Abstract
Ionizing radiation is an established source of chromosome aberrations (CAs). Although double-strand breaks (DSBs) are implicated in radiation-induced and other CAs, the underlying mechanisms are poorly understood. Here, we show that, although the vast majority of randomly induced DSBs in G2 diploid yeast cells are repaired efficiently through homologous recombination (HR) between sister chromatids or homologous chromosomes, ≈2% of all DSBs give rise to CAs. Complete molecular analysis of the genome revealed that nearly all of the CAs resulted from HR between nonallelic repetitive elements, primarily Ty retrotransposons. Nonhomologous end-joining (NHEJ) accounted for few, if any, of the CAs. We conclude that only those DSBs that fall at the 3–5% of the genome composed of repetitive DNA elements are efficient at generating rearrangements with dispersed small repeats across the genome, whereas DSBs in unique sequences are confined to recombinational repair between the large regions of homology contained in sister chromatids or homologous chromosomes. Because repeat-associated DSBs can efficiently lead to CAs and reshape the genome, they could be a rich source of evolutionary change.
Keywords: ectopic recombination, gamma radiation, genome rearrangements, nonallelic homologous recombination, retrotransposon
From the time that H. J. Muller discovered that x-rays increased mutation rates (1) and Barbara McClintock first identified chromosome abberations (CAs) that correspond to specific phenotypes (2), ionizing radiation has been used as a powerful tool for mutagenesis and exploration of genome organization. Despite the long-known connection between CAs and x-rays, the underlying mechanisms that give rise to rearrangements remain unclear. In Saccharomyces cerevisiae, various types of DNA damage result in elevated levels of chromosome rearrangements including deletions, duplications, and translocations (3). These studies usually involve genetic methods that select for one type of event at specific loci. For example, Fasullo et al. (4) showed that DNA-damaging agents stimulated homologous recombination between ectopic repeats (resulting in translocations) by selecting for histidine prototrophs in strains with his3 alleles located at sites on chromosomes II and IV. Myung and Kolodner (5) showed that a variety of DNA-damaging agents stimulated the frequency of chromosome rearrangements associated with loss of markers located near the end of chromosome V; most of these rearrangements reflected nonhomologous end-joining or telomere addition to the broken end.
In our study, we took advantage of genomic tools to analyze a large number of unselected CAs arising from randomly induced double-strand breaks (DSBs) across the entire genome. We showed that most of the CAs result from homologous recombination between retrotransposons located at nonallelic sites. Although interactions between transposable elements have been proposed as sources of genome rearrangements after chromosomal damage (6), our findings provide a direct demonstration that DSBs within these elements can reshape the genome.
Results and Discussion
Chromosomal Damage and Repair.
We chose to examine the outcome of randomly induced DSBs on the stability of the genome under conditions where opportunities for homologous recombination (HR) repair of DSBs were maximal. In S. cerevisiae, repair of DSBs by HR is highly favored over repair by NHEJ, particularly in diploid cells (7). Breaks were introduced into the yeast S. cerevisiae genome by ionizing radiation, and the resulting CAs were characterized at the molecular level. Before irradiation, the diploid cells were arrested in the G2 stage of the cell cycle with nocodazole; this arrest was maintained during the irradiation [Fig. S1 in supporting information (SI) Appendix]. This treatment allowed efficient HR repair between sister chromatids (8) or homologous chromosomes. DSB induction was assessed by analyzing changes in full-length chromosomal molecules using pulsed-field gel electrophoresis (PFGE) (Fig. 1a). Cells were exposed to 80 krad (800 Gray), corresponding to 7% and 28% survival in two independent experiments (JW and A sets, respectively; Table S1 in SI Appendix). Using Southern blots to quantify loss of full-length molecules, we showed (Fig. S2 A–F in SI Appendix) that this dose produced ≈250 DSBs per diploid G2 cell.
