Abstract
Alpha-actinins are critical components of the actin cytoskeleton. Here we show that α-actinins serve another important biological function by binding to and competitively inhibiting calcium-dependent activation of endothelial NOS (eNOS). α-actinin-2 was found to associate with eNOS in a yeast two-hybrid screen. In vascular endothelial cells, which only express α-actinin-1 and -4, α-actinin-4 interacted and colocalized with eNOS. Addition of α-actinin-4 directly inhibited eNOS recombinant protein, and overexpression of α-actinin-4 inhibited eNOS activity in eNOS-transfected COS-7 cells and bovine aortic endothelial cells (BAECs). In contrast, knockdown of α-actinin-4 by siRNA increased eNOS activity in BAECs. The α-actinin-4-binding site on eNOS was mapped to a central region comprising the calmodulin-binding domain, and the eNOS-binding site on α-actinin-4 was mapped to the fourth spectrin-like rod domain, R4. Treatment of endothelial cells with a calcium ionophore, A23187, decreased α-actinin-4-eNOS interaction, leading to translocation of α-actinin-4 from plasma membrane to cytoplasm. Indeed, addition of calmodulin displaced α-actinin-4 binding to eNOS and increased eNOS activity. These findings indicate that eNOS activity in vascular endothelial cells is tonically and dynamically regulated by competitive interaction with α-actinin-4 and calmodulin.
Keywords: endothelium, nitric oxide, ionophore
Endothelium-derived NO is an important mediator of vascular function (1). NO is generated from the conversion of L-arginine to L-citrulline by endothelial NOS (eNOS or NOS3). Because of its critical role in vascular homeostasis, eNOS is tightly regulated post-translationally by several factors. Lipid modifications such as myristoylation or palmitoylation are important to anchor eNOS at the cell membrane, in close proximity to its cofactors and modulators (2). Upstream modulators include protein kinases, which activate eNOS through phosphorylation at Ser-1177 and Ser-635 or inhibit eNOS through phosphorylation at Thr-495 and Ser-1163. In particular, Ser-1177 phosphorylation of eNOS by protein kinase B/Akt enhances both basal and agonist-mediated eNOS activity (4, 5). In contrast, S-nitrosylation of eNOS renders eNOS inactive under basal conditions (6, 7).
For maximal activity, all NOS isoforms require binding to cofactors such as NADPH, tetrahydrobiopterin, and calmodulin (3). The mechanistic basis underlying the calcium dependency of eNOS activation results from its requirement for direct binding to calmodulin and subsequent allosteric conformational changes in the NOS protein, permitting efficient binding to other cofactors and catalysis of L-arginine (8). Indeed, localization of eNOS within plasma membrane caveolae by caveolin appears to be necessary for the efficient release of NO in response to external stimuli (9). However, caveolin, which competes with and displaces calmodulin from eNOS, inhibits eNOS activity in the caveolae of endothelial cells and cardiomyocytes (10, 11). Other eNOS interacting proteins such as heat shock protein 90 and a voltage-dependent anion/cation channel, porin, also regulate eNOS activation through protein stabilization, sub-cellular localization, and cofactor availability (12, 13). Thus, proteins that interact with eNOS are important physiological regulators of eNOS activity.
To further elucidate the regulation of eNOS through protein-protein interaction, we performed yeast two-hybrid assays to find novel proteins, which could associate with and regulate eNOS. We found that α-actinin-4 binds eNOS and inhibits basal eNOS activity by competitively displacing calmodulin from eNOS.
MATERIALS AND METHODS
Yeast two-hybrid screening
A human heart cDNA library cloned into the yeast vector pACT2 was obtained from Clontech (Mountain View, CA, USA) and prepared according to the manufacturer’s instructions. The yeast strain AH109 (Clontech) was made competent by the lithium acetate method.
Approximately 5 × 109 yeast cells were transformed with 100 μg of the eNOS bait plasmid pBDGAL4-eNOS [1–530 amino acids (aa)] and 100 μg of the library plasmid, followed by plating on sd medium (Clontech) lacking adenine, tryptophan, leucine, and histidine. Surviving clones were then inoculated on plates lacking adenine, tryptophan, leucine, and histidine but containing X-α-Gal (Clontech), and screened for α-galactosidase activity. Clones that remained positive were then grown in liquid medium, and plasmid DNAs from positive clones were prepared as described. The recovered plasmids were used to transform ultracompetent XL-1 blue cells (Stratagene, La Jolla, CA, USA). The cDNA inserts were sequenced by the dideoxy chain termination method. The interactions were verified by the X-α-galactosidase indicator plate assay as described in the manufacturer’s protocol (Clontech).
Plasmid constructs
The eNOS-bait plasmids were obtained using reverse transcription-polymerase chain reaction (PCR) by Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA, USA) on total RNA derived from human saphenous endothelial cells. The PCR products were cloned into the TA cloning vector pCRII-Blunt II-TOPO vector (Invitrogen) and then subcloned into pBDGAL4 Cam (Stratagene). The cDNA of human α-actinin-4 was amplified and cloned into the glutathione S-transferase (GST) gene fusion vector pGEX-4T-1 (GE Healthcare, Piscataway, NJ, USA). The mammalian expression vector for eNOS was made by full-length eNOS cDNA and pCDNA3 (Invitrogen). The ACTN1 (α-actinin) cDNA on pAD was provided by Dr. Wolfgang Siess (Institut für Prophylaxe und Epidemiologie der Kreislaufkrankheiten, München, Germany). The ACTN2 constructs (Δ1–894, Δ273–394, Δ394–505, and Δ506–757) on pBD and the α-actinin-4 mammalian expression plasmid ACTN4/pCDNA3 have been described (14).
