Abstract
The obligate intracellular bacterium Wolbachia pipientis strain wPip induces cytoplasmic incompatibility (CI), patterns of crossing sterility, in the Culex pipiens group of mosquitoes. The complete sequence is presented of the 1.48-Mbp genome of wPip which encodes 1386 coding sequences (CDSs), representing the first genome sequence of a B-supergroup Wolbachia. Comparisons were made with the smaller genomes of Wolbachia strains wMel of Drosophila melanogaster, an A-supergroup Wolbachia that is also a CI inducer, and wBm, a mutualist of Brugia malayi nematodes that belongs to the D-supergroup of Wolbachia. Despite extensive gene order rearrangement, a core set of Wolbachia genes shared between the 3 genomes can be identified and contrasts with a flexible gene pool where rapid evolution has taken place. There are much more extensive prophage and ankyrin repeat encoding (ANK) gene components of the wPip genome compared with wMel and wBm, and both are likely to be of considerable importance in wPip biology. Five WO-B–like prophage regions are present and contain some genes that are identical or highly similar in multiple prophage copies, whereas other genes are unique, and it is likely that extensive recombination, duplication, and insertion have occurred between copies. A much larger number of genes encode ankyrin repeat (ANK) proteins in wPip, with 60 present compared with 23 in wMel, many of which are within or close to the prophage regions. It is likely that this pattern is partly a result of expansions in the wPip lineage, due for example to gene duplication, but their presence is in some cases more ancient. The wPip genome underlines the considerable evolutionary flexibility of Wolbachia, providing clear evidence for the rapid evolution of ANK-encoding genes and of prophage regions. This host–Wolbachia system, with its complex patterns of sterility induced between populations, now provides an excellent model for unraveling the molecular systems underlying host reproductive manipulation.
Keywords: endosymbiont, Wolbachia, mosquito, cytoplasmic incompatibility, prophage, ankyrin
Introduction
Wolbachia pipientis, a very widespread endosymbiont of invertebrates, is an α-proteobacterium within the order Rickettsiales, related to other intracellular bacteria such as the Anaplasma and Ehrlichia. A number of supergroups have been described within the single-species designation W. pipientis: A and B in arthropods, C and D in filarial nematodes, plus at least 3 further primarily arthropod supergroups that are less common (Lo et al. 2002, 2007; Baldo and Werren 2007). It has been estimated that A and B Wolbachia diverged around 60–70 MYA based upon synonymous substitution rates of the ftsZ gene (Werren et al. 1995). To date, complete genome sequences have been published of the wMel strain, a supergroup A Wolbachia found in Drosophila melanogaster (Wu et al. 2004), and the supergroup D wBm strain, a mutualist of the nematode Brugia malayi (Foster et al. 2005).
In insects, where more than 20% of all species have been estimated to carry Wolbachia (Werren and Windsor 2000), and other arthropods, Wolbachia manipulates host reproduction in a remarkable variety of ways, including feminization, parthenogenesis, male killing, and cytoplasmic incompatibility (CI). All are selected due to the uniparental inheritance of Wolbachia—it is solely transmitted through the eggs and not from males. CI, patterns of sterility classically seen when infected males mate with uninfected females, can allow Wolbachia to spread rapidly to very high frequencies in populations because females that do carry the bacterium can mate successfully with both infected and uninfected males. Wolbachia are also obligate mutualists of filarial nematodes, where they seem to play a role in the pathogenesis of lymphatic filariasis (Taylor, Cross, and Bilo 2000). The interactions of Wolbachia with its host provide model systems for studies on the evolution of symbiosis, parasitism, and mutualism. They have also attracted considerable interest as a tool for pest population suppression (Laven 1967; Dobson et al. 2002), the modulation of insect-borne disease transmission through replacement or population age structure modification (Sinkins and O'Neill 2000; Brownstein et al. 2003; Dobson 2003; Sinkins and Gould 2006), and as a target for antifilarial drug development (Bandi et al. 1999; Hoerauf et al. 2000, 2003; Taylor, Bandi, et al. 2000; Taylor, Bandi, and Hoerauf 2005; Taylor, Makunde, et al. 2005).
Wolbachia pipientis was first discovered and described in Culex pipiens mosquitoes (Hertig and Wolbach 1924; Hertig 1936) and thus the strain now designated wPip, a member of supergroup B, is the type strain of the species. It is the only strain reported from this group of sibling species of mosquito and is found at close to 100% frequency in all populations examined, except for one in South Africa (Cornel et al. 2003). The C. pipiens group includes Culex quinquefasciatus, the primary vector of lymphatic filariasis. Wolbachia induces an unusually complex series of crossing types in the C. pipiens group, including partial or complete CI that can be unidirectional or bidirectional (Laven 1967; Barr 1980; Irving-Bell 1983; Magnin et al. 1987; O'Neill and Paterson 1992; Guillemaud et al. 1997), which contrasts with the very low degree of genetic diversity observed in wPip (Guillemaud et al. 1997; Sinkins et al. 2005). The most parsimonious hypothesis to explain the complex CI patterns between C. pipiens populations is the presence of different variants of the (as yet unidentified) Wolbachia genes that control CI, such that the molecules produced by one wPip variant which control sperm modification in males and rescue in females would be able to act independently of those produced by another wPip variant. Given the practical difficulties in research on these intractable intracellular bacteria, genomics provides an important foundation for understanding the molecular basis of Wolbachia–host interactions.