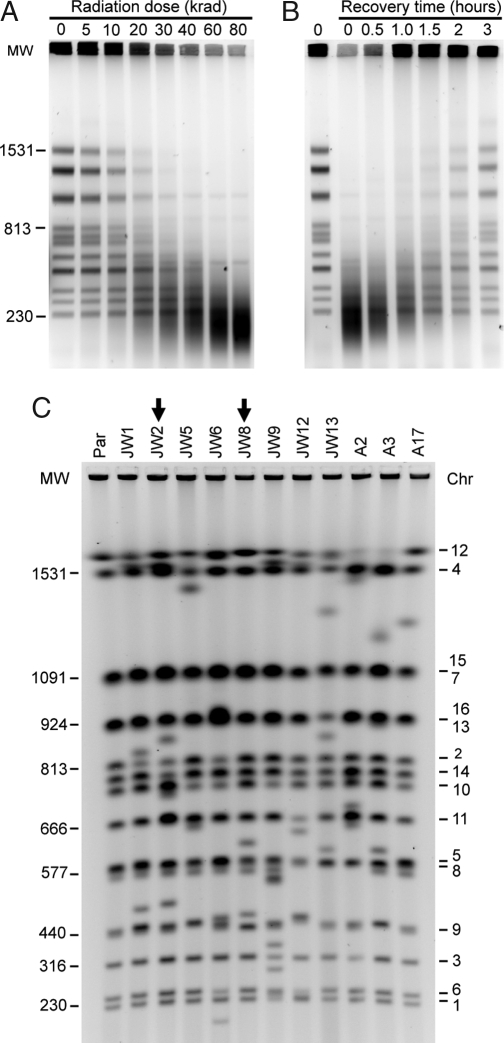
Fig. 1.
DNA DSB induction, chromosomal restoration, and identification of rearrangements. (A) PFGE showing fragmentation of chromosomes in nocodazole-arrested (G2) diploid cells after the indicated dose of γ-radiation. (B) PFGE showing a time course of chromosomal restoration after exposure to 80 krad. (C) PFGE molecular karyotyping of the parental diploid strain (Par) and of the 11 radiation-survivor isolates that were investigated in detail. Molecular weight in kilobases is indicated to the left and specific chromosomes (numbers) to the right. Arrows emphasize the lanes with the JW8 and JW2 isolates.
As shown in Fig. 1B, the G2 diploid cells have a remarkable ability to repair a shattered genome, as shown for haploid G2 cells (8). Repair of specific chromosomes was detected by 1 h postirradiation by using PFGE, and by 3 h, most of the chromosomal bands were restored (Fig. 1B, Fig. S2 G–I in SI Appendix). These results reflect the cumulative repair in the irradiated cell population but do not reveal CAs that may be present in individual cells. To visualize CAs, we analyzed chromosomes from individual colonies that arose on rich media after irradiation, a condition in which the only selection was for viability (Table S1 in SI Appendix). Because the cells were diploid, they could tolerate a wide assortment of CAs, including large heterozygous deletions. This approach differs from a selection system for elaborating the genetic control of gross chromosomal rearrangements (9), where isolation of CAs relies on selecting events that originate in a small nonessential region of single-copy DNA in the haploid genome.
Nearly two-thirds of the colonies (54 of 71) contained at least one novel chromosomal band. The molecular karyotypes of 11 such colonies are shown in Fig. 1C. In contrast, no CAs were found among 24 clones derived from unirradiated cultures (except for occasional expansions/contractions of the ribosomal DNA cluster on Chr 12; Fig. S12 in SI Appendix and data not shown). Because γ-radiation produced ≈250 DSBs per cell, most DSBs were repaired by mechanisms that did not result in a CA. These results differ markedly from findings with haploid cells (10), where only a few percent of colonies contained a CA even at high radiation doses, presumably because many CAs would alter gene dosage and adversely affect growth.
Genome-Wide Detection of CAs.
Microarray-based comparative genomic hybridization (CGH array) was used to analyze the CAs observed in 37 survivors (legend to Table S1 in SI Appendix; see examples in Fig. 2B and Fig. 3B). This analysis (summarized in Table 1, Table S2, and Fig. S14 in SI Appendix) identifies contiguous genomic segments in which there are genomic amplifications or deletions. The sites where gene-dosage transitions from normal to altered, termed chromosome aberration breakpoints (CABs), are presumed to have been involved in the recombination event that gave rise to the CAs. CABs are considered the repair outcome of a DSB and might not represent the actual site of a precursor lesion. With our tiled full-coverage genomic microarrays, the CABs could be estimated with a resolution of one or two ORFs. In addition to imbalanced CAs, CGH arrays also detect aneuploidy. It is important to note that the CGH-array analysis can accurately detect only rearrangements that span regions of unique DNA. Although expansions and contractions of tandemly repeated DNA such as ribosomal DNA and CUP1 (Chr 8) were often observed among survivor colonies in PFGE/Southern blot analysis, they were not detected by CGH arrays and are not shown in Table 1.