Cell culture and transfections
Human saphenous vein endothelial cells (HSVECs) and bovine aortic endothelial cells (BAECs) were harvested as described previously (15). The protocol to isolate and culture HSVECs was approved by the Committee on Human Studies at Harvard Medical School. The cells were cultured at 37°C in a growth medium containing Dulbecco modified Eagle medium (DMEM) supplemented with 5 mM L-glutamine, 10% FBS (Hyclone, Logan, UT, USA), and an antibiotic mixture of penicillin (100 U/ml), streptomycin (100 mg/ml), and fungizone (250 ng/ml). Relatively pure (>95%) endothelial cell cultures were confirmed by immunofluorescence staining with anti-factor VIII antibody (Vector Laboratory, Inc., Burlingame, CA, USA), and only endothelial cells of less than 6 passages were used. COS-7 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM containing 10% FBS. Cells were transfected with the indicated plasmids using FuGENE6 transfection reagent according to the manufacture’s protocol (Roche Diagnostics, Indianapolis, IN, USA).
Western blot analysis
Cells were washed 2× with ice-cold PBS (Invitrogen) and incubated with 500 ml of lysis buffer (1% Triton X-100; 20 mM Tris, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1 mM EGTA; 2.5 mM sodium pyrophosphate; 1 mM β-glycerolphosphate; 1 mM phenylmethylsulfonyl fluoride; and 1 mM sodium or-thovanadate). Insoluble materials were removed by centrifugation at 12,000 g for 10 min at 4°C. Forty micrograms of proteins was separated by SDS-PAGE, blotted onto nitrocellulose membranes (GE Water & Process Technologies, Trevose, PA, USA), and probed with the indicated antibody at 1000× dilution in Tris-buffered saline (10 mM Tris, pH7.4, 100 mM NaCl) with 0.1% Tween 20 and 5% nonfat dry milk. Anti-α-actinin isoform specific antibodies were previously described (16, 17), and anti-α-tubulin antibody and anti-β-tubulin antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) and Lab Vision Corporation (Fremont, CA, USA), respectively. After washing, membranes were incubated with either goat anti-mouse or donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody (GE Healthcare). Detection of protein bands was performed using enhanced chemiluminescence substrate (GE Healthcare).
Immunoprecipitation
For co-immunoprecipitation experiments, cells were washed 2× with PBS, resuspended in radio-immunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 8.0; 150 mM NaCl; 1.0% Triton X-100; 0.1% deoxycholate; 5mM EDTA) with protease inhibitors (Sigma), and incubated on ice for 60 min. Cells were centrifuged at 12,000 g for 10 min at 4°C, and lysates were precleared with Protein G Sepharose (GE Healthcare), following incubation with anti-eNOS antibody, anti-α-actinin-4 antibody, control mouse IgG, or control rabbit IgG at 4°C overnight. Protein G Sepharose was added for 2 h. The immune complexes were washed 3× with RIPA buffers.
GST pull-down assays
GST-α-actinin-4 proteins were expressed in Escherichia coli DH5α strain, affinity-purified, and immobilized on glutathione-sepharose 4B beads (GE Healthcare). Purified immobilized GST fusion proteins were incubated with recombinant bovine eNOS protein (Cayman Chemical, Ann Arbor, MI, USA) in NTEN buffer (20 mM Tris, pH 8.0; 100 mM NaCl; 1 mM EDTA; 0.1% Nonidet P-40; 10% glycerol; 2 mM phenylmethylsulfonyl fluoride; 1 mM dithiothreitol) at 4°C overnight, followed by 3× washes. The bound fractions were separated on SDS-PAGE gel and subjected to immunoblotting with anti-eNOS antibody.
Immunofluorescence study
HSVECs were plated onto gelatin-coated cover slips. Cells were fixed with 2% paraformaldehyde in PBS for 15 min and rinsed 3× with PBS, following permeation by 0.2% Triton X-100 plus 1% normal goat serum in PBS. Cells were washed with PBS with 1% normal goat serum 3×. Cells were incubated with 400× diluted mouse monoclonal anti-eNOS antibody and/or 400× diluted anti-α-actinin-4 antibody, following the secondary staining with Alexa Fluor 488 goat anti-mouse IgG (H+L) and/or Alexa Fluor 568 goat anti-rabbit IgG (H+L) from Invitrogen. Cells were examined with Leica TCS SP5 system (Leica, Wetzlar, Germany).
eNOS activity assay
COS-7 cells were transfected with pCDNA3-eNOS with or without pCDNA3-ACTN4. eNOS activity was determined by measuring the conversion of [3H] L-arginine to [3H] L-citrulline using the NOS assay kit from EMD Bioscience, Inc. (San Diego, CA, USA) according to the manufacturer’s manual. Lysates incubated with 1 mM of the eNOS inhibitor, nitro-L-arginine methyl ester (L-NAME) served as blanks. The eluted GST-α-actinin-4 variants or GST alone was incubated with 2 μg (15 pmol) of recombinant bovine eNOS protein (Cayman Chemical).
BAECs were transiently transfected with either pCDNA3, pCDNA3-ACTN1, pCDNA3-ACTN4 in phenol red-free DMEM containing 10% charcoal-stripped FBS for 24 h, following 8 h starvation in phenol red-free DMEM only. Cells were then treated in Krebs-Ringer’s solution (111.8 mM NaCl, 3.4 mM KCl, 2.5 mM CaCl2, 0.8 mM MgSO4, 1.2 mM KH2PO4, 25 mM NaHCO3, and 11.1 mM glucose), 1 mM arginine, and 10 μM diaminofluorescein (DAF-2, EMD Biosciences, Inc., La Jolla, CA) with or without stimulants for 2 h (18). Overall fluorescence measured by a Victor 3 spectrofluorimeter (PerkinElmer, Waltham, MA, USA) and confirmed by counting the percentage of individually labeled fluorescent cells per high-power field by confocal microscopy (Leica TCS SP5 System).