Materials and Methods
Wolbachia Purification
All Wolbachia genome projects face substantial difficulties in obtaining sufficient quantities of pure DNA because Wolbachia is obligately intracellular and cannot be cultured in cell-free media. Filter purification from preblastoderm embryos proved to be successful because the ratio of Wolbachia to host DNA is then at its highest (as confirmed by quantitative polymerase chain reaction, data not shown). This approach is likely to be applicable to other insect Wolbachia genome projects as long as sufficient quantities of early embryos can be obtained. wPip genomic DNA in large quantities and at high enough quality for genetic analysis was extracted from preblastoderm embryos of the Pel strain (origin Sri Lanka) of C. pipiens mosquitoes, reared using standard rearing procedures in insectary conditions (26 °C, 70% relative humidity). Preblastoderm embryos were selected as the source for purification as the majority of contaminating host genomic DNA was predicted to be contained within the multinuclei single-cell embryos. Quantitative polymerase chain reaction confirmed that a significantly greater ratio of Wolbachia DNA (wsp gene) to Culex DNA (S7 ribosomal protein gene) was extracted from preblastoderm embryos compared with adult female mosquitoes. For each individual extraction, approximately 50 preblastoderm embryo rafts, oviposited within a 30-min period, were gently homogenized in phosphate-buffered saline (PBS) buffer and the resulting homogenate centrifuged at 300 × g (4 °C) for 5 min to pellet large debris including host nuclei. A speed of 200 g was previously shown to pellet Drosophila embryo nuclei (Sun et al. 2001). The resulting supernatant was centrifuged at 4,000 × g (4 °C) for 5 min to pellet the Wolbachia bacteria. The pellet was resuspended in PBS buffer by gently pipetting the solution and then centrifuged for 5 min at 300 × g (4 °C) for a second time to remove any remaining debris. The supernatant was passed through a 5 μm pore size filter (Millipore, Bedford, MA) under unit gravity filtration. The filtrate was centrifuged at 5,000 × g (4 °C) for 15 min to pellet the Wolbachia. As sequencing required unfragmented DNA, vortexing at any stage of the purification was omitted to prevent shearing of DNA fragments. DNA was extracted from the Wolbachia pellet using a modified version of the Livak buffer method with ethanol precipitation (Collins et al. 1987). As the mean total amount of DNA resulting from an individual extraction was 408.8 ± 102.6 ng, 50 extractions were pooled to produce sufficient wPip DNA for genome sequencing. wPip DNA for gap closure was obtained from total DNA extracted from preblastoderm embryos.
Sequencing and Closure
Genomic DNA was sheared by sonication, size fractionated, and used to generate several shotgun libraries. Clones were sequenced from these libraries as follows: 7,357 paired reads from a pUC19 library with an insert size of 1.4–2 kb; 12,165 paired reads from a pUC19 library with an insert size of 2–4 kb; and 5,633 paired reads from a pUC19 library with an insert size of 2–5 kb. This shotgun generated approximately 8.3-fold coverage of the genome, after taking into account contamination from Culex chromosomal DNA, which contributed about 16% of the reads. To scaffold these sequences, 1,407 paired reads were generated from a pBACehr library with an insert size of 30–70 kb, generating around 12-fold genome coverage in bacterial artificial chromosome clones. A further 10,378 directed sequences were generated during the gap closure and finishing phase. DNA was sequenced using ABI BigDye terminator reactions and run on AB3730 capillary sequencers. Sequence assembly was performed by PHRAP and finished to standard criteria using GAP4 as previously described (Parkhill et al. 2000). In essence, these criteria state that all bases must be covered by at least one read in each direction and by reads from at least 2 independent clones; all repeats must be bridged by 2 read pairs or by an end-sequenced PCR product. Around 500 reads proved to be from the Culex mitochondrial DNA and assembled into a single circular molecule of 15,587 bp that is similar in sequence and structure and identical in gene content to that of Aedes albopictus (GenBank accession number: AY072044).
Annotation
Protein-coding genes were identified using Orpheus (Frishman et al. 1998) and GLIMMER (Salzberg et al. 1998) and manually inspected and curated. FastA searches against the UniProt database and Blast searches against the 2 previously sequenced Wolbachia genomes of strains wMel and wBm and GenBank were performed using the predicted gene and protein sequences. Protein domains and motifs were identified using InterProScan (Zdobnov and Apweiler 2001). tRNA genes were identified by tRNAscan-SE (Lowe and Eddy 1997), and rRNA genes were identified using Blast. Pseudogenes contain one or more in-frame stop codons or frameshifts. DNA repeats were identified by the REPuter program (Kurtz et al. 2001). Artemis (Rutherford et al. 2000) was used to compile the data and facilitate the annotation. The sequences and annotation have been submitted to the EMBL/GenBank/DDBJ database with the accession number AM999887.
Ortohologs between wPip, wMel, and wBm were identified by reciprocal best Blast hits and an additional cutoff of E value lower than 1 × 10−5 and Blast alignment covering at least 80% of the shortest sequence. The dot plot between the genome sequences of wPip and wMel was generated using MUMmer3 (Kurtz et al. 2004).
Results and Discussion
General Features of the Genome of wPip
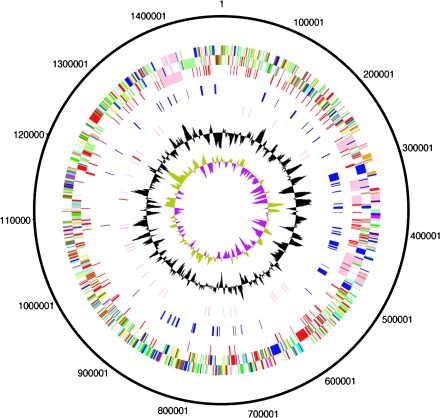
The genome of wPip presented here is a single circular chromosome consisting of 1,482,355 bp and encodes 1386 coding sequences (CDSs). The general features of the genome are presented in table 1 and figure 1. Although the genome sequence was finished to accepted standards (see Materials and Methods), it was not possible to completely contiguate the sequence likely due to sequence polymorphisms, possibly because of high recombination rates and relaxed selection. Several structural variants of the chromosome appeared to exist in the DNA used for sequencing, including large-scale inversions and several different arrangements and variants of the 5 prophages that exist in the genome of Wolbachia wPip in the C. quinquefasciatus colony Pel. The final genome was artificially joined between WP0322 and WP0323, both genes associated with prophage regions (see section on WO prophages). Base pair 1 was assigned to the first nucleotide in the gene dnaA as for Wolbachia strain wMel (Wu et al. 2004). As observed for wMel, there is no clear GC skew identifying a probable origin of replication in wPip (fig. 1, inner circle). It was notable that there appeared to be variants of several DNA sequences that did not assemble into the whole-genome sequence. On analysis, these were seen to be variants of prophages in the assembled genome and most probably represent the natural variation in the prophage regions within the Wolbachia population, indicating that they evolve rapidly.