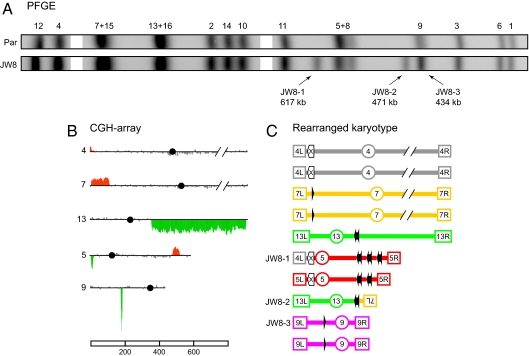
Fig. 2.
Molecular dissection of CAs in the JW8 isolate. (A) Cropped alignment of the PFGE profiles from Fig. 1C. (Par) Parental diploid strain. JW8-1, -2, and -3 indicate the CAs characterized in JW8. (B) CGH-array data for chromosomes involved in CAs. Chromosome numbers are shown to the left of each plot and the horizontal lines correspond to the genomic position of microarray probes from the left to the right telomeres; black circles indicate the position of centromeres. Vertical bars correspond to the average signal of seven consecutive probes. Coloring indicates gene dosage as follows: gray. no significant change; red, gene amplifications; green, gene deletions. (C) Schematic representation of CAs and parental chromosomes with the respective genomic sites involved in rearrangements. Terminal boxes with internal labeling represent the left (L) and right (R) telomeres, and labeled circles represent centromeres. Each chromosome is drawn in a different color. Solid black arrows represent full-length Ty elements with their respective LTRs; arrowheads represent solitary LTR insertions. Empty box arrows with an internal “X” label represent the HXT loci. Chromosomes in B and C were scaled according to the reference bar in kilobases, except for Chr 4 and 7, which are truncated.
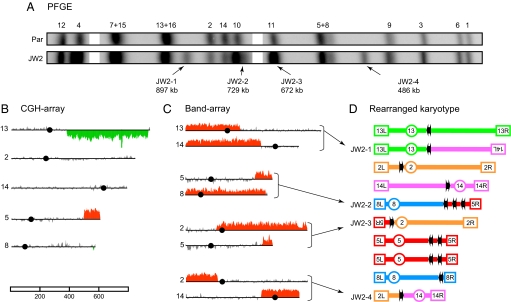
Fig. 3.
Molecular dissection of CAs in the JW2 isolate. All numbers and drawings are presented according to the legend in Fig. 2. (A) Cropped alignment of the PFGE profiles. (B) CGH-array data for chromosomes involved in CAs. (C) Band-array data for CAs. The plots for the specific chromosomes involved in the CAs are shown, with red rising bars, indicating the genomic segments enriched in each band. Background signal from comigrating parental chromosomes are not shown. (D) Schematic representation of the CAs and of the parental chromosomes with the respective genomic sites involved in rearrangements. The JW2-1, -3, and -4 CAs resulted from tripartite recombination and were structured as follows: JW2–1 was composed of a region of Chr 13 from the left telomere, passing through the centromere (CEN13) up to YMRCTy1-4, and a region of Chr 14 from YNLWTy1-2 to the left telomere; JW2-3 was a translocation including Chr 5 sequences from the right telomere to YERCTy1-2, and Chr 2 sequences from YBLWTy1-1, passing through CEN2 and including the entire right arm; finally JW2-4 was a translocation involving Chr 2 sequences from the left telomere to YBLWTy1-1 and Chr 14 DNA from YNLWTy1-2 passing through CEN14 to include the entire right arm. The remaining CA, JW2-2, was a complex nonreciprocal translocation involving the Chr 8 sequences from the left telomere, passing through CEN8 and including most of the right arm up to YHRCTy1-1, combined with sequences from Chr 5 represented by an interstitial duplication between YERCTy1-1 and YERCTy1-2 and a single copy of the distal region up to the right telomere.
