Abstract
The yeast Saccharomyces cerevisiae proliferates rapidly in glucose-containing media. As glucose is getting depleted, yeast cells enter the transition from fermentative to nonfermentative metabolism, known as the diauxic shift, which is associated with major changes in gene expression. To understand the expression evolution of genes involved in the diauxic shift and in nonfermentative metabolism within species, a laboratory strain (BY), a wild strain (RM), and a clinical isolate (YJM) were used in this study. Our data showed that the RM strain enters into the diauxic shift ∼1 h earlier than the BY strain with an earlier, higher induction of many key transcription factors (TFs) involved in the diauxic shift. Our sequence data revealed sequence variations between BY and RM in both coding and promoter regions of the majority of these TFs. The key TF Cat8p, a zinc-finger cluster protein, is required for the expression of many genes in gluconeogenesis under nonfermentative growth, and its derepression is mediated by deactivation of Mig1p. Our kinetic study of CAT8 expression revealed that CAT8 induction corresponded to the timing of glucose depletion in both BY and RM and CAT8 was induced up to 50- to 90-folds in RM, whereas only 20- to 30-folds in BY. In order to decipher the relative importance of cis- and trans-variations in expression divergence in the gluconeogenic pathway during the diauxic shift, we studied the expression levels of MIG1, CAT8, and their downstream target genes in the cocultures and in the hybrid diploids of BY–RM, BY–YJM, and RM–YJM and in strains with swapped promoters. Our data showed that the differences between BY and RM in the expression of MIG1, the upstream regulator of CAT8, were affected mainly by changes in cis-elements, though also by changes in trans-acting factors, whereas those of CAT8 and its downstream target genes were predominantly affected by changes in trans-acting factors.
Keywords: cis-regulation, trans-regulation, diauxic shift, expression evolution
Introduction
The significance of regulatory evolution has been recognized since 1970s. The morphological and behavior differences between humans and chimpanzees were suggested to be mainly due to regulatory differences between the 2 species (King and Wilson 1975). However, regulatory evolution has not been well studied in the past mainly because of technical difficulties in obtaining suitable data. Recent technology advances provide new tools to pursue this fundamental question. Especially, microarray has been used to study genome-wide patterns of the evolution of gene expression within and between species (Ranz and Machado 2006). However, although these studies provided large amounts of data about genes that are differentially expressed, most did not investigate the actual genetic changes that are responsible for the expression variation.
Differences in gene expression may arise from cis- or trans-regulatory changes. cis-regulatory changes affect transcription initiation, elongation rate, and/or transcript stability in an allele-specific manner, whereas trans-regulatory changes modify the activity or expression levels of transcription factors (TFs) that interact with cis-regulatory elements (Carrol et al. 2001; Davidson 2001). The relative contributions of cis- and trans-regulatory changes to gene expression variations have been studied (Brem et al. 2002; Yvert et al. 2003; Wittkopp et al. 2004; Wang et al. 2007), but much remains to be explored. Using real-time polymerase chain reaction (PCR), pyrosequencing analysis, and promoter swapping, we studied regulatory evolution in the yeast Saccharomyces cerevisiae.
Saccharomyces cerevisiae proliferates rapidly in media containing fermentable sugar, such as glucose, with the production of ethanol. This stage is known as fermentative (anaerobic) growth. As glucose is depleted, yeast cells turn to ethanol as carbon source for the stage of aerobic growth. The transition from fermentative growth to respiratory growth is known as the diauxic shift, which is associated with remarkable changes in the expression of genes involved in carbon metabolism, protein synthesis, as well as carbohydrate storage (Johnston and Carlson 1992). One key feature of entering the diauxic shift is that cells release glucose repression and express a large set of genes involved in the utilization of alternate carbon sources, gluconeogenesis, the tricarboxylic acid (TCA) cycle, respiration, and peroxisomal functions.
Genomic scale profiling of gene expression by microarray analyses revealed that the diauxic shift involves reprogramming of several regulatory pathways, affecting the expression of at least one-quarter of the genes in the entire yeast genome (DeRisi et al. 1997). Among the highly induced genes are those involved in the TCA cycle, oxidative phosphorylation, gluconeogenesis, β-oxidation, and ethanol and glycerol utilization. These genes can be classified into 5 different regulatory pathways mainly controlled by 11 TFs (see Schuller 2003). The Hap2p/3p/4p/5p complex, which is heme-activated or glucose-repressed, is a transcriptional activator and global regulator of respiratory gene expression (Pinkham and Guarente 1985; Olesen and Guarente 1990). The Rtg3p/Rtg1p complex, activated under nonfermentative or some mitochondrial dysfunctional conditions, is involved in retrograde gene regulation of some respiratory genes (Jia et al. 1997; Rothermel et al. 1997). Adr1p, a carbon source-responsive zinc-finger TF, is required for ethanol, glycerol, and fatty acid utilization and derepression of some glucose-repressed genes (Simon et al. 1991; Tachibana et al. 2005). Oaf1p, acting alone or with Pip2p, activates genes involved in oxidation of fatty acids and peroxisome organization (Rottensteiner et al. 1997; Karpichev and Small 1998). Cat8p, together with Sip4p, is necessary for activating many genes involved in gluconeogenesis, ethanol utilization, and the glyoxylate cycle during the diauxic shift (Haurie et al. 2001; Tachibana et al. 2005). Some of the genes regulated by Cat8p and Sip4p were also coregulated by Adr1p (Tachibana et al. 2005).
Besides these TFs, which regulate specific pathways of carbon source utilization, some other regulators show multiple functions and thus influence a great number of target genes. The Snf1p complex functions as a global positive regulator of carbon source utilization, including upregulating activities of several TFs, such as Cat8p/Sip4p and Oaf1p/Pip2p, during the diauxic shift. The Snf1p complex comprises of protein kinase Snf1p and its regulatory subunits Snf4p, Sip1p, Sip2p, and Gal83p (Jiang and Carlson 1997). On the other hand, Mig1p functions as a general negative regulator involved in glucose repression, though it was originally identified as a repressor of genes in sucrose, maltose, and galactose metabolic pathways (Carlson et al. 1984; Nehlin and Ronne 1990). Snf1p-dependent phosphorylation of Mig1p triggers its nuclear export and deactivates Mig1p within the nucleus (Ostling and Ronne 1998).
Although the diauxic shift is known to be associated with major changes in gene expression in the pathways mentioned above, the regulatory evolution of these pathways has not received much attention. A microarray analysis of global patterns of gene expression in a laboratory strain (BY) and a wild strain (RM) followed by a genetic linkage analysis showed that over 1,500 genes were differentially expressed between these 2 strains, suggesting that the regulatory variation was mainly characterized by the trans-acting effects when cells were at the exponentially growing stage (Brem et al. 2002; Yvert et al. 2003). However, little is known about how the genetic polymorphisms affect the variation of gene expression in nonfermentative metabolism. The well-characterized transcriptional regulatory pathways during the diauxic shift and the difference in efficiency in glucose utilization between the BY and RM strains provide us with an opportunity to investigate mechanisms of transcription evolution.