Table 1.
General Features of the Wolbachia pipientis Genomes
| wPip | wMel | wBm | |
| Size (bp) | 1,482,355 | 1,267,782 | 1,080,084 |
| G + C content (%) | 34.2 | 35.2 | 34.2 |
| Coding sequences | 1,386 | 1,270 | 903 |
| Coding densitya | 0.82 | 0.80 | 0.67 |
| Average gene size (bp)a | 944 | 851 | 899 |
| rRNA | 1 of each | 1 of each | 1 of each |
| tRNA | 34 | 34 | 34 |
| Pseudogenes | 97 | 94 | 98 |
Pseudogenes are excluded.
FIG. 1.—
Circular representations of the genome of Wolbachia pipientis. The circles represent from the outside in: 1 + 2, all genes (transcribed clockwise and counterclockwise); 3, mobile elements (pink: prophage, red: IS elements); 4, ankyrin repeat genes (blue); 5, RNA genes (red: rRNAs, purple: tRNAs); 6, G + C content (plotted using a 10-kb window); 7, GC deviation ([G − C]/[G + C] plotted using a 10-kb window; khaki indicates values >1, purple <1). Color coding for genes in circles 1 + 2: dark blue, cell processes/adaptation/pathogenicity; black, energy metabolism; red, information transfer; dark green, surface associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, mobile elements; and gray, miscellaneous.
Lack of clonality in an inbred laboratory line of mosquitoes suggests that chromosomal variation in Wolbachia is being generated at a fast rate. It is possible that the observed lack of clonality is the consequence of a very labile chromosome in wPip and furthermore that these rapid changes might underpin the phenotypic complexity of CI seen in the C. pipiens group, for example, by generating expression differences in key genes.
Comparative Genomics
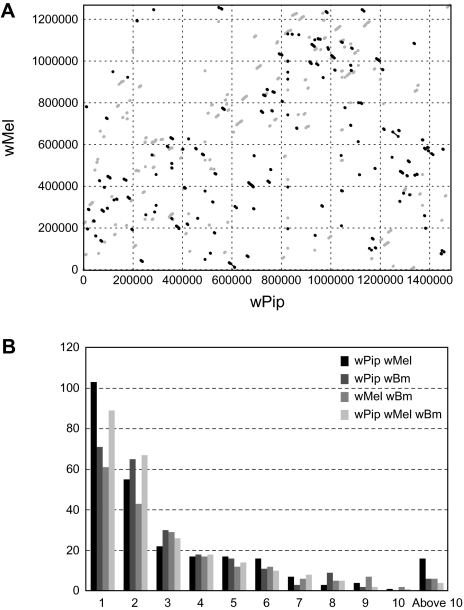
When the genome sequences of wPip and wMel are compared, it is immediately apparent that a very high degree of rearrangement has occurred between them, illustrated in figure 2. A similar pattern was seen when wMel was compared with the wBm strain from filarial nematodes (Foster et al. 2005), although wMel (supergroup A) is more closely related to wPip (supergroup B) than it is to wBm (supergroup D) (Lo et al. 2007). MUMmer dot plots between pairs of bacteria with a similar estimated time since divergence to that of wPip and wMel often have largely colinear genomes with only a small number of rearrangements that tend to be symmetrical around the terminus (or origin), as, for example, seen in comparisons between Rickettsia typhi and Rickettsia prowazekii/Rickettsia conorii (McLeod et al. 2004). Extensive intrachromosomal rearrangement has clearly taken place since the divergence of wPip and wMel, and no long-range gene order has been maintained. The high number of repeats, particularly the considerable expansion in number of several transposable elements, is likely to be responsible by providing numerous sites for homologous recombination. The frequent occurrence of rearrangements could contribute to genome plasticity and provide a mechanism for rapid gene evolution (Foster et al. 2005; Brownlie and O'Neill 2006).
FIG. 2.—
Synteny in Wolbachia between-genome comparisons. (A) Dot plot of the wPip genome sequence versus the wMel genome sequence. Coordinates were generated using nucmer with parameters—maxgap = 500—mincluster = 100, and the plot was generated by mummerplot. Gray dots represent forward matches, and black dots represent reverse matches. (B) Sizes (in number of genes) of blocks of genes found in synteny between the 3 different Wolbachia genomes. The x axis shows the number of genes in a block, and the y axis shows the number of blocks with a certain size. The bars correspond to comparisons between wPip–wMel, wPip–wBm, wMel–wBm–wPip, and wMel–wBm, respectively. All comparisons were based on reciprocal best Blast hits from BlastP searches (see Materials and Methods).
There are a number of blocks of genes where conservation of gene order has been maintained between wMel, wBm, and wPip. Given the extensive rearrangements that have occurred between these lineages, it seems likely that some of these blocks, particularly the larger ones, have been maintained because of functional relationships between the encoded products and/or cotranscription. An example that has already been noted in comparisons between wMel and Rickettsia species is the maintenance of 2 blocks of genes encoding the type IV secretion system (T4SS), also maintained in wPip and shown to be coregulated operons (Wu et al. 2004). Genome comparisons between the 3 Wolbachia from divergent supergroups provide a starting point for interpreting functional interrelationships between Wolbachia genes.
There are 116 insertion sequence (IS) elements belonging to 8 different families (table 2) and 8 additional CDSs similar to transposases present in single copies. The distribution of IS elements between wMel and wPip is almost nonoverlapping, and most of the IS elements from the same families that are seen in both strains (IS110 and IS5, group IS1031) are not very similar to each other, indicating that recent lineage-specific expansions in IS elements have occurred. This suggests that the IS elements present in the 2 genomes have invaded the genomes quite recently, or alternatively, there have been waves of expansion and loss of IS elements leaving different families in the different Wolbachia strains. A total of 44 genes are disrupted by insertion of an IS element, 15 of these are inserted within a gene of which 6 are other transposases, and interestingly, 2 are wsp (Wolbachia surface protein) paralogs, one of which has previously been reported (Sanogo et al. 2007).