Table 1.
Summary of CGH-array analysis
| Number of events (%) | |
|---|---|
| Survivor isolates analyzed | 37 |
| Numerical chromosomal aberrations: | 13 |
| Monosomy | 4 (30.8) |
| Trisomy | 9 (69.2) |
| Structural chromosomal aberrations: | 78 |
| Terminal deletions | 28 (35.9) |
| Terminal amplifications | 27 (34.6) |
| Interstitial deletions | 13 (16.7) |
| Interstitial amplifications | 10 (12.8) |
| Breakpoint positions: | 97 |
| Full-length Ty insertions | 64 (66.0) |
| Solo LTRs insertions | 17 (17.5) |
| Other repetitive DNA | 9 (9.3) |
| Uncharacterized | 7 (7.2) |
| Most frequent breakpoints: | |
| YERCTy1-1 | 9 (9.3) |
| YERCTy1-2 | 5 (5.2) |
| YCRWdelta8 | 4 (4.1) |
| YHRCTy1-1 | 4 (4.1) |
| YJRCdelta19 | 4 (4.1) |
A complete description of the CGH-array analysis is provided in Table S2 in SI Appendix. The numbers in parentheses are percentages within each category.
Despite the random induction of DSBs (Fig. S2 in SI Appendix), 91% of the 97 CABs were found at dispersed repetitive DNA sequences. Eighty-one were located at Ty retrotransposon sequences, either full-length element insertions of Ty1 or Ty2 (≈6 kb) or at solo delta elements (≈0.3-kb LTRs of Ty1 and Ty2). Retrotransposons and LTRs comprise 3% of the genome and represent the most abundant class of dispersed repetitive DNA in S. cerevisiae (11). Another nine breakpoints were found in diverged gene families such as HXT and FLO. These genes are frequently located near yeast telomeres and have been identified as sites of genome rearrangements between closely related yeast species (12).
There were seven CABs that appeared to be in single-copy DNA regions, based on the published yeast sequence. Because our strain is not identical to the sequenced strain, such CABs could represent homologous recombination between repeats not present in the sequenced strain or could represent NHEJ events. CABs in this class are termed “uncharacterized” in Table 1. Subsequent analysis of two such CABs showed that one was associated with a previously unidentified Ty, and the other was likely due to DSB healing by telomere addition. Thus, at most only five of the radiation-induced CABs could involve NHEJ.
Molecular Characterization of Recombination Products.
To understand completely the events leading to chromosomal rearrangements, we sought to define all of the CAs (excluding rDNA) within each of the 11 strains in Fig. 1C using a combination of Southern blot, PCR, and Band-array analysis. Band-array analysis involves excision of specific chromosomal bands from PFGE that are then examined in a second round of CGH-array (13). Molecular characterization of 32 CAs (3 by Southern analysis, 2 by PCR, and 27 by Band-array) enabled us to account for all novel chromosomes in nine of the isolates.
This molecular autopsy approach revealed a variety of chromosomal changes involving repetitive DNA sequences. The CAs in the JW8 and JW2 isolates (shown in Figs. 2 and 3, respectively) are examples of the recombination events induced by ionizing radiation. Detailed analysis of eight other isolates is available in SI Appendix. There were three categories of rearrangements in JW8: interstitial duplication, nonreciprocal translocation, and a potential loss of heterozygosity (LOH) event. The JW8-1 chromosome aberration resulted from two independent recombination events in the same DNA molecule. The interstitial duplication on the right arm of Chr 5 between two Ty1 insertions (YERCTy1-1 and -2) presumably reflects an unequal cross-over between the Ty elements. The second event was a nonreciprocal translocation between the HXT13 (Chr 5) and HXT15 (Chr 4) loci, which share 90.7% sequence identity over a 1,670-bp homology region. Sequencing of this translocation product showed that exchange occurred inside identical 26-bp regions (Fig. S8E in SI Appendix). Another CA was a nonreciprocal translocation, JW8-2, which involved a 197-bp homologous region in a solo LTR on Chr 7 (YGLWdelta2) and an LTR associated with a Ty1 element on Chr 13 (YMRCdelta8). One mechanism for generating this nonreciprocal translocation is break-induced replication (BIR) (14) repair of a DSB that may have occurred in YMRCdelta8 using the YGLWdelta2 as a template. Because of size polymorphisms found in Chr 8 and 9, it was also possible to identify events that may have been due to radiation-induced recombination between homologues. One such event (CA JW8-3) resulted in a sharp deletion peak on Chr 9 through loss of a hemizygous Ty3 insertion (Fig. 2B; detailed in Fig. S8 in SI Appendix). This event could be an LOH event (reflecting either gene conversion or mitotic crossing-over proximal to the heterozygous insertion) or a “pop-out” of the Ty3 element.