Small interfering RNA (siRNA) of ACTN4
Two pairs of siRNA oligonucleotides for bovine ACTN4 (sense strand, 5′-GGA GAA GCA GCU AGA GAC CAU CGA C-3′ (siACTN4-A) and 5′-CUU CGA AGU GGC UGA GAA AUA CCT C-3′ (siACTN4-B)) and a pair of control siRNA oligonucleotides (5′-CAG AGA GGA GGA AAG GAG ACG CAG G-3′) were synthesized by Integrated DNA Technologies (Coralville, IA, USA). BAECs grown to ~50% confluence in 6-well plates were transfected with Gene Silencer (Gene Therapy System, San Diego, CA, USA) transfecting agent with target-specific siRNA (20 nM) and control siRNA (20 nM) in serum-free DMEM according to the manufacturer’s recommendation. Approximately 3 h post-transfection, 1 ml of fresh complete DMEM medium containing 10% FBS was added, and the cells were cultured for an additional 72 h for protein analyses of ACTN4 and assay for eNOS activity.
Statistical analysis
Differences between groups were determined by the 1-way ANOVA test. Values of P < 0.05 were considered statistically significant.
RESULTS
Association of eNOS with α-actinins in yeast
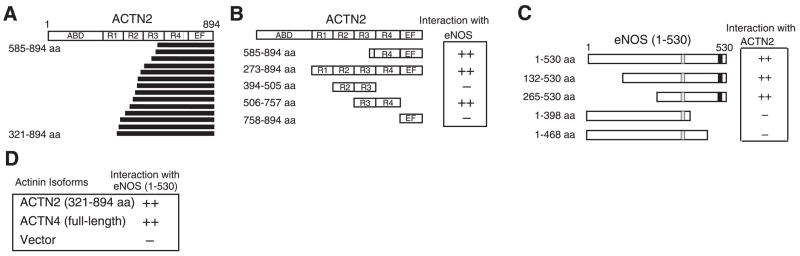
We screened 2.5 × 106 yeast colonies transformed with a human heart cDNA library fused to the Gal4 transcriptional activation domain with a bait construct encoding the N-terminal region (1–530 aa) of eNOS fused to the Gal4 DNA binding domain. The N-terminal region of eNOS was selected to avoid identifying known proteins, which could interact with the FMN/FAD/NADH C-terminal domain that is homologous with various cytochrome P450 monooxygenase enzymes (3). The bait protein was expressed in the yeast and did not have transcriptional activity. One hundred forty-four clones grew, and 65 galactosidase-positive clones were obtained. Twenty-two clones representing 14 independent prey constructs encoded the actin-binding protein, α-actinin-2, encoded by the ACTN2 gene (Fig. 1A). All positive clones contained the entire C-terminus but lacked varying portions of the N-terminal domains. The shortest ACTN2 clone (585–894 aa) encoded a small portion of spectrin-like repeat 3 (R3), all of R4, and the EF hand domain, suggesting that the eNOS-binding motif was contained within this region.
Figure 1.
Characterization of binding between α-actinin and eNOS. A) The structure of α-actinin-2 and 14 independent clones isolated from a human heart cDNA library using yeast two-hybrid screen. α-Actinin-2 has an actin-binding domain (ABD), 4 spectrin-like repeats (R1-R4), and an EF hand domain (EF). Minimal clone that was obtained contained a small part of R3, R4, and an EF domain. B) Mapping of eNOS-binding domains on α-actinin-2 by X-α-galactosidase plate assay. The shortest isolated clone of ACTN2 (585–894aa) and a deletion mutant (506–757 aa) containing R4 were sufficient to enable binding to the N-terminal domain of eNOS (1–530 aa). C) Mapping of α-actinin-binding domains on eNOS by X-α-galactosidase plate assay. Lack of 469–530 aa of eNOS, which includes the calmodulin-binding domain (■), and the caveolin-binding domain (□), resulted in loss of binding ability to α-actinin-2. D) Binding of α-actinin isoforms to eNOS in yeast two-hybrid system. Binding of vector served as control, α-actinin-2 and -4, but not α-actinin-1, interacted with eNOS.
To confirm and refine the location of the eNOS-binding region of α-actinin-2, we used a reporter assay to examine the interaction of four ACTN2 deletion constructs with eNOS. Two ACTN2 constructs (273–894 and 506–757 aa) exhibited strong interaction with eNOS, whereas the other two constructs (394–505 and 758–894 aa) did not (Fig. 1B). These findings suggest that the R4 domain contains the putative eNOS-binding region of α-actinin-2. Interestingly, the R4 domain is also important for α-actinin-2 binding to other cytoskeletal proteins such as calsarcin-2/myozenin (14).
To determine the α-actinin-binding region of eNOS, we made 4 N-terminal deletional constructs of eNOS and examined their interaction with α-actinin-2 (585–894 aa). Compared with the N-terminal construct of eNOS (1–530 aa), more deletion of amino acids 469–530 (1–468 aa construct) resulted in no interaction with α-actinin-2, indicating that this 62-amino-acid region, which contains the calmodulin-binding site, is essential for binding to α-actinin (Fig. 1C). Because calmodulin binds and activates eNOS in response to an increase in intracellular calcium concentrations, these findings raise the possibility that α-actinin may compete with calmodulin for binding to eNOS. Such competitive binding between α-actinin-2 and calmodulin has been shown for the N-methyl-D-aspartate (NMDA) -type glutamate receptor in neural synapses (19).