Materials and Methods
Yeast Strain Constructions
The yeast strains used in this study are listed in table 1. The laboratory strain BY4741 (BY) was derived from S288C and was one of the parental strains for the international systematic S. cerevisiae gene disruption project. The wild isolate RM11-1a (RM) was kindly provided by Lee Hartwell (Fred Hutchinson Cancer Research Center). It is a haploid strain derived from Bb32(3), a natural isolate collected by Robert Mortimer (Mortimer et al. 1994). The pathogenic isolate YJM789 (YJM), kindly provided by L. M. Steinmetz, is isogenic with YJM145, which is a segregant of a clinical isolate of S. cerevisiae (McCusker et al. 1994; Gu et al. 2005). BY 4742 and BY 4741 are isogenic but with different mating types and so are RM11-1a and RM11-1α.
Table 1.
List of Saccharomyces cerevisiae Strains Used in This Study
| Strain | Genotype | Source | Note |
| YL6 | MATa HO::kan ura3Δ leu2Δ | Lee Hartwell | RM11-1a |
| YL7 | MATa his3 Δ 1 leu2Δ met15Δ ura3Δ | Open Biosystems | BY4741 |
| YL8 | MATα his3 Δ 1 leu2Δ lys2Δ ura3Δ | Open Biosystems | BY4742 |
| YL51 | MATα lys2Δ ura3Δ | Lee Hartwell | RM11-1α |
| YL123, 124 | MATa/α HIS3/his3Δ 1 LEU2/leu2Δ ura3Δ/ura3Δ MET15/met15Δ LYS2/lys2Δ | This study | RM 11-1α × BY4741 |
| YL125, 126 | MATa/α HO::kan HIS3/his3 Δ 1 leu2Δ/leu2Δ ura3Δ/ura3Δ LYS2/lys2Δ | Mei-Yeh Lu | RM 11-1a × BY4742 |
| YL129 | MATa HO::kan ura3Δ leu2Δ | This study | CAT8 promoter swapped |
| YL130 | MATa HO::kan ura3Δ leu2Δ | This study | CAT8 promoter swapped |
| YL135 | MATa/α HO::kan HIS3/his3Δ 1 leu2Δ/leu2Δ ura3Δ/ura3Δ LYS2/lys2Δ | This study | YL129 × BY4742 |
| YL136 | MATa/α HO::kan HIS3/his3Δ 1 leu2Δ/leu2Δ ura3Δ/ura3Δ LYS2/lys2Δ | This study | YL130 × BY4742 |
| YL140 | MATα HO::hisG lys2 cyh | Winzeler et al. (1998) | YJM789α |
| YL141, 142 | MATa/α HO::kan/hisG LEU2/leu2Δ URA3/ura3Δ LYS2/lys2Δ CYH/cyh | This study | YJM789α × RM11-1a |
| YL146, 147 | MATa/α HO::hisG/HIS3/his3Δ LEU2/leu2Δ URA3/ura3Δ LYS2/lys2Δ CYH/cyh | This study | YJM789α × BY4741 |
| YL184 | MATα his3 Δ 1 leu2Δ lys2Δ ura3Δ | This study | BY42-72 |
| YL200 | MATa/α HO::kan HIS3/his3 Δ 1 leu2Δ/leu2Δ ura3Δ/ura3Δ LYS2/lys2Δ | This study | RM 11-1a × YL184 |
| YL208 | MATa his3 Δ 1 leu2Δ met15Δ ura3Δ | This study | BYa-2-51 |
| YL213 | MATa/α HIS3/his3Δ 1 LEU2/leu2Δ ura3Δ/ura3Δ MET15/met15Δ LYS2/lys2Δ | This study | RM 11-1α × YL208 |
The swapped strains YL129 and YL130 were both constructed in RM11-1a by replacing the nucleotide sequence from −412 to +10 of the CAT8 promoter region with the same region from BY4741. The swapped strain YL184 was constructed in BY 4742 by replacing the nucleotide sequence from −176 to −151 of the MIG1 promoter region, which includes 3 substitution sites, with the same region of RM11-1a. Another swapped strain YL208 was constructed in BY4741 by replacing the nucleotide sequence from −508 to −151 of the MIG1 promoter region, which includes 4 substitution sites, with the same region of RM11-1a. The 2-step site-specific genomic mutagenesis at each indicated promoter region was conducted by PCR and homologous recombination, following Storici et al. (2003). Hybrid diploid strains were constructed by pairwise mating of BY4742 and RM11-1a, BY4741 and RM11-1α, RM11-1a and YJM789α, BY4741 and YJM789α, the swapped strain YL184 and RM11-1a, or YL208 and RM11-1α as indicated in table 1.
Growth Conditions and Glucose Measurement
Yeast cells were grown in standard yeast-rich growth media, which has 1% (w/v) Bacto-yeast extract, 2% of Bacto-peptone, 1% of adenine-HCl, and 2% (w/v) glucose, at 30 °C and at 200 rpm shaking. The cultures were typically started at a density of 0.1 OD600 units, and the growth rate was monitored by measuring the absorbance with a spectrophotometer at 600 nm wavelength. Yeast cells were harvested at different time points, and the glucose concentration of the media was determined by a Glucose Assay Kit from Sigma (St Louis, MO).
PCR Amplification and Sequencing of DNA Segments
PCR and sequencing primers were designed based on the sequences from the Saccharomyces Genome Database. PCR amplification and sequencing reactions were carried out following the condition described in Yu et al. (2004).
RNA Isolation and Real-Time PCR Analyses
Yeast cells were harvested at different time points, and the total RNA was prepared by the hot phenol extraction method (Ausubel et al. 1995). The rRNA levels were monitored by visual inspection after staining the gel with ethidium bromide. The reverse transcription was carried out by SuperScript III First-Strand Synthesis System for quantitative real-time polymerase chain reaction (qRT-PCR) from Invitrogen (Carlsbad, CA) with random hexamer primers. qRT-PCR was performed in triplicate using the Applied Biosystems 7500 Real-Time PCR System. Independent PCRs were performed using the same cDNA for the gene of interest and using cDNA for18S rRNA, encoded by RDN18, to serve as an endogenous control, with the SYBR Green PCR Master Mix from Applied Biosystems (Foster City, CA). The 18S rRNA levels were used as internal loading controls for each sample during the analysis. For the allele-specific quantification, PCRs were performed using the allele-specific primers and TaqMan MGB probes, with the TaqMan Universal PCR Master Mix from Applied Biosystems. Gene-specific primers and probes were designed using Primer Express software from Applied Biosystems.
