Abstract
We have tested the impact of tags on the structure and function of indirect flight muscle (IFM)-specific Act88F actin by transforming mutant Drosophila melanogaster, which do not express endogenous actin in their IFMs, with tagged Act88F constructs. Epitope tagging is often the method of choice to monitor the fate of a protein when a specific antibody is not available. Studies addressing the functional significance of the closely related actin isoforms rely almost exclusively on tagged exogenous actin, because only few antibodies exist that can discriminate between isoforms. Thereby it is widely presumed that the tag does not significantly interfere with protein function. However, in most studies the tagged actin is expressed in a background of endogenous actin and, as a rule, represents only a minor fraction of the total actin. The Act88F gene encodes the only Drosophila actin isoform exclusively expressed in the highly ordered IFM. Null mutations in this gene do not affect viability, but phenotypic effects in transformants can be directly attributed to the transgene. Transgenic flies that express Act88F with either a 6x histidine tag or an 11-residue peptide derived from vesicular stomatitis virus G protein at the C terminus were flightless. Overall, the ultrastructure of the IFM resembled that of the Act88F null mutant, and only low amounts of C-terminally tagged actins were found. In contrast, expression of N-terminally tagged Act88F at amounts comparable with that of wild-type flies yielded fairly normal-looking myofibrils and partially reconstituted flight ability in the transformants. Our findings suggest that the N terminus of actin is less sensitive to modifications than the C terminus, because it can be tagged and still polymerize into functional thin filaments.
INTRODUCTION
Actins, a highly conserved family of cytoplasmic proteins, are among the most abundant proteins in eukaryotic cells. As a major component of the cytoskeleton, they control shape and motility in nonmuscle cells. In muscle, actin assembles into thin filaments, which together with interdigitating myosin thick filaments provide the framework for muscle contraction.
Many organisms synthesize multiple isoforms of actin that are very similar in amino acid sequence even within the same cell. The differential expression of distinct actins as well as the high conservation of specific isoforms across species emphasize the functional importance of isoforms. In the case of actin, the question of how structure determines function appears to be particularly challenging. Considerable efforts have been made to understand how the different isoforms fulfill their various functions despite their extremely high sequence identity, and yet the basis of their functional diversity remains elusive.
Studying the specific role of a particular actin isoform has always been hampered by the difficulty of discriminating between the introduced and the endogenous actins. Several experimental strategies have been designed to overcome this problem. For example, fluorescent labeling of actin was used to trace the fate of a distinct actin isoform after its microinjection into living cells (Sanger et al., 1984), but this technique requires that significant amounts of actin be purified. Other labeling techniques such as the specific interaction of fluorescent phalloidin derivatives with filamentous actin (F-actin)1 (Estes et al., 1981; Collucio and Tilney, 1984) are dependent on the conformational state of the protein, because this toxin only binds to F-actin but not to monomeric actin. Alternatively, specific antibodies have been used to identify particular actins (Lubit and Schwartz, 1980). However, because actins are highly conserved, only a few isoform-specific antibodies devoid of cross-reactivity with homologous isoforms exist (Skalli et al., 1986; Gimona et al., 1994).
Epitope tagging has become a widely used approach of tracking different proteins with antibodies directed against the tag. A viral epitope such as the 11-amino acid peptide derived from vesicular stomatitis virus G protein (VSV-G) (Soldati and Perriard, 1991) decreases the risk that the antibody recognizing the tag cross-reacts with cellular components. Insertion of this particular tag at the C terminus of different actin isoforms has been used to study their distribution relative to the endogenous actins in fibroblasts and cardiomyocytes (von Arx et al., 1995), smooth muscle, endothelial and epithelial cells (Mounier et al., 1997), and hippocampal neurons (Kaech et al., 1997). For the interpretation of these experiments it has been assumed that the tag does not interfere with the correct folding and function of the protein. Accordingly, heterologous muscle actins tagged at their C terminus with the 11-mer were found to coassemble with purified rabbit α-skeletal actin and did not perturb the sarcomeric organization when expressed in adult rat cardiomyocytes (von Arx et al., 1995; Mounier et al., 1997). However, in these experiments the large excess of unmodified endogenous actin is likely to overpower the properties of the modified recombinant actin. To rule out any dominant effects of unmodified endogenous over introduced actin, we have taken advantage of the indirect flight muscle (IFM) of Drosophila melanogaster, which allowed us to unambiguously analyze the consequences of epitope tagging the IFM-specific Act88F actin on muscle structure and function in its bona fide environment.
Of the six actin genes in Drosophila, Act88F is expressed only in the IFM, where it is the sole actin isoform found (Fyrberg et al., 1983; Ball et al., 1987). Null mutations of the Act88F gene have yielded strains, for example, KM88 (Okamoto et al., 1986), which because of the lack of endogenous Act88F actin in the IFM are flightless but otherwise perfectly viable. Valuable information on the significance of specific amino acids in myofibrillar assembly and function has been obtained from P element–mediated germ line transformation of such null mutants with mutated or truncated Act88F genes (Hiromi et al., 1986; Reedy et al., 1989; Drummond et al., 1990, 1991). The different mutations produce a wide range of phenotypes, ranging from antimorphic effects (Karlik et al., 1987; Sakai et al., 1990) to a virtually normal IFM sarcomere organization (Drummond et al., 1991).