The human α-actinin gene family encodes 4 isoforms: 2 nonmuscle (cytoskeletal) forms, α-actinin-1 and α-actinin-4 (20–22), and 2 striated muscle (sarcomeric) forms, α-actinin-2 and α-actinin-3 (22, 23). Although eNOS is present in cardiomyocytes where it likely interacts with α-actinin-2, eNOS is predominantly expressed in vascular endothelial cells, which express α-actinin-1 and -4, but not -2. Indeed, similar to α-actinin-2, we found that α-actinin-4, but not α-actinin-1, was able to interact with eNOS in the yeast two-hybrid assays (Fig. 1D).
Expression of α-actinin isoforms in vascular endothelial cells
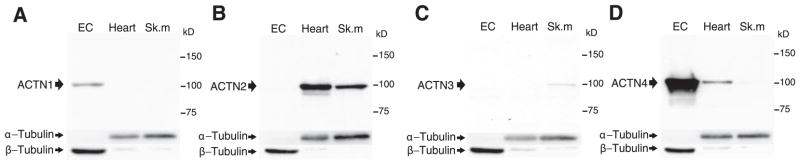
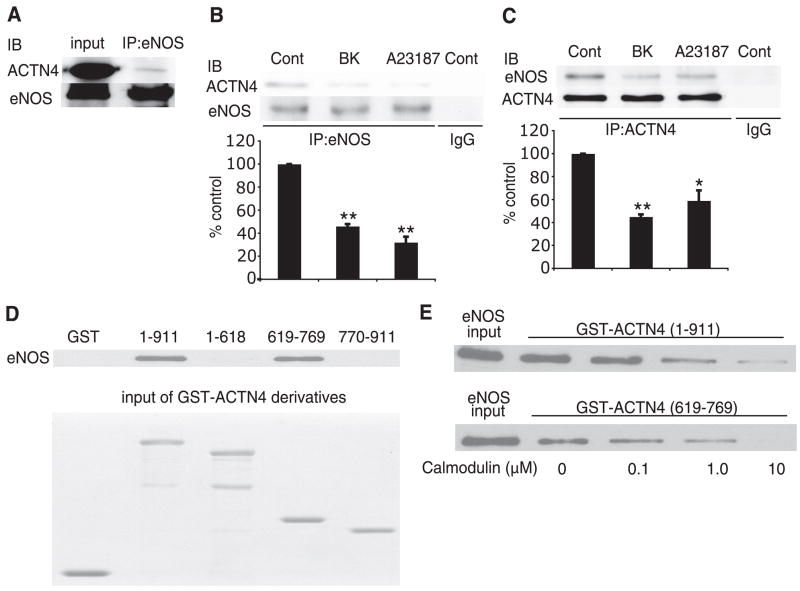
Using α-actinin isoform-specific antibodies (16, 17), we confirmed that endothelial cells express α-actinin-1 and α-actinin-4 (Fig. 2A, D) but do not express the striated muscle α-actinin isoforms, α-actinin-2 or α-actinin-3 (Fig. 2B, C). Interestingly, α-actinin-4, which is generally more abundant in highly motile cells than α-actinin-1 (24), is robustly expressed in endothelial cells, suggesting that endothelial cells may possess a high-motility phenotype. Indeed, α-actinin-4 co-immunoprecipitated with eNOS in vascular endothelial cells (Fig. 3A–C). This is consistent with the interaction of α-actinin-4 with eNOS observed in the yeast reporter assay (Fig. 1D).
Figure 2.
Expression of α-actinin isoforms in vascular endothelial cells. Western blot of ACTN isoforms in human saphenous vein endothelial cell (EC), mouse heart (Heart), and skeletal muscle (Sk.m) using isoform-specific antibodies to α-actinin-1 (A), α-actinin-2 (B), α-actinin-3 (C), and α-actinin-4 (D). For internal control, blots were reprobed with antibodies to α- and β-tubulin. Three separate experiments yielded similar results.
Figure 3.
Interaction of α-actinin-4 with eNOS. A) Immunoprecipitation (IP) of eNOS from human endothelial cell lysates with anti-eNOS antibody followed by immunoblotting with α-actinin-4 antibody. Experiments were performed 3 times with similar results. B) Lysates from untreated endothelial cells (Cont) or endothelial cells stimulated with bradykinin (BK, 1 μM) or the calcium ionophore A23187 (10 μM) were immunoprecipiated (IP) with eNOS antibody followed by immunoblotting with α-actinin-4 antibody. Experiments were performed 3 times with similar results. C) Lysates from untreated endothelial cells (Cont) or endothelial cells stimulated with bradykinin (BK, 1 μM) or the calcium ionophore A23187 (10 μM) were immunoprecipiated (IP) with α-actinin-4 antibody followed by immunoblotting with eNOS antibody. Densitometry analysis in bar graph. *P < 0.05 and **P < 0.01 compared with control. Experiments were performed 3 times with similar results. D) GST-ACTN4 pull-down assay using various deletional constructs of ACTN4 and recombinant bovine eNOS protein. Experiments were performed 3 times with similar results. E) Dose-dependent effects of calmodulin on eNOS interaction with GST-ACTN4 constructs. Note that calmodulin inhibited the interaction of eNOS with either GST-ACTN4 (1–911 aa) or GST-ACTN4 (619–769 aa). Experiments were performed 3 times with similar results.
