Abstract
Background
DNA-methyltransferase-3B (DNMT3B), which plays a role in DNA methylation, is usually aberrant expression involved in carcinogenesis. Polymorphisms of the DNMT3B gene may influence DNMT3B activity on DNA methylation in several cancers, thereby modulating the susceptibility to cancer.
Methods
DNMT3B -579G>T genotypes and -149C>T were determined by PCR-RFLP and sequencing in 137 colorectal cancer patients and 308 controls matched for age and sex, who did not receive radiotherapy or chemotherapy for newly diagnosed and histopathologically confirmed colorectal cancer. The association between two SNPs of the DNMT3B promoter and the risk of the development of colorectal cancer was analyzed in a population of Chinese.
Results
The allele frequency of -149C >T among patients and controls was 0.73% versus 0.65%, respectively. The allele frequency of -597G>T for patients and controls was 6.57% versus 11.53%, respectively. Individuals with at least one -149C>T allele were no at a significantly increase risk of colorectal cancer compared with those having a -149TT genotype. However, Individuals with at least one 579G>T allele were decreased risk of colorectal cancer compared with those having a -579TT genotype.
Conclusion
The relative distribution of -149C>T DNMT3B SNPs among a Chinese population can not be used as a stratification marker to predict an individual's susceptibility to colorectal cancer. However, the DNMT3B -579G>T polymorphism may contribute to the genetic susceptibility to colorectal cancer.
Background
Single nucleotide polymorphisms (SNPs) are the most common form of human genetic variation, and may contribute to an individuals' susceptibility to cancer. Some studies have suggested that some variants in the promoter regions of genes may affect either the expression or activity levels of enzymes [1-3] and therefore may be mechanistically associated with cancer risk. DNA methylation is a major epigenetic modification involving the addition of a methyl group to the 5' position of a cytosine in a CpG dinucleotide. A number of studies have suggested that aberrant DNA cytosine methylation may play an important role in carcinogenesis [4-8]. DNMT3a and DNMT3b are required for the establishment and maintenance of genomic methylation patterns and proper murine development [9-12]. Both genes are up-regulated to differing degrees in some malignancies, including esophagus carcinoma, lung cancer and colorectal cancer [13-18]. Recently, several studies showed that some of SNPs in the DNMT3B gene may influence DNMT3B activity on DNA methylation, thereby modulating the susceptibility to lung cancer, breast cancer and gastric cardiac adenocarcinoma [14,19,20]. The DNMT3B gene contains a single C>T transition polymorphism (C46359T, GenBank accession no. AL035071) in the promoter region of the DNMT3B gene, -149 base pairs from the transcription start site, is reported to greatly increase promoter activity [21]. Some reports have shown that the C/T polymorphism is associated with an increased risk for lung cancer and carcinoma of the head and neck. Carriers of T allele, particularly heterozygotes, have a significantly increased risk for such cancers[2,22-24]. However, C/T polymorphism is not associated with the increased risk of hapetocellular carcinoma (HCC) and gastric cancer, especially in Chinese. Another G>T SNP of DNMT3B gene, in the transcription start site of the promoter region (-579 bp from exon 1B, GenBank accession no. NT_028392) and this probably affects gene function [2]. Some studies have suggested that the DNMT3B -579 G>T may modify susceptibility to tumors, although conflicting results have been reported in different tumor types, the heterozygous genotype have been reported to have a significantly reduced risk of developing lung and colorectal cancer [25-27], but DNMT3B genetic polymorphism is variable in different races, ethnic groups or geographic areas. In the present study, we evaluated the association of _149C>T and _579G>T polymorphisms with colorectal cancer in Chinese population.
Methods
Study population
This case-control study included 137 colorectal cancer and 308 healthy controls, and the informed consent was obtained. The 137 case subjects were patients who had undergone surgery and been histopathologically confirmed at the Zhongda Hospital of Southeast University and Tumor Hospital, Nanjing, China. The control subjects were selected from a pool of cancer-free subjects who visited the same hospital for a regular physical examination and who volunteered to join the epidemiology survey during the same period. We defined a healthy subject as a person free of disease (including no history of cancer) on health check-up. They were matched with the case patients by age and sex (Table 1). All cases and controls were ethnically Chinese and resided in Jiangsu province or in the surrounding regions.