Real-time PCR analyses were carried out in a final volume of 25 μl containing 40 ng of the cDNA sample, 50 nM of each gene-specific primer, and 40 nM of allele-specific TaqMan MGB probe if needed. The primer and probe sequences are listed in supplementary materials, Supplementary Material online. The PCR conditions included enzyme activation at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified using the software provided by the Applied Biosystems 7500 Real-Time PCR System. The amplification efficiency of each pair of gene-specific primers was tested, and a negative control reaction in the absence of template cDNA (no template control) was also routinely performed for each primer pair. The change in intensity of the fluorescence dye in every cycle was monitored by the 7500 System software, and the threshold cycle (CT) above background for each reaction was calculated. Through out all the time points we monitored in the real-time PCR analyses, the CT values for RDN18 were around 9–12, whereas the Ct values for the TFs we analyzed were around 20–25 or even more, indicating that rRNAs were considerably more abundant than TF RNAs and the rRNA levels were consistent in our total RNA samples. The CT value of 18S rRNA was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of an arbitrary calibrator (e.g., the culture sample collected at the reference time point hour 4 in the case of a particular gene) was subtracted from the ΔCT value of certain time points to obtain a ΔΔCT value. The gene expression level relative to the reference time point T4, the calibrator, was expressed as 2−ΔΔCT.
Pyrosequencing Analyses
Genomic DNA was extracted according to the standard yeast DNA isolation procedure (Burke et al. 2000). Both the genomic DNA and cDNA samples from the same strain were subjected to pyrosequencing analysis. All the primers were designed using the Pyrosequencing software, Assay Design version 1.0. A single nucleotide difference that distinguished between the allele-specific transcripts of BY4741 and RM11-1a, or YJM789 and RM11-1a, was identified for each gene tested. Gene-specific primers annealing to conserved sequences of each gene were used to amplify regions of sequence including the divergent sites. PCR was conducted using Taq DNA polymerase from Invitrogen in a reaction mixture containing 20 ng of genomic DNA or cDNA template, 20 nM of tagged gene-specific primer, 180 nM of biotin-labeled primer, and 200 nM of the other gene-specific primer. The PCR cycling program consisted of denaturation at 95 °C for 5 min, followed by 50 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 15 s. The designed primers for the studied genes are available in supplementary materials (Supplementary Material online).
Each pyrosequencing reaction was performed using 20–25 μl of PCR products in accordance with the manufacturer's instructions (http://www.pyrosequencing.com). Biotinylated single-stranded DNA was obtained, and an extension primer, also matching a conserved sequence for both BY and RM alleles, was then annealed. The relative abundance of the 2 alleles in genomic DNA or cDNA samples from BY–RM, YJM–RM, BY–YJM, YL184-RM, or YL208-RM cocultures or hybrid diploid cultures was measured by Pyrosequencing, model PSQ 96MA from Biotage AB (Uppsala, Sweden). The reaction couples DNA synthesis with a series of enzymatic reactions that produce stoichiometric quantities of luminous signals. The amount of signal generated by the incorporation of allele-specific bases was directly proportional to the number of molecules incorporated into the growing DNA chain during the reaction. The ratio of signal intensity of the 2 alleles (BY/RM, YJM/RM, or BY/YJM) was calculated, which corresponds to the relative abundance of the 2 alleles in the sample. cDNA ratios were normalized with genomic DNA measurements as described in Wittkopp et al. (2004).
Results and Discussion
Sequence Data
Genes essential for nonfermentative metabolism can be classified into 5 different regulatory pathways mainly controlled by 11 TFs. We determined the sequences of the promoters and coding regions of these TF genes and several other regulator genes in the RM strain and compared them with the corresponding genes in the BY strain as shown in table 2. Because it is difficult to define the promoter regions of each gene, and because the intergenic regions of each gene in yeast genome are relatively short, we referred the promoter region of each gene as its upstream intergenic region.
Table 2.
Sequence Variations in Coding Sequences (CDs) and in Promoter Regions between the BY and RM Strains for Key Regulators in Nonfermentative Growth
| Locus | Gene Name | ORF Length (bp) | Promoter Length (bp) | CDs (Nonsynonymous:Synonymous) | Promoter Regions |
| Key TFs | |||||
| YGL237C | HAP2 | 798 | 229 | 5:4 | 4 |
| YBL021C | HAP3 | 435 | 307 | 0:0 | 1 |
| YKL109W | HAP4 | 1,665 | 1,000a | 3:7 | 11 |
| YOR358W | HAP5 | 729 | 449 | 0:0 | 0 |
| YDR216W | ADR1 | 3,972 | 1,108 | 2:1 | 5 |
| YOL067C | RTG1 | 534 | 303 | 1:3 | 0 |
| YBL103C | RTG3 | 1,461 | 563 | 1:3 | 4 |
| YAL051W | OAF1 | 3,144 | 314 | 6:19 | 0 |
| YOR363C | PIP2 | 2,991 | 249 | 1:1 | 2 |
| YMR280C | CAT8 | 4,302 | 1,010 | 4:6 | 2 |
| YJL089W | SIP4 | 2,490 | 875 | 1:2 | 1 |
| YGL035C | MIG1 | 1,515 | 1,000a | 4:1 | 13 |
| The Snf complex | |||||
| YDR477W | SNF1 | 1,902 | 1,300 | 0:11 | 20 |
| YGL115W | SNF4 | 969 | 392 | 0:0 | 5 |
| YER027C | GAL83 | 1,254 | 460 | 3:2 | 5 |
| YDR422C | SIP1 | 2,592 | 129 | 3:1 | 1 |
| YGL208W | SIP2 | 1,248 | 332 | 3:11 | 0 |
| Snf1p-activating kinases | |||||
| YGL179C | TOS3 | 1,683 | 1,000a | 1:2 | 10 |
| YKL048C | ELM1 | 1,923 | 327 | 0:0 | 0 |
| YER129W | SAK1 | 3,249 | 812 | 5:4 | 4 |
Sharing promoter region with its neighboring gene.
The genes encoding these 11 TFs, except HAP5, RTG1, and OAF1, showed single-nucleotide polymorphisms (SNPs) in the promoter regions and 9 of them (except HAP3 and HAP5) showed nucleotide differences in the coding region, resulting in at least one or more nonsynonymous changes in each of these 9 genes. None of these nucleotide polymorphisms change the length of the open reading frames (ORFs), except that OAF1 has 6 nonsynonymous substitutions plus 2 nucleotide deletions near the 3′ end of its ORF resulting in a frameshift at the carboxyl end of the ORF. Interestingly, HAP5, which encodes a protein that mediates interaction with other Hap proteins in the CCAAT-binding complex, is the only TF that has the identical promoter and coding sequence between BY and RM. The absence of any SNP in 1.1 kb of HAP5 coding and noncoding regions is significant (P < 0.05), given the expectation of 0.4% average divergence in genic regions between the RM and S288C strains (Broad Institute, http://www.broad.mit.edu/annotation/genome/saccharomyces_cerevisiae.3/Info.html). The sequence polymorphisms of these TFs raise the possibility that the regulatory function of these factors may differ between BY and RM when cells enter the diauxic shift.