Competitive interaction between α-actinin-4 and calmodulin for eNOS
On increases in intracellular calcium concentration, the calcium-calmodulin complex binds to eNOS via the calmodulin-binding domain of eNOS (1). Because the calmodulin-binding region on eNOS overlaps with the α-actinin-binding site, we investigated whether calmodulin can compete with α-actinin-4 for binding to eNOS. When endothelial cells were stimulated with bradykinin (1 μM) or the calcium ionophore A23187 (10 μM), co-immunoprecipitation studies showed decreased α-actinin-4-eNOS association (Fig. 3B, C). To further confirm α-actinin-4-eNOS interaction and to refine the binding region on α-actinin-4, we performed GST pull-down assay using GST fusion proteins containing full-length and 3 deletion mutants of α-actinin-4. The fusion proteins were expressed, purified, and incubated with recombinant eNOS protein. The full-length protein (1–911 aa) and R4 domain (619–769 aa) of α-actinin-4 bound to eNOS protein; however, the actin-binding domain, Rl, R2, and R3 (1–618 aa), and EF domain (770–911 aa) did not (Fig. 3D). These findings suggest that eNOS can directly bind to α-actinin-4 in vitro and confirm that the R4 domain of α-actinin-4 contains the eNOS-binding site. Finally, the interaction between eNOS and α-actinin-4 was inhibited in a concentration-dependent manner by addition of recombinant calmodulin (Fig. 3E). Similar findings were observed with the interaction between eNOS and R4 domain of α-actinin-4 (619–769 aa).
Dynamic subcellular localization of eNOS and α-actinin-4 in endothelial cells
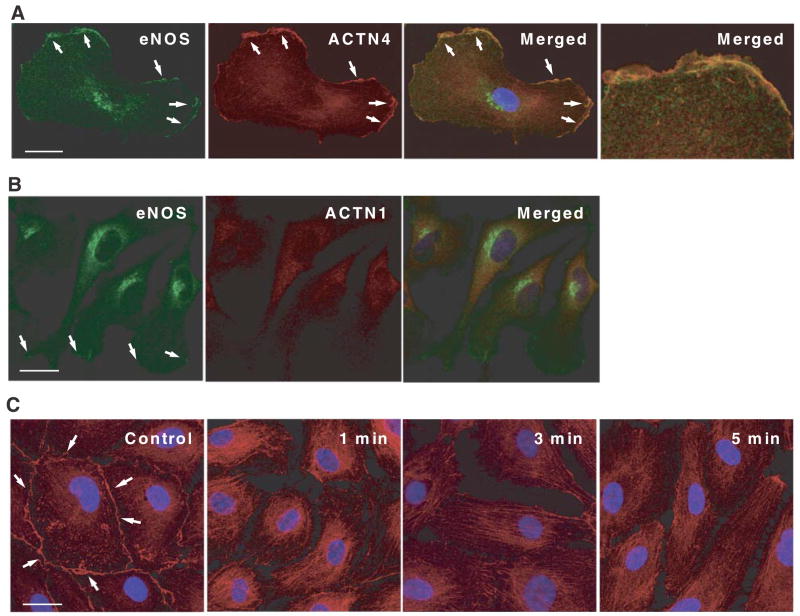
Cellular localization of cytoskeletal α-actinins and α-actinin-1 and -4 is quite distinct from the sarcomeric isoforms, α-actinin-2 and -3. For example, α-actinin-1 typically localizes to focal adhesion plaques and adherens junction, whereas α-actinin-4 is preferentially localized to moving structures, such as dorsal ruffles, lamellipodia, and filopodia and participates in circular ruffling, macropinocytosis, and phagocytosis (20, 25). To determine the subcellular localization α-actinin-4 and whether it colocalizes with eNOS, we performed double immunofluorescence staining of vascular endothelial cells using anti-eNOS and anti-α-actinin-4 antibodies. Although most of the α-actinin-4 resides in the cytoplasm under basal cell condition, substantial amounts of α-actinin-4 also reside at the cell membrane, where it colocalizes with eNOS (Fig. 4A). In contrast, α-actinin-1 localizes predominantly to the cytoplasm with little, if any, present at the cell membrane (Fig. 4B). Treatment with the calcium ionophore A23187, which is a known activator of eNOS through increases in calcium-calmodulin binding, decreased the amount of α-actinin-4 staining at the cell membrane within 1 min (Fig. 4C). After 3 min, most of the α-actinin-4 was detected in the cytoplasm. These findings indicate that α-actinin-4 dynamically regulates the rapid calcium-dependent basal activation of eNOS and suggest that eNOS subcellular localization, in part, may be dependent on interaction with α-actinin-4 and the actin cytoskeleton.
Figure 4.
Dynamic subcellular localization of α-actinin-4 and eNOS in endothelial cells. A) Immunofluorescence localization of eNOS and α-actinin-4 on HSVECs. The cells were double stained for eNOS (green) and α-actinin-4 (red), and analyzed with a confocal microscope. Merged image shows a colocalization of eNOS and α-actinin-4. Right panel: magnification of colocalized area. B) Immunofluorescence localization of eNOS and α-actinin-1. The cells were double stained for eNOS (green) and α-actinin-1 (red). Merged image does not show colocalization of eNOS and α-actinin-1. C) Dynamic movement of α-actinin-4 induced by Ca2+ ionophore. Small amount of α-actinin-4 was localized at cell membrane before stimulation. Stimulation with A23187 (10 μM) decreased membrane localization of α-actinin-4 within 1 min. Scale bars = 25 μm.