Table 1.
Characteristics of the study population.
| Variables | Case (n = 137) | Control (n = 308) |
| Ages (years)* | 65 | 71 |
| Sex | ||
| Male | 91 | 206 |
| Female | 46 | 102 |
* p > 0.05
DNA extraction
5 milliliters of venous blood from each subject was drawn in vacuum tubes containing EDTA and stored at 4°C. Genomic DNA was extracted within one week after sampling by using proteinase K digestion followed by a salting out procedure [28].
DNMT 3B genotyping
The transition of C/T of DNMT3B SNP creates a BlnI restriction site and the transition from G to T of the DNMT3B SNP creates a PvuII restriction site, which could be exploited for genotyping by PCR and subsequent restriction fragment length polymorphism (RFLP) analysis. PCR was performed in a volume of 25 μL containing 100 ng of DNA template, 10 × PCR master mix (Promega, USA), and 10 pmol/L each of sense primer (5'-TGCTGTGACAGGCAGA2GCAG-3') and antisense primer (5'-GGTAGCCGGGAACTCCACGG-3') for _149C>T, and sense primer (5'-GAGGTCTCATTATGCCTAGG-3') and antisense primer (5'-GGGAGCTCACCTTCTAGAAA-3') for _579G>T. For PCR amplification, an initial denaturation step at 94°C for 5 min was followed by 30 cycles at 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, and a final extension step at 72°C for 7 min. Subsequently, the PCR products were digested overnight with Blnl (TaKaRa) 4 U for 149C>T and 5 units of PvuII (New England Biolabs, Beverly, Mass.) for 579G>T at 37°C, respectively, and the products separated on 2% agarose gels. RFLP bands were visualized by ethidium bromide staining under UV light. The 149C>T polymorphism depend upon the existence of the Blnl recognizing site, the DNMT3B T/T genotype was expected to show two DNA bands at the positions of 207 and 173 bp, the C/C genotype was expected to show a single band (380 bp), while the heterozygote was expected to have three bands (380, 207 and 173 bp). The 579G>T polymorphism was determined by PvuII. The wild-type G allele has only one band, while the polymorphic T allele has two bands (132 and 93 bp). For quality control, genotyping analysis was performed blind with respect to case/control status and repeated twice for all subjects. The results of genotyping were 100% concordant. To confirm the genotyping results, selected PCR-amplified DNA samples (n = 3, respectively, for each genotype) were examined by DNA sequencing, and the results were also 100% concordant.
Statistical analysis
Cases and controls were compared using Student's t-test for continuous variables and the X2 test for categorical variables. Hardy – Weinberg equilibrium was tested with a goodness-of-fit X2 test with one degree of freedom to compare the observed genotype frequencies with the expected genotype frequencies among the subjects. Comparison of the DNMT3B genotype and allelotype distribution in the study groups was performed by means of two-sided contingency tables using X2 test or Fischer's exact test. The odds ratio (OR) and 95% confidence interval (CI) were calculated using an unconditional logistic regression model and adjusted by age and gender accordingly. P < 0.05 was considered statistically significant.
Results
The demographics of the cases and controls enrolled in this study are shown in Table 1. There were no significant differences in the mean age and sex distribution between cases and controls, suggesting that the matching based on these two variables was adequate. There was no evidence of a deviation from Hardy-Weinberg equilibrium among the case or control subjects. The mean age was 65 years (range, 34 – 80 years) for the case patients and 71 years (range, 32 – 80 years) for the control subjects (Table 1).