We next compared the sequences for genes encoding components of the Snf1p complex, including the protein kinase Snf1p, and its regulatory subunits Snf4p, Sip1p, Sip2p, and Gal83p (table 2). There are 11 nucleotide differences within the ∼2-kb coding region of SNF1 between BY and RM, but all of them are synonymous changes, implying that nonsynonymous sites are functionally constrained. Surprisingly, the SNF1 coding sequence in YJM 789, a pathogenic isolate of S. cerevisiae, is identical to the BY (data not shown). Nonetheless, there are 20 nucleotide differences within the 1.3-kb promoter region of SNF1 between the BY and RM alleles (P < 0.01, given the expected divergence for promoter regions is about 0.52% between RM and S288C). In contrast, the SNF4 coding region is identical in BY and RM, though the promoter region shows 5 nucleotide differences between BY and RM. These sequence data imply that the function of the Snf1 complex might have been relatively well conserved among these strains.
It has been shown that there are 3 upstream Snf1-activating kinases with partially redundant function: Sak1p, Tos3p, and Elm1p (Hong et al. 2003; Hedbacker et al. 2004). Our sequence data showed that there are several nucleotide polymorphisms in SAK1 and TOS3, but the sequence of ELM1 is identical between BY and RM (table 2), suggesting that ELM1 might have been evolutionarily better conserved. It also implies that, although these 3 kinases are partially functionally redundant, Elm1p might play a more distinct role than the other 2 kinases in the regulation of Snf1p activity.
Different Growth Patterns between RM and BY
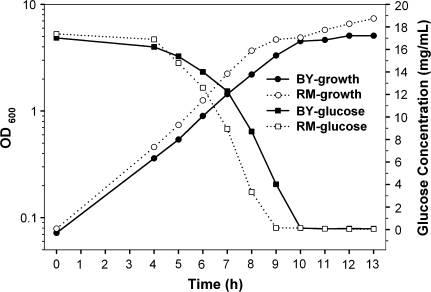
To compare the growth patterns, RM and BY were grown separately in rich media containing 2% glucose as the carbon source, and the cell density and glucose concentration in the media were monitored. Our data showed that the RM strain grew at a faster rate than the BY strain (fig. 1). However, once glucose was exhausted, each strain encountered a transient arrest, during which cells shift from fermentative to nonfermentative metabolism in order to use ethanol as the carbon source produced by glucose fermentation. Our data also showed that the glucose in the media of the RM culture was depleted nearly 1 h earlier than that of the BY culture, which is likely the result of a faster growth rate of RM, indicating that RM entered the diauxic shift earlier than BY.
FIG. 1.—
The growth and glucose concentration curves of the laboratory (BY) and wild (RM) strains. The cultures were started in yeast-rich growth media, YPAD, which has 2% glucose. The culture density is presented as the absorbance at 600 nm. At least 3 biological replicates were performed, and the growth and glucose concentration curves were similar to the ones shown here.
Expression Profiles of CAT8, MIG1, MLS1, and SNF1 Genes in BY, RM, and YJM
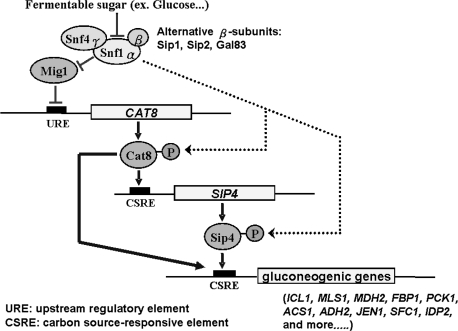
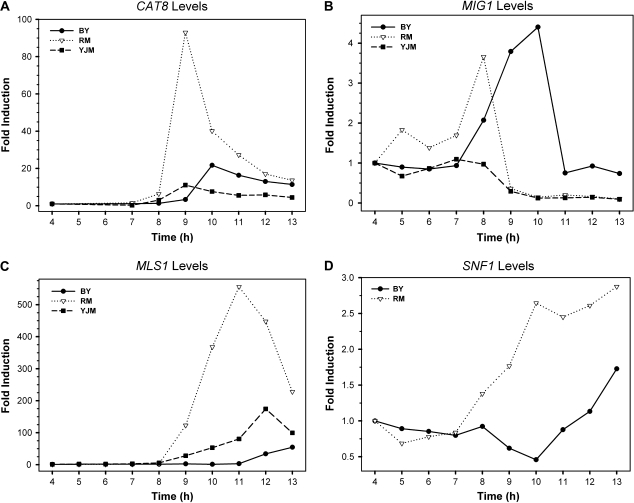
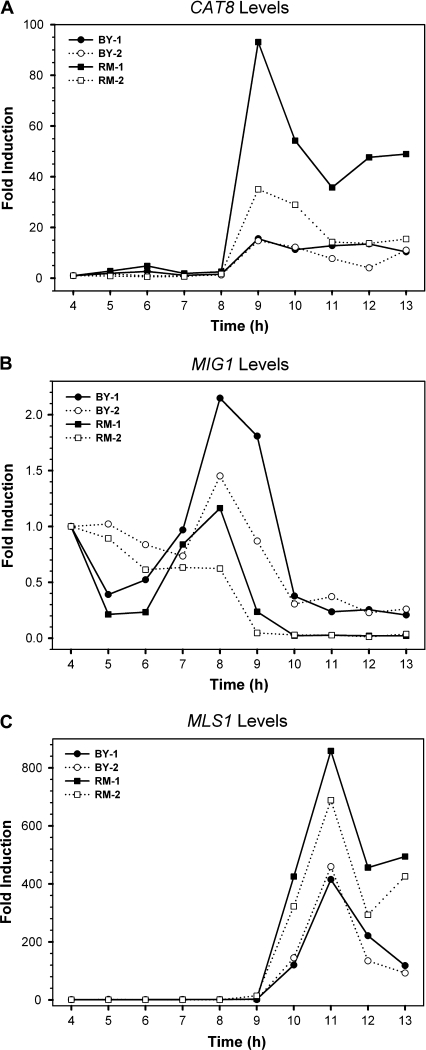
To investigate the regulatory evolution of the TFs involved in nonfermentative metabolism, the expression profiles of these factors were examined. Each culture sample was harvested at different times for RNA extraction, and the mRNA levels of each TF at different times were quantified by qRT-PCR. We focused on the well-studied gluconeogenesis pathway regulated by TFs Cat8p and Sip4p (fig. 2). At a low level of fermentable carbon sources, the Ser/Thr-specific protein kinase Snf1p is activated. The phosphorylation of Mig1p by Snf1p leads to the deactivation of Mig1p repressor activity, allowing the biosynthesis of Cat8p. The posttranslational modification of Cat8p by Snf1p subsequently stimulates the expression of TF Sip4p and further downstream target genes such as MLS1. We found that the expression levels of ADR1, HAP2, and RTG1 were induced within 2-folds (data not shown) but CAT8 and SIP4 showed dramatic induction in both strains during the diauxic shift (fig. 3). The induction of CAT8 and SIP4 occurred earlier by 1 h in RM than in BY, corresponding to their respective timing of glucose depletion. The CAT8 expression was induced up to 50- to 90-folds in RM but only up to 20- to 30-folds in BY (fig. 3). Interestingly, in the YJM strain, CAT8 transcription was only induced up to 10- to 20-folds, even lower than that in the BY strain. We also examined the expression levels of MLS1, which encodes malate synthase for the utilization of nonfermentable carbon sources and is one of the well-studied downstream target genes of Cat8p. MLS1 expression was induced dramatically, up to 400- to 500-folds, in RM after the diauxic shift, but up to only 100-folds in BY, correlating with the observed differential induction levels of CAT8 between RM and BY. The induction folds of these genes in YJM were more similar to those in BY than those in RM. In summary, the induction peak and timing of these genes are consistent with the growth patterns for the BY and RM strains, indicating that RM enters the diauxic shift stage and induces the expression of genes required for the nonfermentative metabolism ∼1 h earlier than does BY.