Using this experimental system, we have examined the impact of epitope tagging on the structural and functional properties of the Act88F isoform in situ without the interference of endogenous normal actin. We have transformed Act88F null mutant KM88 flies lacking resident Act88F actin with Act88F constructs that bear either the 11-mer tag from VSV-G or a tag of six consecutive histidines (6xHis) at the C terminus or at the N terminus. Expression of the recombinant actin was demonstrated by means of the tag. Furthermore, by modifying either end of the molecule, we could examine how the position of the tag affects the processing, accumulation, and sarcomere assembly of tagged Act88F actin. The ultrastructural IFM morphology of N- and C-terminally tagged transformants was examined to assess the competence of tagged Act88F actin to polymerize and assemble into ordered myofibrillar structures. In parallel, by testing the flight ability of the corresponding transformants, we analyzed the consequences of epitope tagging Act88F actin on IFM function in vivo. Our studies demonstrate that addition of 6xHis at the N terminus does not abrogate the intrinsic property of actin to polymerize and therefore provides a valuable tool to isolate recombinant actin for in vitro studies.
MATERIALS AND METHODS
Plasmid Constructions
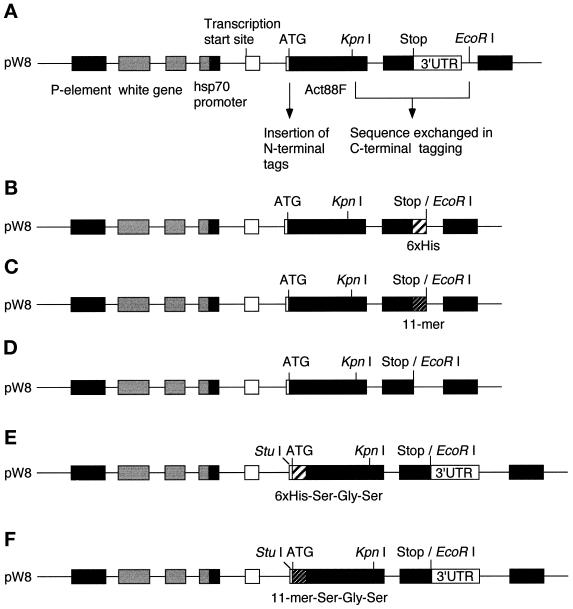
A PstI–EcoRI fragment encoding the Act88F gene, 2-kb regulatory sequences, and the 3′ untranslated region (3′ UTR) was excised from the P[ry+;CSB] plasmid (Hiromi et al., 1986) and cloned into the pW8 Drosophila transformation vector (Klemenz et al., 1987), which contains the selectable white (w) marker gene (Figure 1A).
Figure 1.
Schematic representation of tagged Act88F transformation constructs. The endogenous Act88F gene inserted into the pW8 vector (A) was modified at the C terminus by PCR and site-directed mutagenesis to contain a sequence corresponding to six consecutive histidines (B) or to an 11-mer derived from vesicular stomatitis virus G protein (C). This alteration resulted in the removal of the 3′ UTR. In the corresponding control construct, the 3′ UTR was removed from the endogenous Act88F (D). For tagging the Act88F at the N terminus, the sequences corresponding to six histidines or the 11-mer were introduced in the Act88F gene following the translation start site (E and F). Act88F coding sequences are represented by black boxes, whereas noncoding sequences are shown as white boxes. The epitope tags are shown as shaded boxes. Elements of the pW8 transformation vector are represented by stippled boxes.
Insertion of a 6xHis Tag at the C Terminus of Act88F.
A C-terminal 6xHis tag was introduced by PCR. Primers were designed to span the KpnI site at the 5′ end (Act88F1, 5′-CGGCGGGTACCACCATGTACCCTGG-3′) and to generate six CAC codons, followed by a TAA translation stop signal, and an EcoRI site at the 3′ end (Act88F2, 5′-GCGAATTCTTAGTGGTGGTGGTGGTGGTGAAAG-CATTTGCGGTGAACGATTCC-3′). Subsequently, the KpnI–EcoRI fragment of the original pW8Act88F construct (Figure 1A) was replaced by the KpnI–EcoRI PCR product (Figure 1B).
Tagging Act88F with a C-terminal 11-Mer from VSV-G Protein.
A VSV-G 11-mer tag (Soldati and Perriard, 1991) encoding the amino acids YTDIEMNRLGK was introduced at the C terminus of the Act88F coding sequence by PCR as described (Horton et al., 1989) using two sets of primers: Act88F1 and 5′-CATCTCTATGTCTGTATAAAAGCATTTGCGGTGAAC-3′, and 5′-GTTCACCGCAAA-TGCTTTTATACAGACATAGAGATG-3′ and 5′-GCGAATTCTTC-TTTCCTGCGTTATCCC-3′. The KpnI–EcoRI fragment of the original pW8Act88F plasmid (Figure 1A) was replaced by the final PCR product (Figure 1C).