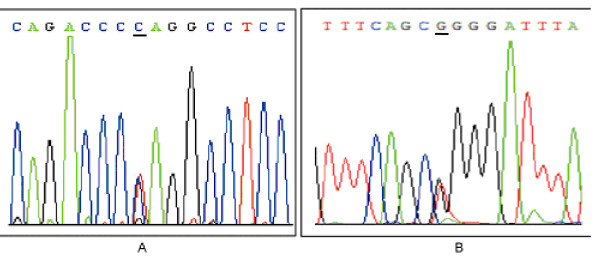
All patients and controls were successfully genotyped for the DNMT3B polymorphism. The genotyping by PCR-RFLP analysis was completely confirmed by DNA sequencing analysis, and the results of PCR-RFLP genotyping and sequencing analysis were also 100% concordant (Fig. 1). The distribution of the 149C>T and the 579G>T polymorphism of DNMT3B were in Hardy-Weinberg equilibrium. The distributions of DNMT3B 149C>T and 579G>T genotypes among controls and cases are shown in Table 2. The allele frequency of 149C>T among patients and controls was 0.73% versus 0.65%, respectively. The allele frequency of 579G>T for patients and controls was 6.57% versus 11.53%, respectively. No significant deviation was observed for the genotype distributions of 149C>T polymorphisms between colorectal cancer cases and controls. However, the distributions of 579G>T genotypes in the colorectal cancer group (GG 0%, GT 13.14%, TT 86.86) were significantly different from those among the controls (GG 1.95%, GT 19.15%, TT 78.90%; 0.01 < P < 0.05).
Figure 1.
Sequencing results for each of the PCR products from -149 C>T (A) and -579 G>T (B). Sequencing results for each of the PCR products from -149 C>T (A) and -579 G>T (B), the SNP sites are as indicated by the arrowhead. These results were completely matched to corresponding results deriving from PCR-RFLP by agarose gel genotyping method.
Table 2.
DNMT3B genotypes and allele frequency among controls and cases
| -149 C > T genotype | -579G.>T genotype | |||||||
| CC | CT | TT | C alleleb frequency (%) |
GG | GT | TT | G allelec frequency (%) |
|
| Control | 0(0.00) | 4(1.30) | 304(98.70) | 0.65 | 6(1.95) | 59(19.15) | 243(78.9) | 11.53 |
| Case | 0(0.00) | 2(1.46) | 135(98.54) | 0.73 | 0(0.00) | 18(13.14) | 119(86.86) | 6.57 |
aNumbers in parentheses, percentage.
bp > 0.05
c0.01 <p < 0.05
The colorectal cancer risk related to the DNMT3B 149C>T and 579G>T genotypes were shown in table 3. ORs and their 95% CIs were calculated using the more common homozygous variant genotype as the reference group (149TT and 579 TT genotypes, respectively). Because of low prevalence of homozygous wild-type genotype, we combined this genotype with heterozygous genotype into one group and compared it with the reference group. As compared with the reference group, the combined 149CC and CT genotype, were associated with non-significant increased of colorectal cancer (OR = 1.13; 95% CI = 0.20–6.22). However, as compared with the reference group, the combined 579 GG and GT genotype, were associated with significant decreased of colorectal cancer (OR = 0.57 95% CI = 0.32–1.00). Then we stratified the results by age, patients and controls were found to be a little different with respect to the genotype distribution. Our data thus reveal that the presence of 579 G>T rather than 149 C>T exists significance likelihood of carcinogenesis for colorectal cancer patients, at least in a Chinese population. The results didn't show that 597 G>T is associated with the onset age of CRC in this study.
Table 3.
Crude and adjusted ORs for colorectal cancer associated with DNMT3B genotypes
| Genotypes | Cases |
| -149 T>C genotype | |
| TT (reference) | 1.00 |
| TC +CC crude | 1.13 (0.20–6.22) |
| -579 G.>T genotype | |
| TT (reference) | 1.00 |
| GT + GG crude | 0.57(0.32–1.00) |
Discussion
Colorectal cancer (CRC) is one of the most common cancers, with an estimated worldwide incidence of more than 570,000 new cases per year [29]. Despite recent surgical, radiotherapy and chemotherapy advances in the treatment protocol, the long-term survival of CRC patients still remains at approximately 50% for the past three decades. Early detection among susceptible populations has been advocated to decrease both the morbidity and the mortality of CRC, but searching for a useful stratification marker to predict its genetic susceptibility still would appear to impose a major challenge for CRC researchers.