Direct inhibition of eNOS activity by α-actinin-4
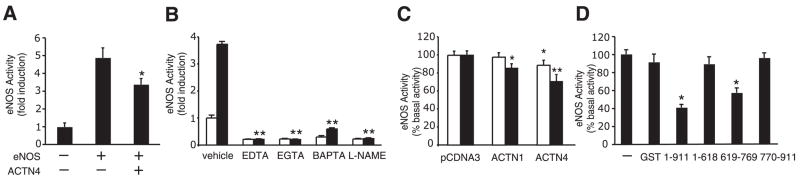
To determine whether α-actinin-4 regulates eNOS activity, we transfected COS-7 cells with expression plasmids containing eNOS, with or without cotransfection with α-actinin-4. Transfection of eNOS alone caused an increase in eNOS activity, measured by the conversion from L-arginine to L-citrulline. Coexpression of α-actinin-4 decreased eNOS activity by ~30% (Fig. 5A). Similarly, cotreatment with calcium chelators, EDTA, EGTA, and l,2-bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid, tetraacetoxymethyl ester (BAPTA-AM) or the NOS inhibitor N-(ù)-nitro-L-arginine methylester (L-NAME) inhibited A23187-induced eNOS activity in BAECs (Fig. 5B). Indeed, overexpression of α-actinin-4 and, to a lesser extent, α-actinin-1 also inhibited A23187-induced eNOS activity in transfected BAECs (Fig. 5C). Transfection efficiency was estimated at 32% using GFP expression plasmid. Overall, transfection of α-actinin-4 resulted in a 28% decrease in eNOS activity of the entire culture dish (transfected and untransfected cells). However, taking into account the transfection efficiency in BAECs that were actually transfected with α-actinin-4, the suppression of eNOS activity by α-actinin-4 was calculated to be ~87%.
Figure 5.
Direct inhibition of eNOS activity by α-actinin-4. A) Reduction in transfected eNOS activity in COS-7 cells cotransfected with ACTN4 cDNA. eNOS activity was measured by arginine-to-citrulline conversion assay. *P < 0.05 compared with transfection with eNOS cDNA alone. B) eNOS activity (fold induction) in BAECs stimulated with (■) and without (□) A23187 (10 μM) in the presence of calcium chelators, EDTA (10 mM), EGTA (10 mM), and BAPTA-AM (10 μM) or NOS inhibitor L-NAME (1 mM). Cells were pretreated with chelators or inhibitors for 30 min before A23187 stimulation. eNOS activity measured using DAF-2. **P < 0.01 compared with vehicle. C) Inhibition of A23187 (1 μM □,10 μM ■) -induced eNOS activity by ACTN1 and ACTN4 cDNA in transfected BAECs. eNOS activity measured using DAF-2. *P < 0.05, **P < 0.01 compared with pcDNA3, respectively. D) The effect of GST-ACTN4 (1–911 aa) or GST-ACTN4 (619–769 aa) on recombinant bovine eNOS protein activity in vitro. eNOS activity measured by arginine-to-citrulline conversion assay. *P < 0.05 compared with GST.
To confirm that the R4 domain of α-actinin-4, which is the putative site of interaction with eNOS, can inhibit eNOS activity, we measured eNOS activity in vitro by incubating recombinant eNOS protein with GST fusion α-actinin-4 protein. The GST alone had no effect on the eNOS activity, whereas full-length α-actinin-4 (1–911 aa) dramatically reduced eNOS activity by ~60% (Fig. 5D). In particular, the α-actinin-4 deletion mutant, 619–769 aa, which comprises the R4 domain, also inhibited eNOS activity. In contrast, the α-actinin-4 deletion mutants, 1–618 aa and 770–911 aa, which do not contain the eNOS-binding site, were unable to inhibit eNOS activity.
To determine whether the loss of α-actinin-4 could affect basal eNOS activity in endothelial cells, we knocked down α-actinin-4 expression using siRNA that is specific for bovine ACTN4. Compared with control siRNA, transfection of bovine ACTN4 siRNA into BAEC resulted in a 90% decrease in α-actinin-4 expression and a 42% increase in eNOS activity (Fig. 6A, B). These findings indicate that α-actinin-4 is critical for basal eNOS activity.
Figure 6.
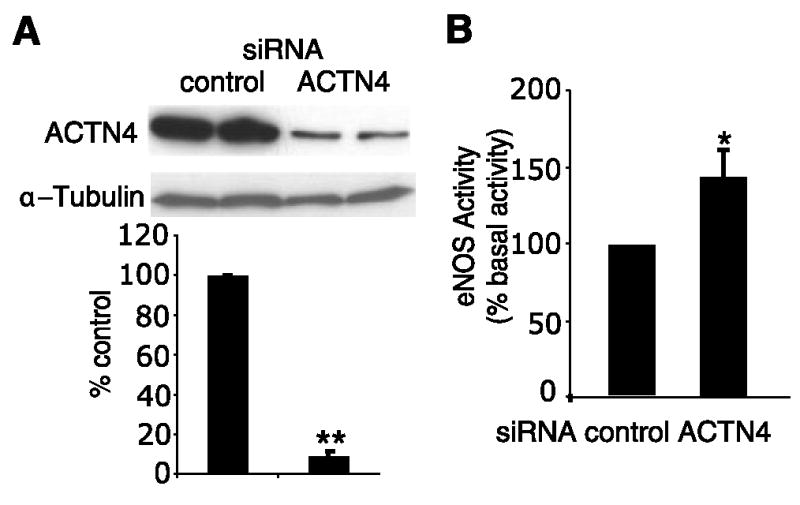
Knockdown ofACTN4 in endothelial cells. A) Effect of transfection of BAECs with control or ACTN4 siRNA on α-actinin-4 expression. **P < 0.01, n = 6. B) Effect of transfection of BAECs with control or ACTN4 siRNA on basal eNOS activity (% basal activity). eNOS activity measured using DAF-2. *P < 0.05, n = 6.
DISCUSSION
As a highly lipophilic and diffusible molecule, the subcellular localization of eNOS is critical for its function, both intercellularly and intracellularly (26). Indeed, eNOS that is restricted to the plasma membrane appears to be more sensitive to calcium- and Akt-dependent agonists than is Golgi-localized eNOS (27). The subcellular localization and activation of eNOS is regulated post-translationally by myristoylation at N-terminal glycine residue 2 (2); palmitoylation at N- terminal cysteine residues 15 and 26 (28); phosphorylation at serine residues 116, 617, 635, 1177, and threonine residue 495 (29); S-nitrosylation at cystein residues 94 and 99 (6, 7); and various protein-protein interactions (3, 30, 31).