The JW2 strain was a good example of the complex events that can occur in a single cell after irradiation. Four new chromosomal bands (JW2–1 to -4) were identified in the PFGE profile of this isolate (Fig. 3A). In addition, the Chr 2 and 14 bands were detected at half the normal intensity, indicating that only a single copy of the parental-sized DNA molecules was present in the diploid. This pattern was more complex than predicted from the CGH array data alone (Fig. 3B), which showed simply a gain of sequences on the right arm of Chr 5 (4× level) and loss of sequences (1× level) on the right arm of Chr 13 and near the right telomere of Chr 8. Because no gene dosage changes were detected for Chr 2 and 14 sequences, these chromosomes must have been involved in conservative chromosomal rearrangements where chromosome structure, but not gene dosage, is altered. Band-array analysis (Fig. 3C) resulted in a complete characterization of the rearranged chromosomes in JW2 (Fig. 3D).
Three of the CAs (JW2-1, -3, and -4) were particularly informative, because they represented interrelated events, which resulted from tripartite recombination between full-length Ty elements located on Chr 2, 5, 13, and 14 (detailed description in Fig. 3 legend). Note that one full copy of Chr 2 and one full copy of Chr 14 were recovered in these three CAs. Because no DNA was lost on Chr 2 or 4, we were able to unambiguously localize the precursor DSB lesions to Tys on those chromosomes. This indicated that a DSB on Chr 2 (at YBLWTy1-1) and a DSB on Chr 14 (at YNLWTy1-2) triggered the formation of these CAs. In both cases, the two DNA ends generated by a DSB each engaged in recombinational repair with independent homologous Ty sequences on other chromosomes. This could have occurred as follows: CA JW2-4 formed as a result of a DSB end from Chr 2 interacting with another DSB end from Chr 14, possibly through a single-strand annealing (SSA) pathway. The remaining DSB end from Chr 2 recombined with a homologous Ty sequence on Chr 5 resulting in CA JW2-3, whereas the second DSB end from Chr 14 engaged a Ty on Chr 13 forming CA JW2-1.
The fourth CA in this isolate, JW2-2, was also complex in structure, because it resulted from two different recombination events on Chr 5 and 8, both involving Ty sequences (see Fig. 3 legend). In summary, a small number of DSBs associated with Tys efficiently triggered nonallelic recombination between repetitive DNA elements and reshaped the karyotype of JW2.
Surprisingly, tripartite recombination was frequent. Repair events analogous to the ones described above were also found in isolates JW6, JW9, and JW13 (Figs. S7, S9, and S10 in SI Appendix). Among the 11 conservative CAs identified in our study, nine were formed by a tripartite mechanism. The participation of both ends in the same exchange event resulting in a reciprocal translocation was found in only 2 of the 11 conservative CAs (isolate A2; Fig. S11 in SI Appendix). Recently, it was proposed that capture of both ends of a DSB by a single D loop in a donor sequence may suppress BIR, thereby making gene conversion a preferential mechanism for accurate repair of DSBs in single-copy DNA and preventing CAs (15). Our results suggest that DSBs in repetitive DNA elements interfere with this mechanism, because both ends are able to find homology independently in the genome rather than being captured by a single a D loop structure.
The predominance of aberrations associated with Tys suggests a strong relationship between CAs, Tys, and DSBs. Using a computational simulation based on DSBs per cell and the portion of the genome occupied by retrotransposons (11), we calculate that the average cell received about seven DSBs within Tys (Fig. S3 in SI Appendix). Thus, although it is possible that DSBs external to Tys could stimulate the frequent Ty-associated CAs, there were enough Ty-associated DSBs to account for the two to three Ty-associated CABs observed per survivor. Overall, ≈2% of all DSBs gave rise to detectable CAs. These results also demonstrate that most DSBs are repaired by HR in a manner that does not result in CAs, presumably using sister chromatids or homologous chromosomes as templates.