DNA methylation has been reported to play a significant role in the development and progression of various cancers. In such cases, DNA methylation is typically mediated by DNA methyltransferases (DNMTs), specifically DNMT1, DNMT3A and DNMT3B. DNMT1 is considered as a maintenance DNA methyltransferase due to its ability to preferentially methylate hemimethylated DNA subsequent to DNA replication [30,31]. DNMT3A and DNMT3B function as de novo methyltransferases, which reportedly methylate unmethylated and hemimethylated DNA with equal efficiencies [32].
A single nucleotide polymorphism (SNP) of the DNMT3B promoter polymorphism -149 C>T, have recently been shown increase susceptibility of an individual to lung cancer [2], breast cancer [23] and significantly associated with increased age-associated risk in HNPCC families [24], but not to head and neck squamous cell carcinoma in Taiwanese [33], HCC [34] and gastric cardiac adenocarcinoma in Chinese [35], and gastric cancer in a Japanese population [36]. Another single nucleotide polymorphism (SNP) of the DNMT3B promoter 579G > T is from exon 1B transcription start site of DNMT3B. Although it is supposed probably affect the gene function [27], the 579 G>T polymorphisms did not significantly change the promoter activity of DNMT3B by luciferase assay [26]. However, studies recent been shown decrease susceptibility of an individual to lung cancer [2] and colon cancer [27]. These data suggested that DNMT3B promoter 579G > T polymorphism can be used a risk factor of cancer to evaluate the population susceptible to tumors. Some other study haven't shown association between polymorphism of 579G > T and head and neck squamous cell carcinoma [33], and esophagus cancer [37].
In the current study, we investigated the influence of DNMT3B polymorphisms -149 C>T and -579G > T on the risk of colorectal cancer in a hospital-based case-control study in a Chinese population. For -149 C>T, CC genotype was not detectable in both CRC patients and controls, while the T allele was predominant just like in a Taiwanese population [33] and a Japanese population [36]. There was no significant difference between CRC patients and control in -149 C>T of DNMT3B polymorphisms. These data implied that, an individual with the -149C>T polymorphism has not an increased susceptibility to CRC cancer in the study Chinese population. However, carriers with 579G allele were at decreased risk of CRC as compared with individuals having 579T alleles. This finding suggests that the 579G>T polymorphism in the DNMT3B gene could be used as a marker of genetic susceptibility to CRC cancer.
Our findings are consistent with those of studies, which showed that individuals carrying the G allele have a significantly lower risk of developing adenocarcinoma of the lung cancer [2] and colon cancer [27]. Carriers with the G allele in the DNMT3B gene were found to have a decreased risk of colon cancer compared with individuals with the T allele. These findings implied that the DNMT3B polymorphism might operate in a tissue-specific manner, especially in colorectal tissue. However, this finding needs to be confirmed by a larger study. The present result, and those from lung and colon cancer which are the only published data on association between DNMT3B SNP and cancer development, can show the different possible roles of DNMT3B in different cell types. Since the different splice variants of DNMT3B which may alter catalytic activity are expressed in a tissue specific manner [10,38-40] and repression of DNMT3B activity does not result in the re-expression of all hypermethylated tumor suppressor genes in some cell system [41-44], it is therefore important to explore the complex interplay of DNMTs in different tumor types. Since genetic polymorphisms often vary between ethnic groups, further studies are needed to clarify the association of the DNMT3B polymorphism with colorectal cancer in diverse ethnic populations. Future studies of other DNMT3B sequence variants and their biologic function are also needed to understand the role of DNMT3B polymorphisms in determining the risk of cancer.