Table 2.
Number of IS Elements in Families
| Family | Group | Number in wPip |
| IS6 | 11 | |
| IS982 | 51 | |
| IS110 | 3 | |
| IS5 | IS903 | 14 |
| IS256 | 23 | |
| IS630 | 7 | |
| IS200/IS605 | 4 | |
| IS5 | IS1031 | 3 |
The wPip genome is substantially larger than that of wMel, but the number of single-copy protein-coding genes is not greatly increased. Excluding WO prophage regions, insertion sequences, and pseudogenes, there are 184 wPip genes that have no clear orthologs in wMel or wBm, which are likely to include rapidly evolving genes, any acquisitions via horizontal transfer and genes that have been lost in the other 2 lineages. Of the 57 that have an assigned putative function or domain, 28 encode ankyrin repeat (ANK) domains; this important category is discussed in more detail below. These wPip-specific genes most commonly occur as singletons, with exceptions such as WP729-33, which consists of 4 adjacent ANK genes. Of special interest are the genes that occur in the 2 reproductive parasites, wMel and wPip, but not in the mutualist wBm because they might provide a clue to what genes might be involved in the expression of CI. There are 46 genes that are found in wPip and wMel that are pseudogenes in wBm and 73 genes that are found in wPip and wMel that are completely absent in wBm. Of the 46 genes with pseudogenes in wBm, 38 have putative functional assignments representing many different functional categories including 4 transporters, 4 non–WO-associated phage genes, and 2 ANK domain containing genes. Twenty of the 73 genes with no homologs in wBm have functional assignments of which 4 contain ANK domains and 5 are transporters. As the majority of these genes have no putative function, it is difficult to speculate about their potential role in CI, but they might provide a starting point for further investigations.
Secreted and Surface Proteins: Host Interactions
The wPip genome contains a number of genes encoding putative surface proteins that might be involved in interactions with the host. Of interest are 7 predicted membrane proteins and 3 predicted outer membrane proteins (WP0743, WP1137, and WP1139) that have no clear orthologues in wMel or wBm. However, all 3 predicted outer membrane proteins have paralogs in wMel and seem to have arisen through tandem duplications in wPip. Fourteen putative membrane protein-encoding genes are found only in the 2 insect Wolbachia strains. The components of various secretion pathways are also present, which are significant because they are likely to be required for host reproductive manipulations. Wolbachia homologs of the T4SS (Masui, Sasaki, and Ishikawa 2000; Wu et al. 2004) consist of 14 Vir genes present in 2 main clusters and 3 sites containing one gene (WP0130, WP0599–WP0604, WP0636, WP0871, and WP1255–WP1259). The conservation of its operon structure despite extensive genome rearrangement suggests that the T4SS system is an important mechanism by which Wolbachia exports proteins. Unfortunately, secretion signals for the T4SS are not well characterized. Specific hidden Markov models were trained using 2 different consensus T4SS signal motifs identified in Bartonella and Agrobacterium (Schulein et al. 2005; Vergunst et al. 2005), but these did not identify significant matches in the Wolbachia proteome; it would appear that these signatures are lineage specific and remain to be identified in Wolbachia.
WO Prophage
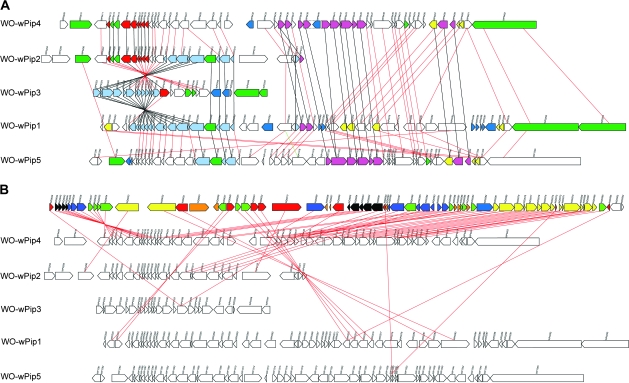
Five copies of the prophage previously named phage WO (Masui, Kamoda, et al. 2000) were identified in the genome of wPip, which represents a major expansion and is a significant contributing factor to the larger overall genome size of wPip. All the copies are, in general, more closely related to the prophage named WO-B than to WO-A in the genome of wMel, but each is also more similar to at least one of the other copies in the wPip genome than to any of the phage sequences previously published from any Wolbachia strain, even though individual genes may show a higher similarity to genes from prophages in other Wolbachia strains. The 5 different copies have been numbered according to their position in the genome, starting with WO-wPip1 closest to the nucleotide designated as 1 in the genome sequence.
Some regions are identical between the different prophage copies in wPip. The largest region of 100% identity is shared between copies WO-wPip1 and WO-wPip2 (10,303 bp in size, light blue in fig. 3A) but is also partly identical to regions in WO-wPip3 (8,302 bp in size) and WO-wPip5 (5,779 bp). Another identical region is found between WO-wPip2 and WO-wPip4 (6,821 bp, red in fig. 3A) and partly contains homologs of the genes found in the identical region between the other copies. These regions seem to contain the most conserved genes between all different WO prophages, both between wPip copies and between WO prophages found in other Wolbachia strains and appear to represent the core of the WO phage gene repertoire. The prophage regions called WO-wPip2 and WO-wPip3 contain parts of the identical regions from both of the described regions, which suggest that there has been recombination between the different copies. WO-wPip2 and WO-wPip3 are located back to back in opposite directions and were the ends of the contig before it was joined up to form the artificially created circular genome; these 2 copies also do not contain any part of the P2-like prophage, described below.