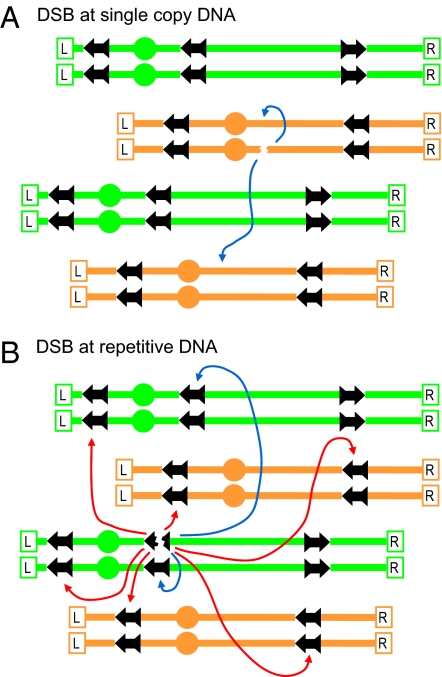
The finding that repetitive elements are the predominant sites of CAs induced by random DSBs suggest a model (Fig. 4) wherein the combination of repetitive DNA sequences and DSBs (and possibly other lesions) play a key role in providing plasticity to an otherwise rigid genome. A DSB in a region of unique DNA provides the genome with a limited choice of repair partners (sister chromatid or homolog; blue arrows), none of which can yield a chromosomal rearrangement (Fig. 4A). Once a DSB is formed inside a repetitive DNA element, the HR system is confronted with the choice of recombining with allelic sequences located on either a sister chromatid or homologous chromosome or of recombining with nonallelic repeats (red arrows). Our results suggest that the ends produced by a DSB within a Ty element (Fig. 4B) open the genome to DSB interactions among essentially all of the chromosomes, often independently, as discerned from the high incidence of tripartite recombination. Considerable sequence divergence between the Ty and delta elements (Table S3 in SI Appendix) might reduce, but does not necessarily prevent repair of, radiation-induced DSBs (16, 17). Although it is formally possible that CAs could arise via resected DNAs that extend to Tys (18, 19), such events would not readily explain the observed tripartite events described here.
Fig. 4.
Model for generation of CAs through the repair of repeat-associated DSBs. Given the random distribution of induced DSBs, most are expected to appear in single-copy DNA sequences as indicated in A, where efficient recombinational repair can occur between a sister chromatid or homolog (blue arrows). In contrast, DSBs that occur within the repetitive DNA sequences shown in B also have numerous opportunities for the ectopic recombination (red arrows), generating the CAs. The two ends formed by a single DSB can act independently in these interactions. The ectopic repair of DSBs in repetitive elements is in competition with the repair involving the sister chromatid or the homologue.
We previously suggested that translocations generated in yeast strains with low levels of DNA polymerase alpha reflected a DSB in one Ty element that was repaired by a BIR event involving a Ty element on a nonhomologous chromosome (18), consistent with the translocations observed in the present study. Alternatively, translocations could be formed by annealing of two Ty elements each containing a DSB (as in CA JW2-4, Fig. 3). Such events have been termed “half crossovers” and have been observed in strains lacking Rad52p (20). Translocations could also result from two consecutive BIR events, using a Ty cDNA to initiate the first BIR event (21). Chromosome rearrangements in which retrotransposon sequences are captured at the translocation breakpoint have been observed in yeast (22). Finally, although in our experiments, most CAs reflect homologous recombination between nonallelic repeats, under different experimental conditions, CAs resulting from nonhomologous end-joining might also occur. Such events have been detected as a consequence of HO (homothallic) endonuclease-induced DSBs in haploids (23).