Deletion of 3′ UTR in pW8Act88F Plasmid.
The KpnI–EcoRI fragment in the original pW8Act88F plasmid (Figure 1A) was replaced by a DNA fragment amplified by PCR using the Act88F1 primer and an antisense primer (5′-CCGGAATTCTTAAAAGCATTTGCGGTG-3′) containing an EcoRI site following the TAA stop codon (Figure 1D).
Tagging Act88F with 6xHis and 11-Mer at the N Terminus.
For inserting tags at the N terminus, the EcoRI site in the original pW8Act88F plasmid was obliterated, and instead a new EcoRI site was generated after the stop codon by PCR. Then, the nucleotide sequence at position −11 to −9 preceding the translation start codon was changed from 5′-CAA-3′ to 5′-GGC-3′ by site-directed mutagenesis to yield a unique StuI site without affecting the splice acceptor sequence or the sequence immediately upstream of the start site. For cloning purposes, a BamHI site (Gly-Ser) was inserted after the ATG by PCR. Finally, an oligonucleotide linker encoding the translational start site, followed by the 6xHis tag and an additional Ser (5′-TAGAAGGCCTGCCAAGATGCACCACCACCACC-ACCACTCCG-3′ and 5′-ATCCGGAGTGGTGGTGGTGGTGGTGCATCTTGGCAGGCCTT-3′; Figure 1E) or the 11-mer tag with an additional Ser (5′-TAGAAGGCCTGCCAGGATGTACACCGACATCGAGATGAACCGCCTGGGCAAGTCCG-3′ and 5′-ATCC-GGACTTGCCCAGGCGGTTCATCTCGATGTCGGTGTACATCTT-GGCGGCGGCCTT-3′; Figure 1F), was inserted using the StuI and BamHI sites. The resulting constructs encoded the respective tag followed by a short sequence encoding a Ser-Gly-Ser tripeptide.
The correct reading frame of the modified Act88F genes was confirmed by DNA sequencing.
Germ Line Transformation
Germ line transformation was carried out essentially as described by Rubin and Spradling (1982) using the helper P element plasmid pπ25.7Δ2-3 wc. The recipient strain for all constructs was the KM88 mutant (w; Act88FKM88) (Hiromi and Hotta, 1985; Okamoto et al., 1986).
The posterior ends of homozygous KM88 embryos were injected with 100 ng/μl helper plasmid and 100–300 ng/μl pW8 constructs. Individual adult Go flies were back-crossed to KM88 flies, and the progeny was scored for red eyes. For each construct at least four independent homozygous lines were established using balancer chromosomes (Lindsley and Zimm, 1992).
Flight Ability Test
Three days after eclosion, flies were tested for flight ability using the flight tester described by Okamoto et al. (1986). The flight tester consists of a transparent plastic cylinder that is 40 cm in diameter and 60 cm high. The bottom and the top are sealed with transparent plastic covers. A funnel with a 17-cm-long duct is inserted at the center of the top cover, and a saucer is hung 3 cm below the funnel. The cylinder is divided into intervals of 5 cm from bottom to top, the ceiling, the bottom, and the saucer. The inner surface is coated with liquid paraffin oil. The flight tester is illuminated from the top to attract flies. Flight ability is scored by releasing 200 flies through the funnel into the flight tester. After 3 min, the number of flies landing in each region was counted.
Electron Microscopy
IFMs were prepared for electron microscopy according to Reedy and Beall (1993) with minor modifications. Twenty-four- to 48-h-old female flies were etherized and mounted in modeling clay. The head and abdomen were removed, and the dorsal half of the thorax containing the dorsal longitudinal IFM was separated from the ventral half with microsurgical scissors. Dissected dorsal thoraces were directly immersed in a freshly prepared fixative consisting of 3% glutaraldehyde and 0.2% tannic acid in MOPS-buffered Drosophila Ringer’s solution (Fyrberg et al., 1990) without phosphate (pH 6.8) for 2 h at room temperature. After primary fixation, half-thoraces were rinsed three times for 15 min in MOPS-buffered Drosophila Ringer’s solution and three times for 2 min in 100 mM phosphate buffer (pH 6.0). Subsequently, the thoraces were immersed for 1 h in ice-cold secondary fixative consisting of 1% osmium tetroxide in 100 mM phosphate buffer and 10 mM MgCl2 (pH 6.0). After three washes in water for 5 min, thoraces were block stained in aqueous 2% uranyl acetate for 1 h at 4°C. After dehydration by a series of increasing ethanol concentrations (50–100%), specimens were embedded in a series of acetone-Epon mixtures and finally in pure Epon.