FIG. 3.—
WO phage in wPip. (A) The 5 prophage copies in the genome of wPip. The colors of the boxes represent: green, ANK genes; dark blue, transposases; and yellow, putative transcriptional regulators. Other colors represent identical genes, that is, red genes are identical to other red genes. Black lines connect genes that are identical, and red lines connect genes that are homologous but not identical. (B) WO phage comparing wPip copies with wMel: number of homologues in wPip to genes in the WO-B prophage region in the wMel genome. The top row represents the genes in the WO-B prophage region in the wMel genome, and the next 5 rows depict the 5 prophage regions in wPip. The genes in wMel are colored according to the number of homologues found in the different copies of the wPip prophages. Black, 5 homologues; blue, 4 homologues; green, 3 homologues; yellow, 2 homologues; red, 1 homologues; and orange, 0 homologues. Lines connect the WO-B gene to one of the homologues.
Three of the WO prophages in wPip include both the part described as WO in wMel and a part similar to the pyocin or P2-like smaller phage as well as the genes located between WO-B and the P2-like phage in wMel. Given the close proximity of the 2 entities and the fact that several of the copies have both parts, including some of the intervening genes, it is likely that the P2-like region, annotated as a separate prophage in wMel, is a part of the mosaic WO phage and not a separate entity. Additionally, because several of the genes in between the 2 prophage regions (WO and P2-like) are conserved, they are also likely to be part of a larger prophage. This hypothesis is strengthened by the fact that several of the intervening genes are identical between 2 of the copies containing the P2-like section. No insertion sites for the prophages have been identified in silico, which indicates that none of the prophages are recent insertions and the surrounding sequence has evolved neutrally or under low selection.
Evolution of Ankyrin Repeat Encoding Genes
Ankyrin repeat (ANK) domain proteins function in protein–protein interactions (Sedgwick and Smerdon 1999), and one of the numerous functions in eukaryotes of genes with these domains is to mediate protein–protein interactions in cyclin-dependent kinase (CDK) inhibitors. In Nasonia wasps, the control of host cell cycle timing at karyogamy appears to be disrupted in CI, and inhibition of CDK1 has been hypothesized to be a possible cause of the CI phenotype (Tram and Sullivan 2002; Tram et al. 2003). Therefore, it is possible that one or more Wolbachia ANK genes could be functioning in the generation of CI. Furthermore, an ANK gene labeled ankA in the related intracellular bacterium Anaplasma phagocytophila was shown to colocalize with nuclei (Caturegli et al. 2000) and to bind both DNA as well as several different nuclear proteins (Park et al. 2004), suggesting that it may play a role in the regulation of host gene expression. Recently, the AnkA protein of A. phagocytophila was shown to be secreted into the host cell by the T4SS and thereafter tyrosine phosphorylated by acting on a tyrosine kinase interactor that activates the corresponding tyrosine kinase. This process was shown to be critical for bacterial infection (Lin et al. 2007).
There are 60 genes in the wPip genome that contain one or more ankyrin repeat (ANK) domains. Because 2 genes are found in identical duplicates and 1 gene is found in identical triplicates, this leaves 56 unique ANK genes. wPip has the largest number of ANK genes of any bacterial species for which information is available. All these genes showed evidence for expression using reverse transcriptase–PCR and 3 of them showed host sex-specific differences in expression (Walker et al. 2007). Twenty-five of the wPip ANK genes encode putative transmembrane domains, of which 1 contains an overlapping predicted signal peptide sequence, increasing the possibility that these genes may be involved in interactions with the host.
Understanding the mechanisms of evolution of ANK genes in Wolbachia and the considerable differences in number of these genes between Wolbachia strains is an important aim. Intergenomic comparisons provide a very useful starting point, especially because the 3 genomes of wPip, wMel, and wBm represent 3 different Wolbachia supergroups and both parasitic and mutualistic lifestyles. Out of the 60 ANK genes only 25 are homologous to any wMel ANK gene, and out of the 23 ANK genes found in wMel 16 show homology to wPip ANKs. The discrepancy in number between comparisons is due to the fact that several of the wMel ANK genes, mainly associated with prophage regions, are duplicated in the genome of wPip (see fig. 3). Seventeen out of the 25 wPip ANKs with homology to wMel ANKs are located in regions of local synteny between the 2 genomes indicating that they are part of a set of ANK genes that were present in Wolbachia before the divergence of supergroups A and B.
There are 6 ANK genes in wPip that have a reciprocal best hit in wBm, of which 3 contain ANK domains. However, generally the similarity is quite low or partial. Additionally, there are 3 annotated pseudogenes that have reciprocal best hits to wPip ANKs, and a few new remnants/pseudogene regions similar to wPip ANKs were found that are located in regions of synteny between wPip and wBm. In 2 of these cases, wMel does not have an ANK or pseudogene in that position (wPip_ANK22 [WP0390] and wPip_ANK45 [WP0774]). In 2 cases, there is a pseudogene in wBm where there is an ortholog in wMel that is not in synteny with wPip on either side (wPip_ANK50 [WP1105] and wPip_ANK53 [WP1275]). This indicates that at least some of the expansion of the ANK gene family in Wolbachia probably happened before the divergence of the supergroups and that different sets of ANK genes have been lost or retained in the different strains.
wPip-Specific ANK Genes
A total of 26 of the ANK genes do not show any significant similarity to any currently available sequences except for some low similarity found in the conserved ankyrin repeat domains of the gene. Five of these are inserted in regions of local synteny with only wMel, 3 are in regions of local synteny with wBm only, and 1 is in a region of local synteny with all 3 genomes. Seven of these are located next to an IS element and 2 are on the border of prophage regions, which could provide a possible mechanism of insertion.