FIG. 2.—
Signaling pathway in the regulation of gluconeogenesis by carbon source-responsive element-binding factors, Cat8p and Sip4p. Adapted from Schuller (2003).
FIG. 3.—
The expression profiles of genes CAT8, MIG1, MLS1, and SNF1 in the BY and RM and YJM separated cultures. Each culture was harvested at different time points for RNA extraction, and the mRNA levels of genes CAT8 (A), MIG1 (B), and MLS1 (C), and SNF1 (D) were quantified by qRT-RCR. At least 3 biological replicates were performed, and the induction folds for CAT8, MIG1, and MLS1 were similar to the ones shown here.
It has been shown that during the diauxic shift, both the derepression of Cat8p by deactivation of Mig1p and the transcriptional activation mediated by Cat8p require an active Snf1p complex (Haurie et al. 2001; Schuller 2003; Young et al. 2003). Because Snf1p controls the most upstream step in this regulatory pathway, we wondered whether it has played an important role in the expression evolution of the downstream genes. Our data, however, showed that there was little change in the SNF1 mRNA level before the diauxic shift in both BY and RM (fig. 3). Although the induction folds fluctuated between 0.5 and 2.7 during the time points we monitored within 5 replicates, this variation might be due to the low expression level of SNF1. The microarray data of DeRisi et al. (1997) also suggested that the SNF1 mRNA levels did not show significant changes (Max increase was 1.2 in the whole data sets) during the diauxic shift. Therefore, we concluded that there is little change in the SNF1 mRNA level during the diauxic shift. This suggested that Snf1p plays no big role in the remarkable expression divergence in downstream genes, such as CAT8 and MLS1. This conclusion was strengthened by the observation of no amino acid difference in Snf1p between BY and RM. It is possible that Snf1p is differentially activated on a nontranscriptional level. Snf1-GFP is mainly localized in the cytoplasm in glucose-grown cells but becomes enriched in the nucleus 10 min after shift to a nonfermentable carbon source, suggesting that the translocation of the Snf1 protein is important in response to glucose limitation (Vincent et al. 2001; Hong and Carlson 2007). It has also been shown that the phosphorylation state of Snf1p at threonine 210 and the Snf1p kinase activity are correlated with glucose limitation and are responsible for the phosphorylation of Mig1 protein (McCartney and Schmidt 2001). These data suggest that rather than the Snf1 protein expression level, phosphorylation, and translocation are required in response to glucose limitation.
Because Mig1p is another key regulator of CAT8 transcription, we examined the expression levels of MIG1 in the BY, RM, and YJM strains. We found that the MIG1 expression level was induced up to 3- to 4-folds in both BY and RM, and the induction peak was observed about 1 h before the depletion of glucose in the media (fig. 3). These results suggested that the expression difference of MIG1 between the 2 strains is largely responsible for the expression divergence of its downstream genes.
Expression Profiles of MIG1, CAT8, and MLS1 Genes in BY and RM Cocultures
We observed that the glucose in the media of RM culture was depleted nearly 1 h earlier than that of BY culture and that the induction peak of CAT8 also occurred 1 h earlier in RM than in BY. We therefore decided to examine whether the difference of the induction timing of CAT8 is due to the differences of the growth patterns between BY and RM. We grew the BY and RM strains together in the same culture (coculture), starting from approximately the same cell number of each strain. We then analyzed the expression patterns of BY-CAT8 and RM-CAT8 alleles by qRT-PCR with allele-specific probes and found that the CAT8 transcripts of both alleles showed an induction peak at the same time, that is, when the glucose was depleted (fig. 4). The expression differences between the 2 replicates for CAT8 might be due to the variation in the cell numbers at the starting time points, which may affect the growth rate and competition of the BY and RM population. However, the expression from the RM allele was induced to a much higher level than that from the BY allele, and the levels of induction were similar to the ones when these 2 strains were cultured separately. These results indicated that the induction time difference between BY and RM when these 2 strains were grown separately was due to difference in glucose consumption rate, whereas the difference in induction peak was due to genetic differences between BY and RM. We also examined the expression levels of MLS1 and MIG1 in this coculture system and found a simultaneous induction of the BY and RM alleles for these 2 genes. Taken together, these results indicated that glucose depletion acts as a signal for the induction of genes involved in the gluconeogenic pathway in both the BY and RM strains.
FIG. 4.—
The expression profiles of genes CAT8, MIG1, and MLS1 in the BY and RM cocultures. The mRNA levels of genes CAT8 (A), MIG1 (B), and MLS1 (C) were quantified by TaqMan qRT-RCR using BY or RM allele-specific probes. The data from 2 replicates of the BY and RM cocultures are represented by solid and dotted lines and indicated as batch 1 and 2, respectively.
