-influenza ASCs from 11 donors (n = 63, 18, 33, 46, 49, 11, 36, 11, 30, 35, 25); IgG GC/memory cells from 14 donors (n = 110, 37, 19, 28, 174, 40, 25, 15, 21, 18, 22, 24, 19, 71); IgM GC/memory from 17 donors (by donor, n = 56, 158,18, 91, 17, 10, 16, 30, 19, 28, 11, 36, 29, 13, 22, 20, 64); and naïve cells from 6 donor (by donor, n = 18, 42, 21, 34, 15, 36). Background mutations rates were insignificant (Sup. Table 2).
Recombinant monoclonal antibody expression and analysis
All assays are further detailed in the supplement at Nature Online. VH or Vk genes amplified from each single cell were cloned into IgG1 or Igκ expression vectors as previously described15,24. Heavy/light chain plasmids were cotransfected into the 293A cell line for expression and antibodies purified with protein A sepharose. The influenza virus strains used for ELISA or HAI were freshly grown in eggs and purified by standard methods23 and included: A/New Caledonia/20/9 (H1N1), A/California/7/2004 (H3N2) for 2005/6 or A/Wisconsin/67/2005 (H3N2) for 2006/7, and B/Shanghai/361/2002-like for 2006/2007or B/Malaysia/2506/2004 for 2006/7. Serum or mAb HAI titers were determined as previously described27. After ELISA screening with a cocktail of all influenza strains and 1/20 dilutions of the vaccines, the affinity and specificity of binding-positive mAbs were determined with the individual influenza viruses. ELISA affinities were calculated by nonlinear regression analysis of curves from 8 dilutions of antibody (10 to 0.125 ug/ml) using GraphPad Prism. Influenza neutralizing activity was detected as inhibition of MDCK cell death by 50% Tissue culture infectious doses of A/Wisconsin/67/2005 or B/Shanghai/361/2002 based on the WHO manual protocol23.
Immunoprecipitation and Western Blot analyses
All assays are further detailed in the supplement at Nature Online. For IP, 8 HAU of virus were lysed and incubated with 10ug/ml of mAb before purification with Protein A-Sepharose (Pierce). mAb was eluted from the protein-A by boiling in Laemmli buffer (Bio-Rad) and analyzed on 12% Tris-Glycine polyacrylamide gels. Protein was detected by staining the gels with sypro-orange (1X, Invitrogen). For Western blots, 8 HAU of virus was diluted and boiled in denaturing/reducing sample buffer, then run on denaturing polyacrylamide gels (as above) followed by electrophoretic transfer to nitrocellulose membranes. The membranes were incubated with each Ab at 5ug/ml and detected with HRP anti-human IgG (Jackson Immunoresearch) and developed with ECL plus reagent (GE health care). IP gels and Western blot membranes were analyzed using a STORM840 system (Molecular Dynamics).
Statistics
Statistical analyses (described in context) were performed using GraphPad Prism: Frequencies of clonal relatedness and somatic mutation were compared by non-paired, 2-tailed student’s t-tests and Chi-square tests were used to compare summed mutation frequencies.
Footnotes
The authors declare no competing financial interests.
References
- 1.Francis Thomas., Jr On the Doctrine of Original Antigenic Sin. Proceedings of the American Philosophical Society. 1960;104(6):572–578. [Google Scholar]
- 2.Ahmed R, Oldstone MB, Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8(11):1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subbarao Kanta, Joseph Tomy. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol. 2007;7(4):267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerhard W, et al. Role of the B-cell response in recovery of mice from primary influenza virus infection. Immunol Rev. 1997;159:95–103. doi: 10.1111/j.1600-065x.1997.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 5.Luke Thomas C., Kilbane Edward M., Jackson Jeffrey L., Hoffman Stephen L. Meta-Analysis: Convalescent Blood Products for Spanish Influenza Pneumonia: A Future H5N1 Treatment? Ann Intern Med. 2006;145(8):599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 6.Puck JM, Glezen WP, Frank AL, Six HR. Protection of infants from infection with influenza A virus by transplacentally acquired antibody. J Infect Dis. 1980;142(6):844–849. doi: 10.1093/infdis/142.6.844. [DOI] [PubMed] [Google Scholar]
- 7.Simmons Cameron P., et al. Prophylactic and Therapeutic Efficacy of Human Monoclonal Antibodies against H5N1 Influenza. PloS Medicine. 2007;4(5):e178. doi: 10.1371/journal.pmed.0040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298(5601):2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 9.Brokstad KA, et al. Parenteral influenza vaccination induces a rapid systemic and local immune response. J Infect Dis. 1995;171(1):198–203. doi: 10.1093/infdis/171.1.198. [DOI] [PubMed] [Google Scholar]
- 10.Brokstad KA, et al. Parenteral vaccination against influenza does not induce a local antigen-specific immune response in the nasal mucosa. J Infect Dis. 2002;185(7):878–884. doi: 10.1086/339710. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki Sanae, et al. Comparison of the Influenza Virus-Specific Effector and Memory B-Cell Responses to Immunization of Children and Adults with Live Attenuated or Inactivated Influenza Virus Vaccines. J Virol. 2007;81(1):215–228. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulsen TR, et al. Kinetic, affinity, and diversity limits of human polyclonal antibody responses against tetanus toxoid. J Immunol. 2007;179(6):3841–3850. doi: 10.4049/jimmunol.179.6.3841. [DOI] [PubMed] [Google Scholar]
- 13.Odendahl M, et al. Generation of migratory antigen-specific plasma blasts and mobilization of resident plasma cells in a secondary immune response. Blood. 2005;105(4):1614–1621. doi: 10.1182/blood-2004-07-2507. [DOI] [PubMed] [Google Scholar]
- 14.Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J Immunol Methods. 2004;286(12):111–122. doi: 10.1016/j.jim.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Koelsch Kristi, et al. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117(6):1558–1565. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng NY, Wilson K, Jared M, Wilson PC. Intricate targeting of immunoglobulin somatic hypermutation maximizes the efficiency of affinity maturation. J Exp Med. 2005;201(9):1467–1478. doi: 10.1084/jem.20042483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng NY, et al. Human immunoglobulin selection associated with class switch and possible tolerogenic origins for C delta class-switched B cells. J Clin Invest. 2004;113(8):1188–1201. doi: 10.1172/JCI20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke SH, et al. Inter- and intraclonal diversity in the antibody response to influenza hemagglutinin. J Exp Med. 1985;161(4):687–704. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook WD, Scharff MD. Antigen-binding mutants of mouse myeloma cells. Proc Natl Acad Sci U S A. 1977;74(12):5687–5691. doi: 10.1073/pnas.74.12.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijer PJ, et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J Mol Biol. 2006;358(3):764–772. doi: 10.1016/j.jmb.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 21.Llewelyn MB, Hawkins RE, Russell SJ. Discovery of antibodies. Bmj. 1992;305(6864):1269–1272. doi: 10.1136/bmj.305.6864.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crotty S, et al. SAP is required for generating long-term humoral immunity. Nature. 2003;421(6920):282–287. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- 23.Webster R, Cox N, Stohr K. World Health Organization Manual on Animal Influenza Diagnosis and Surveillance. WHO; Geneva: 2002. [Google Scholar]
- 24.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.