ANK Genes with Similarity to Non-ANK Genes
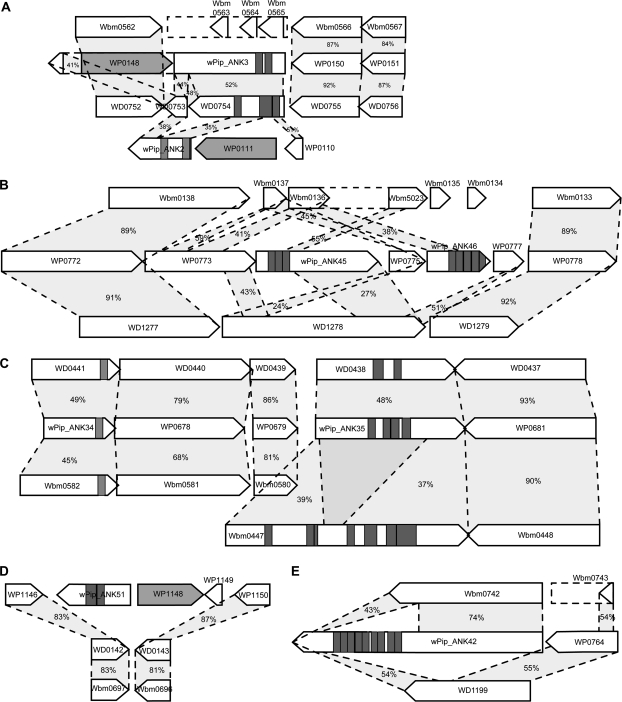
Six of the ANK genes show sequence similarity with genes that do not contain any ankyrin repeats in wMel (4) or in both wMel and wBm (2). One of these genes, wPip_ANK42 (WP0763), has a hit to the hypothetical protein WD1199 at the 3′ end, and the neighboring gene also has a hit against WD1199. wPip_ANK42 is even more similar to Wbm0742 in the genome of wBm, a probable extracellular metollopeptidase, sharing 74% identity over 550 amino acids, leaving a gap in the alignment where the ANK domains are located, and having a lower similarity at the end of the gene where it is also similar to WD1199 (fig. 4E). Because the wMel and wBm genes are similar to different regions/genes, it appears more likely that the ANK domains have been lost in 2 separate events in the 2 lineages in this particular example. Further, in the example shown in figure 4B, the wBm pseudogene contains ANK domains, whereas the wMel gene does not, which suggests that the ANK domains were an original feature of this gene that has later been lost in the lineage leading to wMel. The complicated pattern presented in figure 4B and E could have arisen through a number of recombination events, resulting in deletions and rearrangements specific to each lineage.
FIG. 4.—
ANK gene evolution: 5 examples of different evolutionary scenarios of ANK genes in wPip. Gray rectangles show the position of ANK domains, and dark gray genes represent transposons. Dotted lines are drawn between homologous genes, and the number represents percent identity on the amino acid level. (A) Example of a possible duplication event in the wPip genome of wPip_ANK2 and wPip_ANK3 with comparison to wMel and wBm. (B) Possible deletions of ANK domains in wMel and wBm, putative duplication in wPip and wPip ANK is homologous to non-ANK genes. (C) Conserved gene order of ANKs between wPip, wMel, and wBm. (D) Example of a wPip ANK gene inserted in a region of local synteny with wMel and wBm. (E) Possible deletions of ANK domains in wMel and wBm, wPip ANK homologies are found to non-ANK genes.
Phage-Associated ANK Genes
A total of 19 (all in fig. 3) of the ANK genes are found within or next to the predicted 5 prophage regions. Three of the ANK genes seem to be part of the core content of the WO prophage as these have unambiguous homologs in both wMel and wKue (and also in the incomplete sequences of other Wolbachia strains in Drosophila). These 3 genes are found in 3 copies that are spread between the 5 different prophages and are identical or highly similar to each other, probably reflecting recent duplications of these genes together with the prophage, which could have occurred either through insertion of a new phage or through recombination between existing prophage copies.
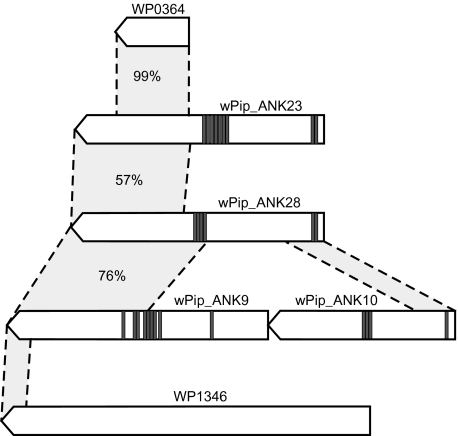
There are 4 very large ANK genes (wPip_ANK9 [WP0292]-2748aa, wPip_ANK10 [WP0293]-1970aa, wPip_ANK23 [WP0407]-2620aa, and wPip_ANK28 [WP0462]-2662aa) that are all located in close proximity to or within the prophage regions of wPip (see figs. 3 and 5). Three of these 4, WP0292, WP0407, and WP0462, show similarity to each other in the C-terminal region, which in turn is also similar to a wMel gene (WD0512) that does not contain any ANK domains. Additionally, there are 2 hypothetical proteins in the wPip genome, WP0364 and WP1346, that show similarity to these ANK-containing proteins, although to different parts (see fig. 5). The C-terminal section of WP1346 is similar to the C-terminal section of WP0292, WP0407, and WP0462 and to WD0512, whereas the rest of the protein is homologous to WD0513 in wMel. Given the large spread in gene size, presence/absence of ANK domains and the partial similarity between these genes, it is possible that they represent a paralogous group of genes that are evolving very rapidly, and it seems likely that some of the copies have evolved through gene fusion or fission. However, the function of these genes is not known, and no additional protein domains have been detected.
FIG. 5.—
Schematic figure of the large ANK genes found in close proximity of the WO prophages in wPip. Gray rectangles show the position of ANK domains, dotted lines are drawn between similar regions, and the number percent represents percent identity on the amino acid level.