Expression Profiles of MIG1, CAT8, and MLS1 Genes in Diploid Hybrid Strains and Promoter-swapped strains
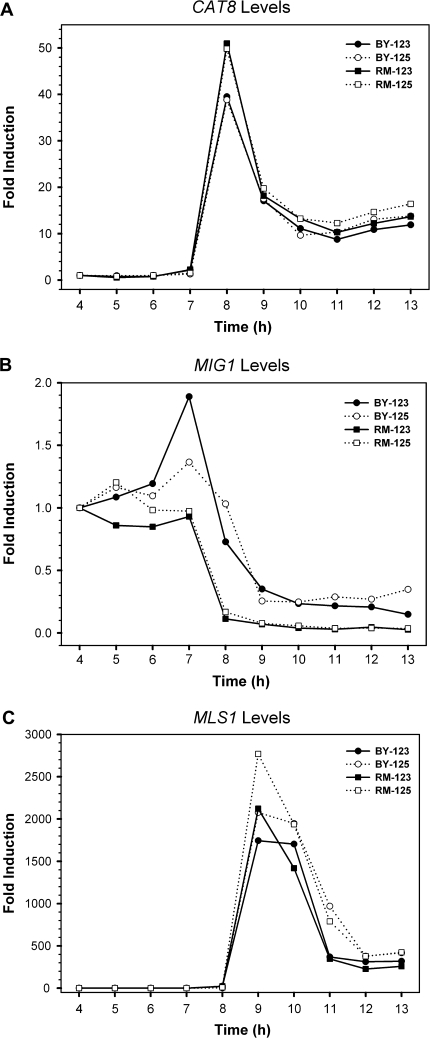
We used the following 2 experiments to determine whether the difference in CAT8 expression between BY and RM is due to the variation in the cis elements or in the trans-acting factors. First, we generated a diploid hybrid strain by mating the haploid BY and RM strains and examined the CAT8 expression in the diploid hybrid strain, in which the expression of both alleles shares the same trans-acting factors under the same genetic background. Therefore, if there is any observed expression difference between the BY and RM alleles, it should be due to differences in the cis-acting elements. The hybrid diploid strains were generated from reciprocal crosses. Our results suggested that the direction of the cross used to generate the hybrids had little effect on the expression levels of the BY and RM alleles (fig. 5). In addition, the transcription levels of BY-CAT8 and RM-CAT8 alleles were, respectively, induced to 40- and 50-folds higher than the basal levels. We found similar situation for the MLS1 expression between the 2 alleles. The expression levels of BY-MLS1 and RM-MLS1 were similar in the hybrid diploid strains. These data implied that the differences in the expression profiles of the CAT8 and MLS1 genes between the haploid BY and RM strains were mainly due to trans-acting factors because once the BY and RM alleles were regulated under the same genetic background, they showed similar expression profiles. In contrast, the expression patterns and induction folds of BY-MIG1 and RM-MIG1 in the hybrid strains were closer to the ones we observed in the BY–RM coculture, suggesting that the expression difference in MIG1 between BY and RM was mainly but not solely due to cis variation.
FIG. 5.—
The expression profiles of genes CAT8, MIG1, and MLS1 in the BY–RM hybrid diploid strains YL123 and YL125. YL123 and YL125 were 2 independent reciprocal crosses that served as the replicates for the experiment. The mRNA levels of genes CAT8 (A), MIG1 (B), and MLS1 (C) were quantified by TaqMan qRT-RCR using BY or RM allele-specific probes. The data from strains YL123 and YL125 are represented by solid and dotted lines, respectively.
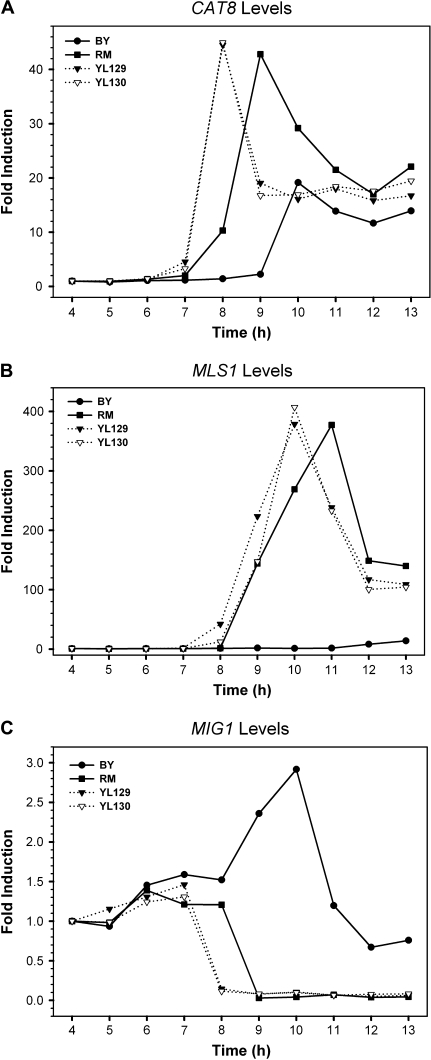
A second approach is to swap a promoter region of interest from one strain into the other and then compare the expression levels between the swapped strain and its parental strain. If differential expression is still observed between the BY and RM alleles after promoter swapping, it should be due to the trans-variation. Applying this approach, we replaced the CAT8 promoter region in the RM strain with the promoter region of the BY strain and examined the expression level of CAT8 in the promoter-swapped strain. Interestingly, the expression profile of CAT8 in this promoter-swapped strain was very similar to the profile of the parental RM strain, suggesting that the change of the cis elements from the RM allele to the BY allele has little effect on the CAT8 expression (fig. 6). Even though both the BY-CAT8 and RM-CAT8 coding regions were driven by the same promoter region from the BY allele but regulated by variant TFs, the differences between BY-CAT8 and RM-CAT8 expression profiles still existed. This observation was consistent with the data we had for the hybrid diploid strains. Furthermore, the expression profiles of MIG1 and MLS1, an upstream and a downstream gene, respectively, of CAT8 in this swapped strain were also similar to those of the parental RM strain. In summary, both approaches suggest that the expressional differences in CAT8 and its downstream target gene MLS1 between BY and RM are mainly due to trans-variations.
FIG. 6.—
The expression profiles of genes CAT8, MLS1, and MIG1 in separated cultures of BY, RM, and RM-swapped strains YL129 and YL130 in which we replaced the CAT8 promoter region of the RM allele by the one from the BY allele. YL129 and YL130 were 2 independent swapped clones that served as the replicates of the experiments. The mRNA levels of genes CAT8 (A), MLS1 (B), and MIG1 (C) were quantified by qRT-RCR. The data from the BY and RM strains are presented by solid lines, whereas those from YL129 and YL130 are presented by dotted lines.