Nonallelic Ty-Ty recombination has been extensively investigated by Kupiec and coworkers (24–26), who used a selection system to detect loss of a genetically marked Ty element by gene conversion with other Tys or intra-Ty recombination between the two flanking delta sequences. Interestingly, these workers showed that Ty-Ty gene conversion and intra-Ty deletion were not stimulated by ionizing radiation (24, 26) but could be induced by a site-specific DSB (25). The absence of detectable radiation-induced Ty-Ty events could be because they used a 100-fold lower dose and the requirement for specific interactions with the Ty reporter being used (26), unlike the present study, which can sample interactions across nearly all Tys. In addition, a site-specific DSB would cut both chromatids, limiting the opportunities for repair, a situation different from randomly induced DSBs in sister chromatids.
Our studies show that, in response to DSBs, repetitive DNA is a major source of genome plasticity. The efficient repair of G2/M-induced DSBs displayed in yeast resembles the extraordinary HR properties of the radioresistant bacterium Deinococcus radiodurans (27). Both organisms have similar amounts of repetitive DNA [3.8% in D. radiodurans (28)]. It would be interesting to determine whether under the highly efficient homology-driven repair of D. radiodurans there is a similar capability for the generation of genome rearrangements.
Chromosomal rearrangements between repetitive DNA sequences have been observed in a variety of laboratory and natural populations (12, 21, 29–31). Although some CAs are selectively advantageous, there are also negative consequences to a mechanism that generates high rates of CAs. Selection against cells with high levels of genome instability, reflecting high levels of transposable elements, may be one mechanism by which the number of such elements per genome is limited (32). In higher eukaryotes such as humans, whose genomes are replete with repetitive DNA, a compromise between opportunities for variation and excessive genome instability could be accomplished by increasing the efficiency of local interactions (end rejoining and sister chromatid recombination) and by shifting the balance of DSB repair from homologous to nonhomologous pathways. Despite the presence of these balancing forces, recent studies of structural genomic variation have uncovered a very significant role for nonallelic HR in reshaping the human genome (33, 34). In these studies, about half of the structural variants reflected nonallelic HR between repetitive DNA sequences such as transposable elements. Taken together, these recent results support the proposal that HR between repetitive DNAs is a major source of genomic variation in humans (6, 35) and a mechanism for disease-associated CAs that might arise from DNA lesions such as DSBs.
Methods
Procedures and Strains.
Standard procedures were used for yeast genetic manipulation and culture. The parental diploid strain used in this study (JW1777) was constructed, as described in SI Appendix, from nearly isogenic derivatives to obtain complete homozygosity. JW1777 was derived from crosses between strains of the S288c background, with a minor contribution from strains of the A364A background (Craig Giroux, personal communication).
Nocodazole Arrest and Irradiation.
A detailed description of the G2 arrest and irradiation is described in SI Appendix. Briefly, nocodazole was added to logarithmically growing cells. By 2 hours, 80–90% of cells were in G2, as determined by cell morphology and flow cytometry. Cells were harvested, washed, resuspended, and kept in ice-cold sterile water throughout the irradiation procedure. Cell suspensions were irradiated in a 137Cs irradiator at a dose rate of 2.38 krads/minute with periodic aeration and cooling intervals after every 10 krads of irradiation. After irradiation, cell suspensions were held on ice, diluted, and plated on yeast extract, peptone, dextrose, adenine (YPDA). Colonies were counted after 3 days at 30°C.
PFGE.
Two types of instruments were used to analyze high-molecular-weight DNA: transverse alternating field electrophoresis gels (Fig. 1 A and B and Fig. S2 in SI Appendix) were run in a Gene Line II apparatus from Beckman, and contour-clamped homogeneous electric field gels (CHEF; Fig. 1C and Fig. S12 in SI Appendix) were run in a BioRad CHEF Mapper XA system. Running conditions were according to the manufacturer's recommendations, with appropriate modifications. Detailed PFGE protocols are available upon request.
Microarray Analysis.
The procedures used to prepare, label, and hybridize genomic DNA for CGH arrays were described in ref. 18. To determine the gene composition of specific chromosomes (Band-arrays), we used a modified version of a previously described protocol (13) (see complete Band-array protocol in SI Appendix). Briefly, the procedure consisted of excising specific ethidium bromide stained bands from PFGE, followed by β-agarose treatment, purification, amplification, and labeling with Cy5. The resulting DNA was competitively hybridized to microarrays in the presence of Cy3-labeled JW1777 total genomic DNA. Genomic regions present in the bands were found as regions of enriched Cy5 signal relative to the Cy3 total DNA background.