We have shown that under basal conditions, eNOS is bound to α-actinin-4 in vascular endothelial cells. Stimulation with either bradykinin or A23187 leads to rapid dissociation of α-actinin-4-eNOS interaction, loss of α-actinin localization in the cell membrane, and increased eNOS activity. Indeed, addition of exogenous calmodulin can competitively disinhibit the effects of α-actinin-4 on eNOS. Overexpression of α-actinin-4 or addition of calcium chelators such as EDTA, EGTA, or BAPTA leads to inhibition of A23187-induced eNOS activity. In contrast, the knockdown of α-actinin-4 increases basal eNOS activity in endothelial cells. These findings suggest that the association of α-actinin-4 with eNOS dynamically regulates basal eNOS activity. Activation of eNOS by agents, which increases intracellular calcium, leads to the displacement of α-actinin-4 from eNOS by calmodulin.
At the cell membrane, caveolin-1 is also known to regulate eNOS activity negatively by competing with calmodulin (32). However, eNOS-caveolin protein-protein interaction appears unnecessary for membrane targeting of eNOS (33), and the caveolin-binding site is distinct and far from the α-actinin-4-binding site (Fig. 1C). Furthermore, targeted disruption of the caveolin-1 gene is not lethal (34), although caveolin-1 appears to be required for calcium-independent eNOS activation by vascular endothelial growth factor (35). Thus, in caveolin-1-deficient endothelial cells, other factors such as, perhaps, α-actinin-4 may be involved in regulating calcium-dependent eNOS activation by calmodulin. Our findings suggest that caveolin-1 and α-actinin-4 may work cooperatively to tonically inhibit basal eNOS activity.
We find that basal eNOS activity in vascular endothelial cells is tonically regulated by competitive interaction between α-actinin-4 and calmodulin for eNOS. α-actinin was isolated as an actin-binding protein that bridges other cytoskeletal proteins such as vinculin and zyxin and cell adhesion proteins such as integrin β1, β2, intercellular adhesion molecule 1 (ICAM1), ICAM2, and L-selectin to the actin network (24). Recently, α-actinin has been shown to bind to signal transduction proteins such as the NMDA receptor and serves as scaffold protein for phosphatidylinositol 3-kinase, MEK kinase 1, and extracellular regulated kinase (24). In particular, a splice variant of α-actinin-4 localizes predominantly in the nucleus (36), where it associates with histone deacetylase 7, resulting in increased myocyte enhancer factor-2A activity. Therefore, actinins are not only cytoskeletal proteins but also serve as important regulators of cellular functions.
α-Actinins form antiparallel dimers with actin-binding domain (composed of 2 calponin-homology domains) at each end, which are used to cross-link actin filaments. α-actinins contain 4 spectrin-like repeats and a calmodulin-like domain consisting of 2 EF-hand motifs. In our study, the eNOS-binding site is mapped to the R4 domain of α-actinin, and this spectrin-like repeat is curved axially and twisted to form high-affinity binding sites for other molecules such as titin, α-catenin, ICAM2, NMDA receptor, and calsarcin-2/myozenin (14, 37, 38). Although α-actinin-1 may also bind to eNOS, albeit to a lesser degree compared with α-actinin-4, its subcellular localization (i.e., cytoplasm) does not make it conducive for inhibiting basal eNOS activity at the cell membrane. Interestingly, alternative splicing of the EF hand renders the cytoskeletal α-actinin isoforms, α-actinin-1 and α-actinin-4, more sensitive to calcium binding compared to the relatively insensitive sarcomeric isoforms (23, 39). Perhaps this process allows α-actinin-4 in endothelial cells, as opposed to α-actinin-2 in cardiomyocytes, to fine-tune eNOS activity on increases in intracellular calcium and competition with calmodulin. Further studies are required to determine whether this dynamic interaction of α-actinin-4 with eNOS is the primary mechanism by which eNOS is localized to the plasma membrane and whether the competition between α-actinin-4 and calmodulin for eNOS is the principle determinant of eNOS sensitivity to changes in intracellular calcium.
Acknowledgments
This work was supported by grants from the U.S. National Institute of Health (HL052233 and AR044345) and the Muscular Dystrophy Association.