Expression Profiles of CAT8 and MLS1 in the Swapped Hybrid Diploid Strain
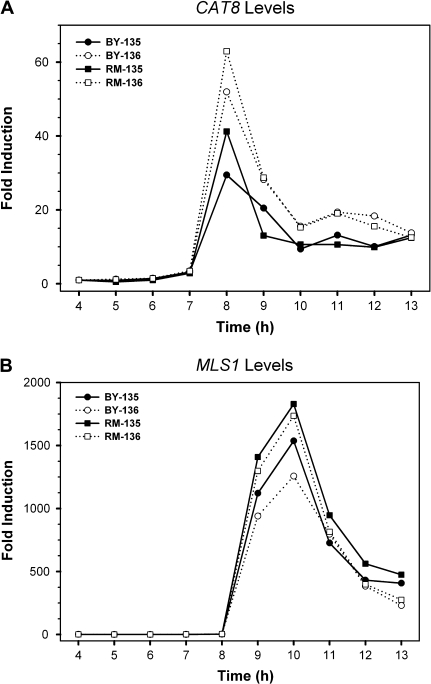
In order to further decipher the effect of cis-regulatory elements in the regulation of CAT8 expression, we generated a hybrid diploid strain by mating the swapped haploid RM strain that carries the BY promoter region to the BY strain. In this hybrid diploid strain, the RM-CAT8 and BY-CAT8 alleles are controlled by the same cis-regulatory element but we found that the differences in expression levels between these 2 CAT8 alleles still existed (fig. 7). Indeed, the induction pattern of both CAT8 alleles in this swapped hybrid diploid resembled the pattern in the parental BY–RM hybrid (fig. 5A). Because the induction folds of BY or RM alleles were calculated from its basal levels separately as described in Materials and Methods, this might be the reason that we still observed some differences in induction peak between the BY and RM alleles. Indeed, as will be discussed below, our pyrosequencing data (supplementary fig. S1, Supplementary Material online) did confirm that the expression levels of BY-CAT8 and RM-CAT8 were almost the same. Therefore, the expressional differences between the BY-CAT8 and RM-CAT8 alleles are not mainly due to differences in the promoter region. However, because there are 10 nucleotide differences between the BY and RM alleles in the coding regions of CAT8, it is possible that these differences affect the accessibility of other regulators due to changes in chromatin structure or the stability of CAT8 transcripts. Therefore, cis variation might have played a role in the expression divergence of CAT8 between BY and RM.
FIG. 7.—
The expression profiles of genes CAT8 and MLS1 in the hybrid diploid strains YL135 and YL136 generated by mating BY and the swapped RM strain. YL135 and YL136 were 2 independent swapped hybrid clones that served as the replicates of the experiments. The mRNA levels of genes CAT8 (A) and MLS1 (B) were quantified by Taqman qRT-RCR using BY or RM allele-specific probes. The data from YL135 and YL136 are represented by solid and dotted lines, respectively.
Pyrosequencing Analysis of MIG1, CAT8, MLS1, IDP2, and SFC1 Allelic Gene Expression
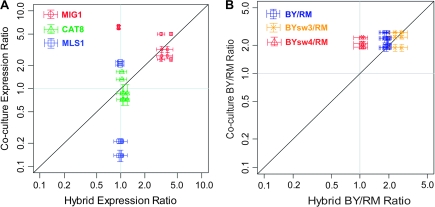
Although our data in the BY and RM hybrid diploid strain indicated that the trans-acting factors are more important than cis elements for the expression divergence of CAT8 and MLS1 between BY and RM, the mild asymmetric allelic expression implied a slight effect of the cis-regulatory divergence. To measure the relative abundance of allele-specific transcripts more accurately, we used pyrosequencing to analyze cDNA samples extracted from the BY–RM cocultures, the hybrid diploid strains, the swapped hybrid diploid strains, as well as the RM–YJM and BY–YJM cocultures and the hybrid diploid strains. If cis-regulatory divergence completely explains the difference between both alleles, the allele ratios of the hybrid diploid strains and the cocultures will be the same (i.e., BY/RMhybrid = BY/RMcoculture). In contrast, if only trans-regulatory differences cause the divergent expression, both alleles will be equally expressed in the hybrid diploid strains (i.e., BY/RMhybrid = 1).
Our pyrosequencing data showed that in the BY–RM cocultures, consistent with our qRT-PCR data, the expression levels of BY-CAT8 were much lower than that of RM-CAT8, making the BY/RM ratio significantly lower than 1 (fig. 8A and supplementary fig. S1 [Supplementary Material online]). On the other hand, in the BY–RM hybrid diploid strains, the expression levels of BY-CAT8 were about 10–20% higher than RM-CAT8, indicating a slight difference in the levels of expression of the 2 alleles. However, during and after the diauxic shift, the differential expression of BY-CAT8 and RM-CAT8 disappeared, and the expression ratio of the 2 alleles was very close to 1. This was also observed in the swapped hybrid diploid strains and the YJM–RM hybrid diploid strains. These data further suggested that CAT8 expressional difference is mainly due to trans variation.
FIG. 8.—
Pyrosequencing results of MIG1, CAT8, and MLS1 from cocultures versus hybrids. The ratios of allele expression levels with the corresponding time pairs of coculture and hybrid are presented in the log scale. The diagonal line represents the 1-to-1 ratio, that is, BY/RMhybrid = BY/RMcoculture. The vertical and horizontal bars on each dot represent the standard deviations from 2 biological and 2 technical repeats. (A) Pyrosequencing results of MIG1, CAT8, and MLS1 between strains. MIG1 is in red, CAT8 in green, and MLS1 in blue. The MIG1 results that are close to the vertical line are for YJM/RM and those near the diagonal line are for BY/RM or BY/YJM. The data points of CAT8 and MLS1 are for BY/RM (below the diagonal line) or YJM/RM (above the horizontal line). (B) Pyrosequencing results of MIG1 between the parental BY strain, the promoter-swapped strains, and the RM strain. The swapped strains were obtained by replacing the MIG1 promoter region in the BY strain that included 3 or 4 substitution sites with the same promoter region of the RM strain as described in Materials and Methods. The number “3” or “4” represents the number of key candidate nucleotides (see text) that were swapped. The data indicated in blue are for the parental BY and RM strains, whereas those indicated in orange are for the swapped strain with 3 substitution sites and the RM strain, and those indicated in red are for the swapped strain with 4 substitution sites and the RM strain.
The Cat8p downstream target genes MLS1, SFC1, and IDP2 showed similar patterns of regulation to that of CAT8 (fig. 8A, supplementary figs. S1 and S2, Supplementary Material online). In the BY–RM cocultures, the expression levels of BY-MLS1 were much lower than that of RM-MLS1, that is, the BY/RM ratio was significantly lower than 1 (P < 0.01). On the other hand, there is no significant difference in expression levels between the BY and RM alleles of MLS1 in the hybrid diploid strains, as in the RM–YJM hybrid diploid strains. These data also indicated that the MLS1 expressional difference between the different S. cerevisiae strains examined is mainly due to trans variation, perhaps largely due to variation in CAT8 expression because when CAT8 is induced to a higher level, so is MLS1.