Supplementary Material
Acknowledgments.
We thank C. Giroux (Wayne State University, Detroit) and A. Gabriel (Rutgers University, Piscataway, NJ) for sharing unpublished data and A. Casper, M. Meselson, A. Gabriel, J. Boeke, D. Gordenin, J. Mason, and M. Shelby for useful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM52319 (to T.D.P.) and by intramural research funds from National Institute of Environmental Health Sciences.
Footnotes
The authors declare no conflict of interest.
Data deposition footnote: The complete set of microarray experiments has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE6991 and GSE6984).
See Commentary on page 11593.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804529105/DCSupplemental.
References
- 1.Muller HJ. Artificial transmutation of the gene. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. Cytological observations of deficiencies involving known genes, translocations and inversions in Zea mays. MO Agric Exp Station Res Bull. 1931;163:1–30. [Google Scholar]
- 3.Kupiec M. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat Res. 2000;451:91–105. doi: 10.1016/s0027-5107(00)00042-7. [DOI] [PubMed] [Google Scholar]
- 4.Fasullo M, Dave P, Rothstein R. DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae. Mutat Res. 1994;314:121–133. doi: 10.1016/0921-8777(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 5.Myung K, Kolodner RD. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2003;2:243–258. doi: 10.1016/s1568-7864(02)00216-1. [DOI] [PubMed] [Google Scholar]
- 6.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 8.Brunborg G, Resnick MA, Williamson DH. Cell-cycle-specific repair of DNA double strand breaks in Saccharomyces cerevisiae. Radiat Res. 1980;82:547–558. [PubMed] [Google Scholar]
- 9.Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. Analysis of gross-chromosomal rearrangements in. Saccharomyces cerevisiae. Methods Enzymol. 2006;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- 10.Friedl AA, Kiechle M, Fellerhoff B, Eckardt-Schupp F. Radiation-induced chromosome aberrations in Saccharomyces cerevisiae: Influence of DNA repair pathways. Genetics. 1998;148:975–988. doi: 10.1093/genetics/148.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 12.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 13.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 15.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 16.Resnick MA, et al. Recombinant repair of diverged DNAs: A study of homoeologous chromosomes and mammalian YACs in yeast. Mol Gen Genet. 1992;234:65–73. doi: 10.1007/BF00272346. [DOI] [PubMed] [Google Scholar]
- 17.Resnick MA, Skaanild M, Nilsson-Tillgren T. Lack of DNA homology in a pair of divergent chromosomes greatly sensitizes them to loss by DNA damage. Proc Natl Acad Sci USA. 1989;86:2276–2280. doi: 10.1073/pnas.86.7.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 19.VanHulle K, et al. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber JE, Hearn M. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell PH, Curcio MJ. Retrosequence formation restructures the yeast genome. Genes Dev. 2007;21:3308–3318. doi: 10.1101/gad.1604707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Gabriel A. Reciprocal translocations in. Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1534/genetics.166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parket A, Kupiec M. Ectopic recombination between Ty elements in Saccharomyces cerevisiae is not induced by DNA damage. Mol Cell Biol. 1992;12:4441–4448. doi: 10.1128/mcb.12.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parket A, Inbar O, Kupiec M. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics. 1995;140:67–77. doi: 10.1093/genetics/140.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kupiec M, Steinlauf R. Damage-induced ectopic recombination in the yeast Saccharomyces cerevisiae. Mutat Res--DNA Repair. 1997;384:33–44. doi: 10.1016/s0921-8777(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 27.Zahradka K, et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature. 2006;443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- 28.White O, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garfinkel DJ. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet Genome Res. 2005;110:63–69. doi: 10.1159/000084939. [DOI] [PubMed] [Google Scholar]
- 30.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 31.Fischer G, et al. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 32.Arkhipova I, Meselson M. Deleterious transposable elements and the extinction of asexuals. BioEssays. 2005;27:76–85. doi: 10.1002/bies.20159. [DOI] [PubMed] [Google Scholar]
- 33.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidd J, Cooper G, Donahue W, Smith D, Eichler E. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.