Electron micrographs were recorded on Kodak (Rochester, NY) SO163 film at a nominal magnification of 10,000× using a LEO (Oberkochen, Germany) 910 transmission electron microscope operated at 80 kV.
Protein Extraction
IFM Dissection. Individual adult flies were anesthetized with ether and mounted in a plasticine mold. The head and abdomen were removed, and a longitudinal incision was made along the dorsal surface of the thorax. The open thorax was then transferred into ice-cold Drosophila Ringer’s solution and cut down the midsagittal plane, exposing the dorsal longitudinal flight muscles at the surface of each half-thorax. Using a fine needle, the IFMs were released from the thorax close to their site of attachment. The IFMs of one fly were transferred to 20 μl of SDS-PAGE sample loading buffer (2.3% SDS, 62.5 mM Tris-HCl, pH 7.0, 15% glycerol, 2.5% β-mercaptoethanol, 0.05% bromphenol blue) and boiled for 5 min.
PAGE and Immunoblot Analysis.
Protein extracts corresponding to the IFMs of half a thorax were separated on 11.5% SDS-polyacrylamide gels (Laemmli, 1970). Gels were electroblotted onto an Immobilon polyvinylidene difluoride membrane (Millipore, Bedford, MA). Blots were rinsed with PBS and transiently stained with Coomassie Brilliant Blue to confirm comparable amounts and equal transfer. After complete destaining with methanol, blots were washed in PBS and 0.1% Tween 20 and blocked for at least 30 min at room temperature in 5% milk powder in PBS and 0.1% Tween 20. Blots were incubated for 2 h at room temperature with a mAb recognizing different actin isoforms (Amersham Pharmacia Biotech, Zürich, Switzerland; diluted 1:7500 in PBS and 0.1% Tween 20), a mouse mAb recognizing the 6xHis tag (Dianova, Milan Analytica AG, La Roche, Switzerland; diluted 1:100) (Zentgraf et al., 1995), or hybridoma supernatant from the mouse mAb P5D4, which recognizes the 11-mer tag from VSV-G (diluted 1:10) (Kreis, 1986). Blots were washed for 15 min each with 5% milk powder in PBS and 0.1% Tween 20, PBS and 0.1% Tween 20, and 1% blocking solution (Boehringer Mannheim, Rotkreuz, Switzerland) in PBS and 0.1% Tween 20, followed by a 2-h incubation with a 1:5000 dilution of a goat anti-mouse IgG alkaline phosphatase–conjugated secondary antibody (Sigma, Buchs, Switzerland). Blots were washed three times for 15 min with PBS and 0.1% Tween 20 and once with 100 mM Tris (pH 9.5), 100 mM NaCl, and 5 mM MgCl2, and then developed with Western Blue stabilized substrate for alkaline phosphatase (Promega, Wallisellen, Switzerland).
RESULTS
Generation of Act88F6xHis and Act88F11-mer Transgenic Flies
To test the effect of epitope tags on the expression and function of actin, we modified the IFM-specific Act88F gene (Figure 1A), as described in MATERIALS AND METHODS, and introduced the recombinant actin into flies lacking endogenous Act88F expression. As shown schematically in Figure 1, B and C, sequences encoding 6 histidine residues or 11 amino acids derived from VSV-G protein (Soldati and Perriard, 1991) were inserted at the C terminus of the endogenous Act88F gene. The resulting pW8 transformation vectors were introduced into KM88, an Act88F null mutant line (Hiromi et al., 1986), by P element–mediated germ line transformation (Rubin and Spradling, 1982). Six Act88F6xHis and five Act88F11-mer homozygous fly lines were independently established. Two lines from each construct had insertions on the X chromosome, one and two had insertions on chromosome 2, and three and one had insertions on chromosome 3 for the Act88F6xHis and Act88F11-mer, respectively. Because addition of the tags at the C terminus of Act88F resulted in the removal of the 3′ UTR, five corresponding control lines were established, which are homozygous for Act88F lacking the 3′ UTR (Figure 1D). These control lines were all able to fly and exhibited an IFM structure that was indistinguishable from wild-type flies (our unpublished results). These findings indicate that the Act88F 3′ UTR is not required for the correct assembly of functional myofibrils.
In contrast to the C-terminal insertion of the epitope tags, addition of the 6xHis or the 11-mer tag at the N terminus (Figure 1, E and F) did not eliminate the 3′ UTR. For constructing N-terminally tagged transformation vectors, the tag sequences were inserted immediately after the ATG start codon in exon 2 of the Act88F gene, followed by a Ser-Gly-Ser tripeptide linker. Four 6xHisAct88F and four 11-merAct88F homozygous fly lines with transgene integration on chromosomes 2 (one and three lines, respectively), 3 (one line each) or X (two 6xHisAct88F lines) were independently established.