Conclusion
The DNMT3b -579G>T polymorphism may contribute to the genetic susceptibility to colorectal cancer. Carriers with the G allele in the DNMT3B gene were found to have a decreased risk of colorectal cancer compared with individuals with the T allele. However, the relative distribution of -149C>T DNMT3B SNP among a Chinese population can not be used as a stratification marker to predict an individual's susceptibility to colorectal cancer.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HF carried out DNMT3B -579G>T polymorphism analysis and wrote the manuscript, FZ collected the samples and patient's clinical data. DSL and JBH performed -149C>T DNMT3B SNP research and analyzed the data. ZJZ contributed to new reagents/analytic tools. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 30470950) and Qinglan project of Jiangsu province of China (2006).
Contributor Information
Hong Fan, Email: fanh@seu.edu.cn.
Feng Zhang, Email: zhangfeng81868@163.com.
Jiabo Hu, Email: hu@ujs.edu.cn.
Dongsheng Liu, Email: lds_16888@tom.com.
Zhujiang Zhao, Email: zhujiangzhao@yahoo.com.cn.
References
- Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol. 2000;183:145–154. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62:4992–4495. [PubMed] [Google Scholar]
- Skoog T, van't Hooft FM, Kallin B, Jovinge S, Boquist S, Nilsson J, Eriksson P, Hamsten A. common functional polymorphism (C-->A substitution at position -863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999;8:1443–1449. doi: 10.1093/hmg/8.8.1443. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Beaulieu N, Morin S, Chute IC, Robert MF, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277:28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Lerner CP, Di Lacio LC, Laird PW, Sharpe AH, Simpson EM. Transgenic mice for the preparation of hygromycin-resistant primary embryonic fibroblast feeder layers for embryonic stem cell selections. Nucleic Acids Res. 1995;23:1273–1275. doi: 10.1093/nar/23.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:1511–1515. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–2287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- Xu W, Fan H, He X, Zhang J, Xie W. LOI of IGF2 is associated with esophageal cancer and linked to methylation status of IGF2 DMR. J Exp Clin Cancer Res. 2006;25:543–547. [PubMed] [Google Scholar]
- Simão Tde A, Simões GL, Ribeiro FS, Cidade DA, Andreollo NA, Lopes LR, Macedo JM, Acatauassu R, Teixeira AM, Felzenszwalb I, Pinto LF, Albano RM. Lower expression of p14ARF and p16INK4a correlates with higher DNMT3B expression in human oesophageal squamous cell carcinomas. Hum Exp Toxicol. 2006;25:515–522. doi: 10.1191/0960327106het649oa. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang J, Sun S, Rodriguez M, Yue P, Jang SJ, Mao L. A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int J Oncol. 2006;29:201–207. [PubMed] [Google Scholar]
- Takeshima H, Suetake I, Shimahara H, Ura K, Tate S, Tajima S. Distinct DNA methylation activity of Dnmt3a and Dnmt3b towards naked and nucleosomal DNA. J Biochem. 2006;139:503–515. doi: 10.1093/jb/mvj044. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Wang YM, Wang R, Wen DG, Li Y, Guo W, Wang N, Wei LZ, He YT, Chen ZF, Zhang XF, Zhang JH. Single nucleotide polymorphism in DNA methyltransferase 3B promoter and its association with gastric cardiac adenocarcinoma in North China. World J Gastroenterol. 2005;11:3623–3627. doi: 10.3748/wjg.v11.i23.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian A, Pharoah PD, Ahmed S, Ropero S, Fraga MF, Smith PL, Conroy D, Luben R, Perkins B, Easton DF, Dunning AM, Esteller M, Ponder BA. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27:1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- Wang L, Rodriguez M, Yue P, Sun SY, Hong WK, Mao L. Polymorphism in DNMT3B6 promoter region and lung cancer risk. Proc Am Assoc Cancer Res. 2001;42:863. [Google Scholar]