Consistent with the results of the qRT-PCR analyses, our pyrosequencing results showed that cis-variations might be responsible for the divergence of MIG1 expression. We observed significant expression differences between the BY and RM alleles of MIG1 in both the hybrid diploid cells and cocultures during the early diauxic shift (P < 0.01) (fig. 8A). When the ratio BY/RMhybrid was plotted against the ratio BY/RMcoculture, we found that our data were relatively close to the diagonal line, which indicates predominantly cis-regulatory divergence. This was also observed in the BY–YJM hybrid diploid strains and the BY–YJM cocultures. Surprisingly, we found that there is a significant difference of expression levels in the RM–YJM cocultures but not between the RM and YJM alleles of MIG1 in hybrid diploid strains. In the promoter region, that is, the 1,500-bp region upstream of MIG1, there are a total of 14 nucleotide substitutions among the 3 strains. Interestingly, there are 10 nucleotide substitutions between RM and YJM but only 4 nucleotide substitutions between BY and YJM at positions −151, −152, −174, and −498; at these 4 positions, the RM and YJM alleles are identical. This observation implies that these 4 nucleotide positions within the MIG1 promoter region were responsible for the expression divergence between the BY and RM (or YJM) alleles.
To investigate whether the cis-variations within the MIG1 promoter region are responsible for the divergence of MIG1 expression, we generated 2 MIG1 swapped strains of a particular promoter region between the BY and RM alleles. In the BY strain, we replaced the MIG1 promoter region including the 3 substitution sites −151, −152, and −174 with the same promoter region of the RM strain. The other swapped strain was also done in the BY background, but all 4 substitution sites −151, −152, −174, and −498 were included. The swapped hybrid diploid strains were also made by mating each of the MIG1 swapped haploid strains to the parental RM strain. Both swapped strains were cocultured with the RM strain, and the cocultured strains were then subjected to pyrosequencing analyses and so were the swapped hybrid diploids. As expected, for the swapped hybrid diploid with all 4 substitution sites, there was no significant difference of expression levels between RM and swapped alleles of MIG1, strongly suggesting that BY-MIG1 and RM-MIG1 differential expression is mainly due to cis-variations (fig. 8B). Surprisingly, for the swapped hybrid diploid with only 3 substitution sites, the MIG1 expression differences between the RM and swapped alleles still remained as in the BY–RM hybrid diploid strains (fig. 8B). These observations strongly suggest that MIG1 differential expression is mainly due to cis variation at the SNP site −498. This position is 2 nucleotides downstream of the Rfx1p putative binding site (Pachkov et al. 2007) and is the same in Saccharomyces paradoxus and Saccharomyces mikatae, though not in Saccharomyces bayanus. It has been shown that Rfx1p might be involved in the DNA damage and replication checkpoint pathway (Emery et al. 1996; Huang et al. 1998). However, whether this single-nucleotide change or the combination of multiple SNPs resulted in the expression differences of MIG1 will require further investigations.
Our data suggested that trans-acting factors may also be involved in the divergence of MIG1 expression. As mentioned earlier, we observed significant difference of expression levels in the RM–YJM cocultures (fig. 8A). However, the difference was not observed between the RM and YJM alleles of MIG1 in the hybrid diploid strain, indicating that the expressional divergence of MIG1 in the cocultures may be due to the trans variation. We also observed similar expression patterns for the RM strain and the MIG1 swapped strain containing the replacement of the 4 substitution sites (fig. 8B). The expression levels of the RM and swapped alleles in hybrid diploid strains were very similar but they showed a 2-fold difference in the cocultures. Altogether, our pyrosequencing data indicated that not only the cis-regulatory elements but also the trans-acting factors have contributed to the variation of MIG1 expression.
Conclusions
Among the pathways reprogrammed during the diauxic shift, we focused on the gluconeogenic pathway mainly regulated by Snf1p, Mig1p, and Cat8p to monitor their expression differences among 3 S. cerevisiae strains, BY, RM, and YJM. Our qRT-PCR data strongly suggested that CAT8 and 3 of its downstream target genes were predominantly regulated by trans-acting elements, with a minor cis-element influence. In contrast, the expression divergence of MIG1 appeared to be due to cis-variations, with a minor effect of trans variation. Our pyrosequencing data further confirmed that the expression evolution of MIG1 was more strongly affected by changes in cis elements than by changes in trans-acting factors. It has been speculated that a gene's position in the regulatory network affects the relative roles of cis- and trans-factors in the expression evolution of the gene. Here we found that for MIG1, which is at a very upstream position in the regulatory network, the expression evolution seems to be mainly due to cis variation, whereas trans variation seems to have played a major role in the expression evolution of its downstream genes. However, because the number of genes we studied is limited, we still cannot draw a general conclusion. Studying additional transcriptional regulatory pathways or other key regulators involved in the diauxic shift will help us further understand evolutionary patterns of gene regulatory network. Our studies also pointed to the importance of combining available molecular tools for analyses of expression evolution. Although microarray is a powerful tool for comparing whole-genome transcription profiles, qRT-PCR and pyrosequencing analyses provide more sensitive and precise quantification of gene expression. In this study of regulatory evolution within species, the expression differences of genes of interest between 2 strains might be subtle. The latter 2 techniques provide better resolution when monitoring the transcription levels along different time points.
Supplementary Material
Supplementary materials and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This study was supported by International Balzan Foundation and National Institutes of Health grants (GM20998 and GM081724) to W.-H.L. We thank Kevin Bullaughey for help on the R program and the 2 anonymous reviewers for valuable comments.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons, Inc; 1995. [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in yeast genetics. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Carlson M, Osmond BC, Neigeborn L, Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984;107:19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrol SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. Oxford (UK): Blackwell Science; 2001. [Google Scholar]
- Davidson EH. San Diego (CA): Academic press; 2001. Genomic regulatory systems: development and evolution. [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, David L, Petrov D, Jones T, Davis RW, Steinmetz LM. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurie V, Perrot M, Mini T, Jeno P, Sagliocco F, Boucherie H. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2001;276:76–85. doi: 10.1074/jbc.M008752200. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Hong SP, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol Cell Biol. 2004;24:8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces gene expression. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- Karpichev IV, Small GM. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6560–6570. doi: 10.1128/mcb.18.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Romano P, Suzzi G, Polsinelli M. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10:1543–1552. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- Nehlin JO, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen JT, Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- Ostling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham JL, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Machado CA. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Rothermel BA, Thornton JL, Butow RA. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- Rottensteiner H, Kal AJ, Hamilton B, Ruis H, Tabak HF. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur J Biochem. 1997;247:776–783. doi: 10.1111/j.1432-1033.1997.00776.x. [DOI] [PubMed] [Google Scholar]
- Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- Simon M, Adam G, Rapatz W, Spevak W, Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991;11:699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sung HM, Wang TY, et al. (12 co-authors) Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 2007;17:1161–1169. doi: 10.1101/gr.6328907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, Davis RW. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li WH. Nucleotide diversity in gorillas. Genetics. 2004;166:1375–1383. doi: 10.1534/genetics.166.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.