Expression of Tagged Act88F Protein in IFM
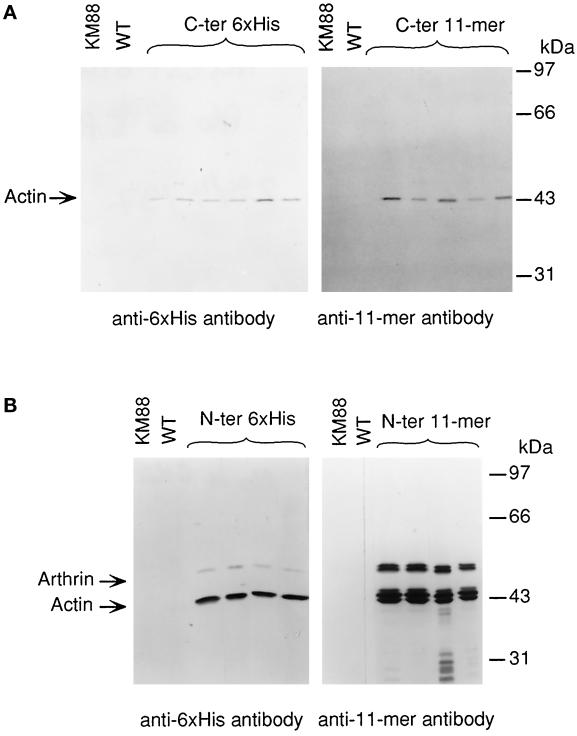
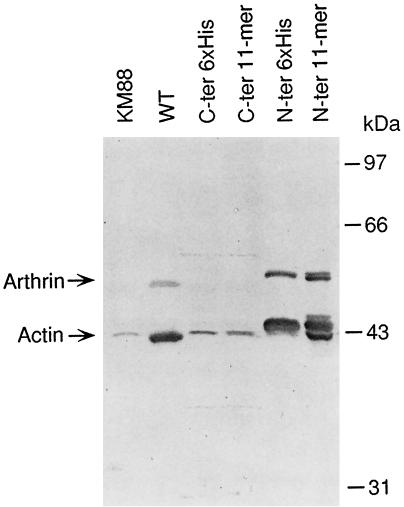
The epitope tags were added to the coding sequence to allow for an unambiguous distinction of the recombinant Act88F from other endogenous isoforms in protein extracts of transformants by immunoblotting with mAbs that specifically recognize the respective tag. In Figure 2, the expression of C-terminally (Figure 2A) and N-terminally (Figure 2B) tagged Act88F in the IFM is shown. Each lane represents a protein extract of dissected IFM corresponding to one half thorax equivalent. Transient staining of the blots confirmed that total amounts of protein were comparable. The mAb against the 6xHis tag (Figure 2A, left) detected a single band, which migrates with an apparent molecular mass of ∼43 kDa in all six transgenic lines. As expected, the band that corresponds to the size of actin is absent in the KM88 mutant and wild-type flies. Likewise, the P5D4 mAb recognizing the VSV-G 11-mer (Kreis, 1986) reacted with a single band of ∼43 kDa in IFM extracts from flies transformed with the 11-mer Act88F construct (Figure 2A, right). C-terminally tagged Act88F protein was expressed in the IFMs of all transgenic lines established with the corresponding construct. We did not observe any significant difference in the expressed levels of tagged Act88F between the individual lines from each construct.
Figure 2.
Expression of epitope-tagged Act88F in transformed KM88 flies. Protein extracts corresponding to an equal number of thoraces from transformants established with C-terminally (A) and N-terminally (B) tagged Act88F constructs were analyzed by immunoblotting. Wild-type (WT) and KM88 protein extracts were included on each blot as controls. Immunblots were incubated with either a mAb recognizing the 6xHis tag (left) or the 11-mer tag (right). (A) Each antibody recognizes a single band in extracts prepared from the transformed lines, which is absent in thoracic extracts from wild-type and KM88 flies. Its apparent molecular mass of 43 kDa corresponds to that of actin. Levels of tagged Act88F protein do not vary significantly among the transformants expressing the same construct. (B) In immunoblots from N-terminally tagged Act88F transformants, an additional band migrating at 55 kDa is detected. This band corresponds to tagged arthrin. In protein extracts from transformants tagged with the 11-mer at the N terminus, multiple actin and arthrin bands are present.
Immunoblotting of IFM extracts from flies transformed with N-terminally tagged Act88F constructs with mAb against the respective tags showed that the constructs are expressed in the IFMs of the transformed flies at similar levels (Figure 2B). The mAb recognizing the 6xHis tag detected not only the prominent band representing tagged Act88F, but an additional tagged protein, which migrates with an apparent molecular mass of ∼55 kDa (Figure 2B, left). Most likely, this band corresponds to 6xHis tagged arthrin, the ubiquitinated form of Act88F, which is typically present in insect IFM at the ratio of one arthrin molecule to six actin molecules (Ball et al., 1987). The P5D4 mAb detected three bands with closely related molecular weights corresponding to actin and two bands representing tagged arthrin (Figure 2B, right). Because the epitope recognized by the P5D4 antibody predominantly consists of the five C-terminal amino acids of the 11-mer (Kreis, 1986) it is possible that partial removal of the N-terminal amino acids accounts for the multiple actin forms. Alternatively, the N-terminal 11-mer tag leads to inefficient posttranslational processing. Whereas the exact origin of the multiple bands is unclear, it appears that the 11-mer tag interferes with the correct processing of the N terminus without, however, abrogating ubiquitination.