References
- 1.Forstermann U, Munzel T. Endothelial nitric oxide synthase in vascular disease: from marvel to menace. Circulation. 2006;113:1708–1714. doi: 10.1161/CIRCULATIONAHA.105.602532. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez E, Kou R, Lin AJ, Golan DE, Michel T. Subcellular targeting and agonist-induced site-specific phosphorylation of endothelial nitric-oxide synthase. J Biol Chem. 2002;277:39554–39560. doi: 10.1074/jbc.M207299200. [DOI] [PubMed] [Google Scholar]
- 3.Dudzinski DM, Igarashi J, Greif D, Michel T. The regulation and pharmacology of endothelial nitric oxide synthase. Annu Rev Pharmacol Toxicol. 2006;46:235–276. doi: 10.1146/annurev.pharmtox.44.101802.121844. [DOI] [PubMed] [Google Scholar]
- 4.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 5.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, Franke TF, Papapetropoulos A, Sessa WC. Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature. 1999;399:597–601. doi: 10.1038/21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci U S A. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erwin PA, Lin AJ, Golan DE, Michel T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 8.Bredt DS, Snyder SH. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc Natl Acad Sci U S A. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair A, Shaul PW, Yuhanna IS, Conrad PA, Smart EJ. Oxidized low density lipoprotein displaces endothelial nitric-oxide synthase (eNOS) from plasmalemmal caveolae and impairs eNOS activation. J Biol Chem. 1999;274:32512–32519. doi: 10.1074/jbc.274.45.32512. [DOI] [PubMed] [Google Scholar]
- 10.Feron O, Belhassen L, Kobzik L, Smith TW, Kelly RA, Michel T. Endothelial nitric oxide synthase targeting to caveolae. Specific interactions with caveolin isoforms in cardiac myocytes and endothelial cells. J Biol Chem. 1996;271:22810–22814. doi: 10.1074/jbc.271.37.22810. [DOI] [PubMed] [Google Scholar]
- 11.Ju H, Zou R, Venema VJ, Venema RC. Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. J Biol Chem. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Liao JK. Functional interaction of endothelial nitric oxide synthase with a voltage-dependent anion channel. Proc Natl Acad Sci U S A. 2002;99:13108–13113. doi: 10.1073/pnas.202260999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada F, Vander Woude DL, Tong HQ, Thompson TG, Watkins SC, Kunkel LM, Beggs AH. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci U S A. 2001;98:1595–1600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laufs U, Fata VL, Liao JK. Inhibition of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase blocks hypoxia-mediated down-regulation of endothelial nitric oxide synthase. J Biol Chem. 1997;272:31725–31729. doi: 10.1074/jbc.272.50.31725. [DOI] [PubMed] [Google Scholar]
- 16.Chan Y, Tong HQ, Beggs AH, Kunkel LM. Human skeletal muscle-specific alpha-actinin-2 and -3 isoforms form homodimers and heterodimers in vitro and in vivo. Biochem Biophys Res Commun. 1998;248:134–139. doi: 10.1006/bbrc.1998.8920. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 18.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and imaging of nitric oxide with novel fluorescent indicators: di-aminofluoresceins. Anal Chem. 1998;70:2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 19.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 20.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millake DB, Blanchard AD, Patel B, Critchley DR. The cDNA sequence of a human placental alpha-actinin. Nucleic Acids Res. 1989;17:6725. doi: 10.1093/nar/17.16.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youssoufian H, McAfee M, Kwiatkowski DJ. Cloning and chromosomal localization of the human cytoskeletal alpha-actinin gene reveals linkage to the beta-spectrin gene. Am J Hum Genet. 1990;47:62–71. [PMC free article] [PubMed] [Google Scholar]
- 23.Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- 24.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 25.Araki N, Hatae T, Yamada T, Hirohashi S. Actinin-4 is preferentially involved in circular ruffling and macropinocytosis in mouse macrophages: analysis by fluorescence ratio imaging. J Cell Sci. 2000;113(Pt 18):3329–3340. doi: 10.1242/jcs.113.18.3329. [DOI] [PubMed] [Google Scholar]
- 26.Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Church JE, Jagnandan D, Catravas JD, Sessa WC, Fulton D. Functional relevance of Golgi- and plasma membrane-localized endothelial NO synthase in reconstituted endothelial cells. Artehoscler Thromb Vasc Biol. 2006;26:1015–1021. doi: 10.1161/01.ATV.0000216044.49494.c4. [DOI] [PubMed] [Google Scholar]
- 28.Shaul PW. Regulation of endothelial nitric oxide synthase: location, location, location. Annu Rev Physiol. 2002;64:749–774. doi: 10.1146/annurev.physiol.64.081501.155952. [DOI] [PubMed] [Google Scholar]
- 29.Bauer PM, Fulton D, Boo YC, Sorescu GP, Kemp BE, Jo H, Sessa WC. Compensatory phosphorylation and protein-protein interactions revealed by loss of function and gain of function mutants of multiple serine phosphorylation sites in endothelial nitric-oxide synthase. J Biol Chem. 2003;278:14841–14849. doi: 10.1074/jbc.M211926200. [DOI] [PubMed] [Google Scholar]
- 30.Sessa WC. eNOS at a glance. J Cell Sci. 2004;117:2427–2429. doi: 10.1242/jcs.01165. [DOI] [PubMed] [Google Scholar]
- 31.Nedvetsky PI, Sessa WC, Schmidt HH. There’s NO binding like NOS binding: protein-protein interactions in NO/cGMP signaling. Proc Natl Acad Sci U S A. 2002;99:16510–16512. doi: 10.1073/pnas.262701999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel JB, Feron O, Sacks D, Michel T. Reciprocal regulation of endothelial nitric-oxide synthase by Ca2+-calmodulin and caveolin. J Biol Chem. 1997;272:15583–15586. doi: 10.1074/jbc.272.25.15583. [DOI] [PubMed] [Google Scholar]
- 33.Prabhakar P, Cheng V, Michel T. A chimeric transmembrane domain directs endothelial nitric-oxide synthase palmitoylation and targeting to plasmalemmal caveolae. J Biol Chem. 2000;275:19416–19421. doi: 10.1074/jbc.M001952200. [DOI] [PubMed] [Google Scholar]
- 34.Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, Di Vizio D, Hou H, Jr, Kneitz B, Lagaud G, Christ GJ, Edelmann W, Lisanti MP. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 35.Sonveaux P, Martinive P, DeWever J, Batova Z, Daneau G, Pelat M, Ghisdal P, Gregoire V, Dessy C, Balligand JL, Feron O. Caveolin-1 expression is critical for vascular endothelial growth factor-induced ischemic hindlimb collateralization and nitric oxide-mediated angiogenesis. Circ Res. 2004;95:154–161. doi: 10.1161/01.RES.0000136344.27825.72. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S, Kao HY. α-Actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J Biol Chem. 2006;281:35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- 37.Tang J, Taylor DW, Taylor KA. The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca(2+)-dependent actin binding. J Mol Biol. 2001;310:845–858. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- 38.Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- 39.Burridge K, Feramisco JR. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature. 1981;294:565–567. doi: 10.1038/294565a0. [DOI] [PubMed] [Google Scholar]