Duplications
Other than the duplications of ANK genes associated with prophage regions in the wPip genome, there are a few other cases where duplications of ANK genes seem to have occurred. However, in all these cases, the sequence similarity between the paralogous ANK genes is small and only partial, which is in contrast to the highly similar, and in some cases, identical paralogous genes seen in the prophage regions. Therefore, it seems likely that the paralogous groups of genes found in the prophage regions are recent duplications and, based on the similarity between different prophage copies, are a consequence of whole prophage regions being duplicated or recombined or of novel prophage insertions in new sites.
In the paralogous groups that are not prophage associated, the higher level of sequence divergence suggests that the duplications are old or that they have evolved very rapidly. The only example of these duplications that has a homolog in wMel is wPip_ANK2 (WP0112) and wPip_ANK3 (WP0149). Interestingly, these genes are not very similar to each other, but instead each is more similar to the wMel ANK gene WD0754 and/or to the neighboring gene in wMel WD0753. Because gene order synteny is maintained near to wPip_ANK3, it is reasonable to believe that this is the site of the original copy. Both wPip_ANK3 and wPip_ANK2 are flanked by IS elements, which possibly could have mediated the original duplication. Other examples of likely duplications are wPip_ANK38-wPip_ANK41 (WP0729, WP0730, WP0731, and WP0733), 4 adjacent ANK genes that are similar to each other in the part of the gene not containing the ankyrin repeats; wPip_ANK17 (WP0346) and wPip_ANK18 (WP0347) and wPip_ANK45 (WP0774) and wPip_ANK46 (WP0776) that seem to have been duplicated as a block because wPip_ANK45 is partly similar to WP0777 and wPip_ANK46 is partly similar to WP0773 and WP0775 is partly similar to WP0773 (see fig. 4B). In general, the duplications found are either associated with mobile elements, such as IS elements and prophages, or are located next to each on the chromosome suggesting that they have arisen through tandem duplication events.
Conclusions
Most of the genome size difference between wMel and wPip comes from repeated and mobile elements. The presence of more mobile genetic elements in the genome of wPip could be due to lower selection pressure leading to slightly deleterious or neutral elements not being purged. The high sequence conservation in some prophage regions between the copies within the genome is probably due to recent recombination, duplication, or insertion from free phage particles. However, the conservation between different strains suggests functionality of these genes, indicating that they may be under selection. Even if there are no free phage particles produced from these prophage sites, it seems likely that the genes encode important functions. It is interesting to note that the WO-B prophage region in wMel is rearranged compared with wPip and wKue, but most of the genes do not show any signs of degradation. The other WO prophage found in wMel, WO-A, is less similar to both the wPip prophages and the wKue phage but has a conserved gene order compared with the other strains. It seems likely that at least one of the prophages in wMel has invaded the genome via horizontal gene transfer.
None of the prophages in the wPip genome show any signs of recent introduction from a foreign source, as also seen in the genome of wMel (Wu et al. 2004), suggesting that these prophages have been resident in Wolbachia genomes for a long time. However, there is a lack of congruence between the phylogenies of Wolbachia strains and their corresponding prophages—indeed Wolbachia strains coinfecting the same host can share identical prophage sequences (Masui, Kamoda, et al. 2000; Bordenstein and Wernegreen 2004), suggesting that WO phages have moved horizontally and provide a mechanism for the movement of genes between strains. WO prophage has likely been very important in the recent evolution of wPip by providing sites for homologous recombination and allowing rapid generation of genetic novelty through interphage reassortment.
It is very likely that ANK genes have evolved through a dynamic combination of insertions, duplications, deletions, and selective divergence in these Wolbachia lineages. Although wPip has an unusually large number of ANK genes, it is not unique in the Rickettsiaceae, with Rickettsia felis having 22 (Ogata et al. 2005) and Orientia tsutsugamushi 50 ANK genes (Cho et al. 2007). The fact that the genome of the mutualistic wBm, which is more distantly related to wPip than wMel to wPip, still contains remnants of some of the wPip ANK genes that are not present in wMel and the conservation of gene order around some of these pseudogenes is a strong indication of their presence before the split of the Wolbachia supergroups. It is therefore highly probable that the large number of ANK genes in wPip is not only a result of expansion in this lineage but that their presence is, in some cases, more ancient. The reasons why particular ANK genes are retained or lost in different strains, and why expansion of ANK genes seems to have occurred in some lineages but not others, should become clearer once their functions—which are probably diverse—begin to be unraveled.
The combination of complex crossing types and very low Wolbachia intrastrain variation in the C. pipiens group provides an excellent system for studies to attempt to elucidate the molecular basis of CI. The wPip genome sequence provides a molecular foundation for these studies, and more broadly for work on B-supergroup Wolbachia, which occur widely in arthropods.
Acknowledgments
This work was supported by the Wellcome Trust. L.K. is an EU Marie Curie Fellow. We gratefully acknowledge the work of Karen Mungall, Zahra Abdellah, Tracey Chillingworth, Kay Jagels, Sharon Moule, and Sally Whitehead on sequencing; Frances Smith, Mark Simmonds, Nathalie Bason, and Ester Rabbinowitsch on library making; Weiguo Zhou on DNA purification; and the Sanger Institute core sequencing and informatics teams.