Immunoblots using the corresponding antibodies revealed an increased amount of the tagged actin in IFM extracts from transformants expressing N-terminally tagged actins compared with the immunoblots of IFM extracts from C-terminally tagged Act88F lines (Figure 2, compare A and B). Together with the detection of tagged arthrin, this finding suggests that N-terminally tagged Act88F is present at higher levels than the C-terminally tagged Act88F.
Effects of Tagged Act88F Expression on Flight Ability
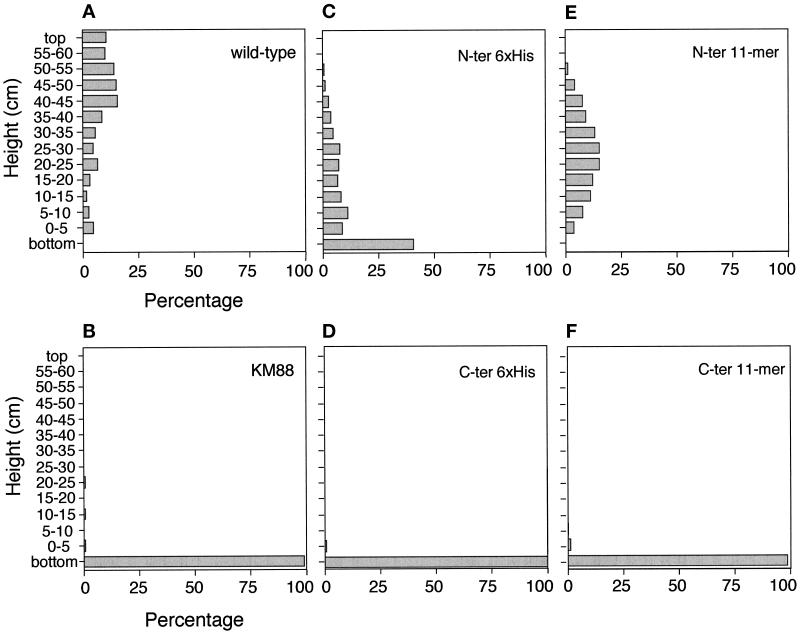
Because the IFM-specific Act88F is absent in the KM88 null mutant, a viable but flightless line ensues (Hiromi and Hotta, 1985; Okamoto et al., 1986). The sarcomeric organization and the myofibrillar structure, as well as the flight ability of transformants expressing the unmodified Act88F gene, were shown to be indistinguishable from wild-type flies (Hiromi et al., 1986; our unpublished results). We tested the effects of the epitope tags on the functional properties of Act88F in a flight test assay (Okamoto et al., 1986; see MATERIALS AND METHODS). Figure 3 displays histograms characterizing the flight ability of the transformants compared with that of wild-type flies and the KM88 null mutant, respectively. Several lines were analyzed for each type of transformant. The results for the different lines were comparable, and a representative histogram is given for each type of transformant. The histograms of the C-terminally tagged transformants (Figure 3, D and F) looked similar to the histogram of the flightless KM88 mutant (Figure 3B). Accordingly, the majority of the flies stayed in the saucer or fell to the bottom of the cylinder. A few flies reached the sidewall of the cylinder near the bottom, probably because of the random trajectories of their falls. The lack of flight ability in both C-terminally tagged transformants indicates that insertion of either six histidines or the 11-mer epitope at the C terminus of Act88F interferes with the assembly and/or function of thin filaments.
Figure 3.
Effects of epitope tagging on IFM function in transformed Drosophila. Flight ability was examined in a flight tester (Okamoto et al., 1986). Accordingly, the “landing sites” of individual flies were scored and are shown as percentages. A consistent fraction of flies (approximately one-third) was found either at the bottom of the cylinder or remained in the saucer and was subtracted from the nonflying flies from each test. Each histogram represents the average of three experiments. (A) Wild-type flies show heterogeneity in their flight ability. (B) The KM88 null mutant is flightless. (C) In N-terminally tagged 6xHis transformants, the percentage of poor fliers is higher than in wild-type flies. (D) C-terminally tagged 6xHis transformants are not able to fly. (E) Transformants with the 11-mer tag at the N terminus have a slightly increased flight ability compared with flies with a 6xHis tag, but they do not fly as proficiently as wild-type flies. (F) Similar to the C-terminally tagged 6xHis transformants, C-terminally tagged 11-mer transformants are flightless.