- Wang L, Rodriguez M, Kim ES, Xu Y, Bekele N, El-Naggar AK, Hong WK, Mao L, Oh YW. A novel C/T polymorphism in the core promoter of human de novo cytosine DNA methyltransferase 3B6 is associated with prognosis in head and neck cancer. Int J Oncol. 2004;25:993–999. [PubMed] [Google Scholar]
- Montgomery KG, Liu MC, Eccles DM, Campbell IG. The DNMT3B C/T promoter polymorphism and risk of breast cancer in a British population: a case-control study. Breast Cancer Res. 2004;6:R390–R394. doi: 10.1186/bcr807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Amos CI, Pande M, Gu X, Chen J, Campos IM, Wei Q, Rodriguez-Bigas M, Lynch PM, Frazier ML. DNMT3b polymorphism and hereditary nonpolyposis colorectal cancer age of onset. Cancer Epidemiol Biomarkers Prev. 2006;15:886–891. doi: 10.1158/1055-9965.EPI-05-0644. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Jeon HS, Jang JS, Park SH, Lee GY, Lee BH, Kim CH, Kang YM, Lee WK, Kam S, Park RW, Kim IS, Cho YL, Jung TH, Park JY. DNMT3B polymorphisms and risk of primary lung cancer. Carcinogenesis. 2005;26:403–409. doi: 10.1093/carcin/bgh307. [DOI] [PubMed] [Google Scholar]
- Hong YS, Lee HJ, You CH, Roh MS, Kwak JY, Lee MJ, Kim JY. DNMT3B 39179GT polymorphism and the risk of adenocarcinoma of the colorectal in Koreans. Biochem Genet. 2007;45:155–163. doi: 10.1007/s10528-006-9047-9. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–150. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Verdine GL. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. Dynamics of DNA methylation pattern. Curr Opin Genet Dev. 2000;10:224–228. doi: 10.1016/S0959-437X(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Chang KP, Hao SP, Liu CT, Cheng MH, Chang YL, Lee YS, Wang TH, Tsai CN. Promoter polymorphisms of DNMT3B and the risk of head and neck squamous cell carcinoma in Taiwan: a case-control study. Oral Oncol. 2007;43:345–351. doi: 10.1016/j.oraloncology.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Wu Y, Lin JS. DNA methyltransferase 3B promoter polymorphism and its susceptibility to primary hepatocellular carcinoma in the Chinese Han nationality population: a case-control study. World J Gastroenterol. 2007;13:6082–6086. doi: 10.3748/wjg.13.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Wang R, Wen DG, Li Y, Guo W, Wang N, Wei LZ, He YT, Chen ZF, Zhang XF, Zhang JH. Single nucleotide polymorphism in DNA methyltransferase 3B promoter and its association with gastric cardiac adenocarcinoma in North China. World J Gastroenterol. 2005;11:3623–3627. doi: 10.3748/wjg.v11.i23.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung PP, Matsumura S, Kuraoka K, Kunimitsu K, Yoshida K, Matsusaki K, Nakayama H, Yasui W. No evidence of correlation between the single nucleotide polymorphism of DNMT3B promoter and gastric cancer risk in a Japanese population. Oncol Rep. 2005;14:1151–1154. [PubMed] [Google Scholar]
- Fan H, Liu DS, Zhang SH, Hu JB, Zhang F, Zhao ZJ. DNMT3B 579 G>T promoter polymorphism and risk of esophagus carcinoma in Chinese. World J Gastroenterol. 2008;14:2230–2234. doi: 10.3748/wjg.14.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Walsh G, Liu DD, Lee JJ, Mao L. Expression of Delta DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 2006;66:8361–8366. doi: 10.1158/0008-5472.CAN-06-2031. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- Soejima K, Fang W, Rollins BJ. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene. 2003;22:4723–4733. doi: 10.1038/sj.onc.1206510. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- Yu Z, Kone BC. Hypermethylation of the inducible nitric-oxide synthase gene promoter inhibits its transcription. J Biol Chem. 2004;279:46954–46961. doi: 10.1074/jbc.M407192200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Park J, Choi MC, Kim HP, Park JH, Jung Y, Lee JH, Oh DY, Im SA, Bang YJ, Kim TY. Zinc-fingers and homeoboxes 1 (ZHX1) binds DNA methyltransferase (DNMT) 3B to enhance DNMT3B-mediated transcriptional repression. Biochem Biophys Res Commun. 2007;355:318–323. doi: 10.1016/j.bbrc.2007.01.187. [DOI] [PubMed] [Google Scholar]