References
- Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol. 2007;55:81–87. doi: 10.1007/s00284-007-0055-8. [DOI] [PubMed] [Google Scholar]
- Bandi C, McCall JW, Genchi C, Corona S, Venco L, Sacchi L. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int J Parasitol. 1999;29:357–364. doi: 10.1016/s0020-7519(98)00200-8. [DOI] [PubMed] [Google Scholar]
- Barr AR. Cytoplasmic incompatibility in natural populations of a mosquito, Culex pipiens L. Nature. 1980;283:71–72. doi: 10.1038/283071a0. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- Brownlie JC, O'Neill SL. Wolbachia genomics: accelerating our understanding of a pervasive symbiosis. In: Bourtzis K, Miller TA, editors. Insect symbiosis. Boca Raton (FL): CRC Press; 2006. pp. 175–183. [Google Scholar]
- Brownstein JS, Hett E, O'Neill SL. The potential of virulent Wolbachia to modulate disease transmission by insects. J Invertebr Pathol. 2003;84:24–29. doi: 10.1016/s0022-2011(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Caturegli P, Asanovich KM, Walls JJ, Bakken JS, Madigan JE, Popov VL, Dumler JS. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NH, Kim HR, Lee JH, et al. (19 co-authors) The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Cornel AJ, McAbee RD, Rasgon J, Stanich MA, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: culicidae) in California and South Africa. J Med Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- Dobson SL. Reversing Wolbachia-based population replacement. Trends Parasitol. 2003;19:128–133. doi: 10.1016/s1471-4922(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Dobson SL, Fox CW, Jiggins FM. The effect of Wolbachia-induced cytoplasmic incompatibility on host population size in natural and manipulated systems. Proc Biol Sci. 2002;269:437–445. doi: 10.1098/rspb.2001.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, et al. (12 co-authors) The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Mironov A, Mewes HW, Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemaud T, Pasteur N, Rousset F. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc Biol Sci. 1997;264:245–251. doi: 10.1098/rspb.1997.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig M. The rickettsia Wolbachia pipientis (gen. et. sp. n) and associated inclusions of the mosquito, Culex pipiens. Parasitology. 1936;28:453–486. [Google Scholar]
- Hertig M, Wolbach S. Studies on rickettsia-like microorganisms in insects. J Med Res. 1924;44:329–374. [PMC free article] [PubMed] [Google Scholar]
- Hoerauf A, Mand S, Fischer K, Kruppa T, Marfo-Debrekyei Y, Debrah AY, Pfarr KM, Adjei O, Buttner DW. Doxycycline as a novel strategy against bancroftian filariasis-depletion of Wolbachia endosymbionts from Wuchereria bancrofti and stop of microfilaria production. Med Microbiol Immunol. 2003;192:211–216. doi: 10.1007/s00430-002-0174-6. [DOI] [PubMed] [Google Scholar]
- Hoerauf A, Volkmann L, Nissen-Paehle K, Schmetz C, Autenrieth I, Buttner DW, Fleischer B. Targeting of Wolbachia endobacteria in Litomosoides sigmodontis: comparison of tetracyclines with chloramphenicol, macrolides and ciprofloxacin. Trop Med Int Health. 2000;5:275–279. [PubMed] [Google Scholar]
- Irving-Bell RJ. Cytoplasmic incompatibility within and between Culex molestus and Cx. quinquefasciatus (Diptera: culicidae) J Med Entomol. 1983;20:44–48. [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature. 1967;216:383–384. doi: 10.1038/216383a0. [DOI] [PubMed] [Google Scholar]
- Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many wolbachia supergroups exist? Mol Biol Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- Lo N, Paraskevopoulos C, Bourtzis K, O'Neill SL, Werren JH, Bordenstein SR, Bandi C. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 2007;57:654–657. doi: 10.1099/ijs.0.64515-0. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin M, Pasteur N, Raymond M. Multiple incompatibilities within populations of Culex pipiens L. in southern France. Genetica. 1987;74:125–130. doi: 10.1007/BF00055223. [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- Masui S, Sasaki T, Ishikawa H. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J Bacteriol. 2000;182:6529–6531. doi: 10.1128/jb.182.22.6529-6531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MP, Qin X, Karpathy SE, et al. (22 co-authors) Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SL, Paterson HE. Crossing type variability associated with cytoplasmic incompatibility in Australian populations of the mosquito Culex quinquefasciatus Say. Med Vet Entomol. 1992;6:209–216. doi: 10.1111/j.1365-2915.1992.tb00608.x. [DOI] [PubMed] [Google Scholar]
- Ogata H, Renesto P, Audic S, Robert C, Blanc G, Fournier PE, Parinello H, Claverie JM, Raoult D. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim KJ, Choi KS, Grab DJ, Dumler JS. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol. 2004;6:743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD, et al. (28 co-authors) Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo YO, Dobson SL, Bordenstein SR, Novak RJ. Disruption of the Wolbachia surface protein gene wspB by a transposable element in mosquitoes of the Culex pipiens complex (Diptera, Culicidae) Insect Mol Biol. 2007;16:143–154. doi: 10.1111/j.1365-2583.2006.00707.x. [DOI] [PubMed] [Google Scholar]
- Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci USA. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, O'Neill SL. Wolbachia as a vehicle to modify insect populations. In: Handler AM, James AA, editors. Insect transgenesis: methods and applications. Boca Raton (FL): CRC Press; 2000. pp. 271–288. [Google Scholar]
- Sinkins SP, Walker T, Lynd AR, Steven AR, Makepeace BL, Godfray HC, Parkhill J. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- Sun LV, Foster JM, Tzertzinis G, Ono M, Bandi C, Slatko BE, O'Neill SL. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J Bacteriol. 2001;183:2219–2225. doi: 10.1128/JB.183.7.2219-2225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf AM, Lazdins J. Wolbachia bacteria of filarial nematodes: a target for control? Parasitol Today. 2000;16:179–180. doi: 10.1016/s0169-4758(00)01661-6. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Cross HF, Bilo K. Inflammatory responses induced by the filarial nematode Brugia malayi are mediated by lipopolysaccharide-like activity from endosymbiotic Wolbachia bacteria. J Exp Med. 2000;191:1429–1436. doi: 10.1084/jem.191.8.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Makunde WH, McGarry HF, Turner JD, Mand S, Hoerauf A. Macrofilaricidal activity after doxycycline treatment of Wuchereria bancrofti: a double-blind, randomised placebo-controlled trial. Lancet. 2005;365:2116–2121. doi: 10.1016/S0140-6736(05)66591-9. [DOI] [PubMed] [Google Scholar]
- Tram U, Ferree PM, Sullivan W. Identification of Wolbachia–host interacting factors through cytological analysis. Microbes Infect. 2003;5:999–1011. doi: 10.1016/s1286-4579(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Tram U, Sullivan W. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science. 2002;296:1124–1126. doi: 10.1126/science.1070536. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, Sinkins SP. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007;5:39. doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, et al. (30 co-authors) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]