Normal flight ability is defined by >50% of the flies reaching the sidewall in the top third of the cylinder, which corresponds to the height of the saucer and higher. Accordingly, the histogram for wild-type flies (Figure 3A) reveals that 64.4% reach the top third. Approximately 30% of the homozygous transformants expressing Act88F with an 11-mer epitope at the N terminus (Figure 3E) and 13% of the lines with an N-terminally 6xHis-tagged Act88F (Figure 3C) were able to land in the top third. However, 35.5% of the 11-mer flies and 51.5% of the 6xHis flies reached the sidewall at a height between 5 and 40 cm. These results suggest that the expression of N-terminally tagged Act88F, although not normal, reconstitutes IFM function to a significant extent.
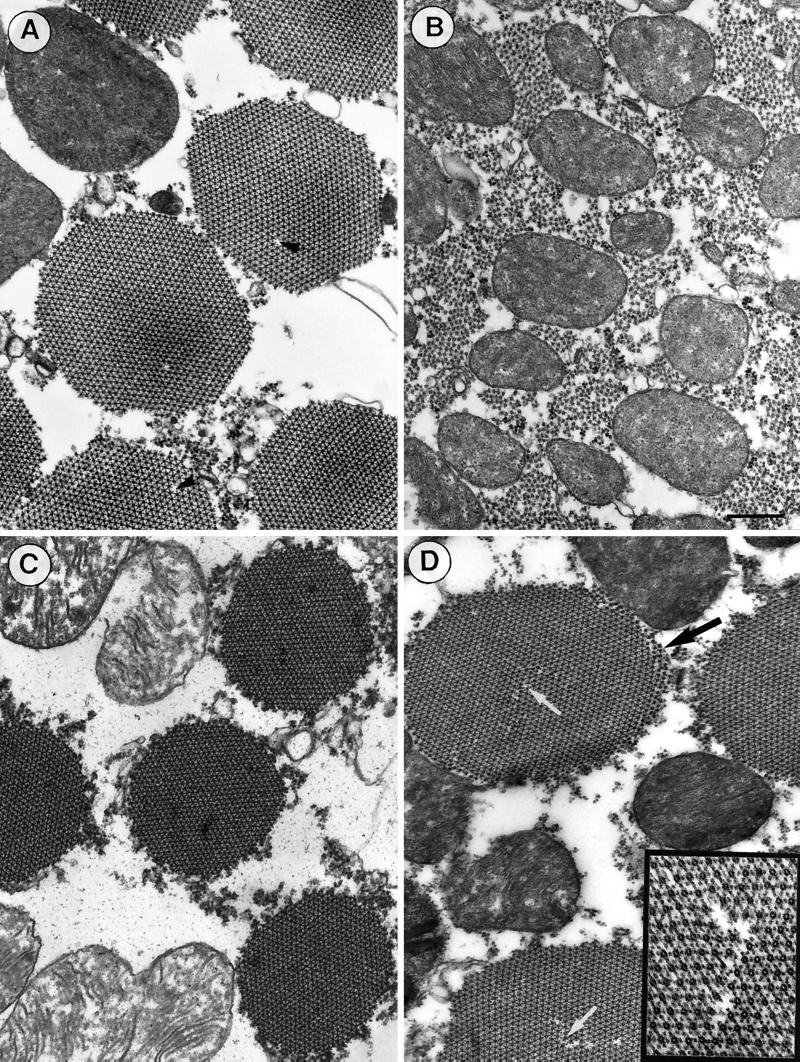
Ultrastructural Analysis of the IFM
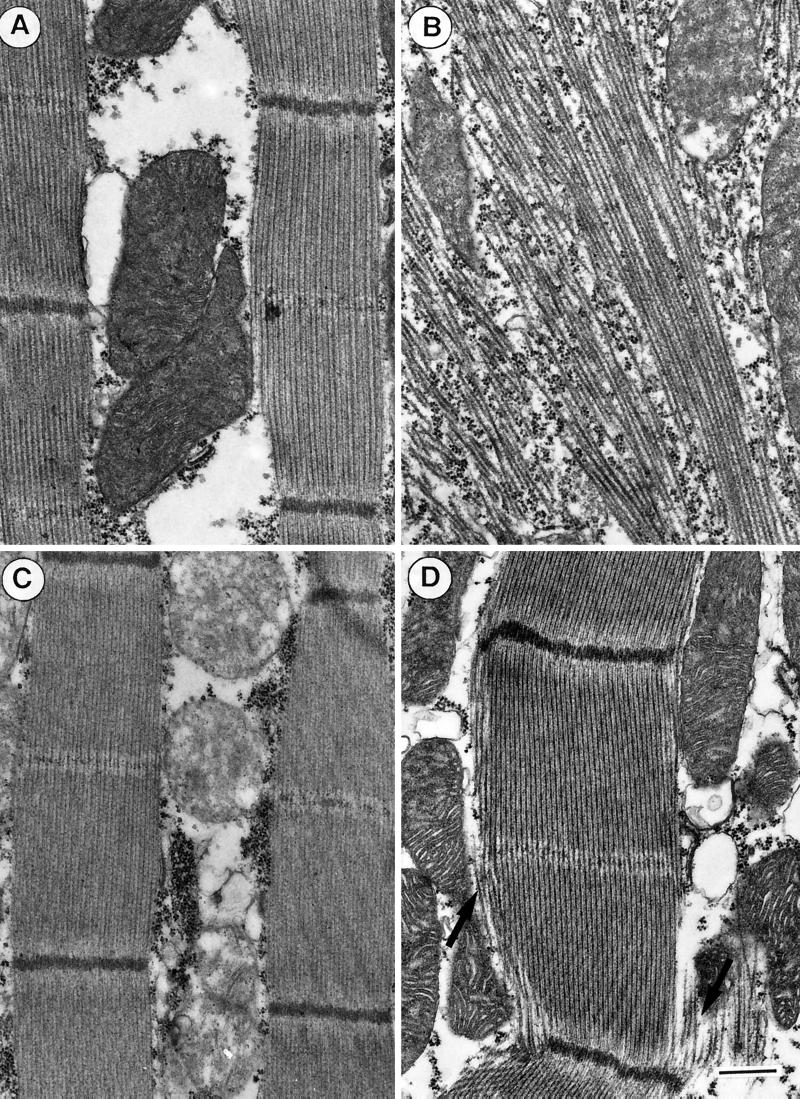
We have used transmission electron microscopy of embedded and thin-sectioned specimens to examine the consequences of epitope tagging on the IFM organization and morphology. For this purpose, the IFMs of adult flies (24–48 h after eclosion) were fixed in situ and embedded and sectioned as described in MATERIALS AND METHODS. Figures 4 and 5 display representative electron micrographs of longitudinal (Figure 4) and transverse (Figure 5) sections of the IFMs from C- and N-terminally tagged transformants in comparison with wild-type flies. In contrast to the well-organized sarcomeres of wild-type flies (Figure 4A), sarcomeric organization is virtually absent on longitudinal sections of C-terminally tagged Act88F flies (Figure 4B). Indications of lateral alignment of thick filaments may still occur in places. The randomly distributed thick filaments and the apparent lack of thin filaments and Z discs are reminiscent of the appearance of the KM88 null mutant IFM (Beall et al., 1989; our unpublished results). In contrast, in longitudinal sections the N-terminally tagged (i.e., with 6xHis or 11-mer) transformants (Figure 4, C and D) display a sarcomeric organization similar to that of wild-type flies. In the transformants expressing 6xHis Act88F (Figure 4C) as well as in those expressing 11-mer Act88F (Figure 4D), thin filaments alternating with thick filaments are evident. However, subtle morphological defects are depicted in the 11-mer Act88F transformants (Figure 4D). Along the periphery of the myofibrils, fraying of thick filaments (Figure 4D, arrows) is often discernible. The imperfections in the lateral register of thick filaments suggest the absence of thin filaments in these peripheral myofibril regions.
Figure 4.
Sarcomeric organization and myofibril morphology are preserved in N-terminally tagged transformants. (A) Electron micrograph of a longitudinal section of wild-type IFM showing the highly ordered sarcomeric organization with alternating thick and thin filaments, Z discs, and central M lines. (B) Longitudinal sections of C-terminally tagged transformants reveal only thick filaments that are randomly distributed. There is no apparent assembly of thin filaments. (C) IFM of N-terminally tagged 6xHis transformants displays myofibrils similar to those of wild-type IFM, with well-defined sarcomeric structures in which M lines and Z discs are clearly discernible. (D) In sarcomeres of N-terminally tagged 11-mer transformants, the register of alternating thick and thin filaments becomes slightly perturbed at the periphery (black arrows). Bar, 500 nm.
Figure 5.
Comparison of transverse sections from IFMs of wild-type and transformed flies. (A) In the wild-type IFM, myofibrils are round, uniform in diameter, and well ordered. They form hexagonal arrays of one thick myosin filament surrounded by six thin actin filaments. As marked by the black arrowheads, a thick filament is occasionally missing in an otherwise undisturbed filament lattice. (B) In transverse sections of C-terminally tagged transformants, there are no myofibrils assembled. As in cross-sections of the KM88 null mutant, thick filaments appear to be more or less randomly distributed and thin filaments are not evident. (C and D) Ordered myofibrillar structures are present in N-terminally tagged transformants. However, subtle effects of the tag on the structure are revealed. (D) In transformants expressing N-terminally 11-mer–tagged Act88F, the edges of the myofibrils appear frayed (black arrow). Occasionally, disturbances are detected in the hexagonal lattice of thick and thin filaments (white arrows). The inset reveals one of these disturbances at a twofold higher magnification. (C) On average, the myofibrils of transformants with an N-terminal 6xHis tag appear to have a slightly smaller (∼10%) diameter than myofibrils of wild-type IFM. Bar, 500 nm.
Cross-sections of wild-type flies (Figure 5A) and N-terminally tagged transformants (Figure 5, C and D) typically reveal myofibrils that are round and rather uniform in diameter. The myofibrils from flies with 6xHis-tagged Act88F (Figure 5C) appear slightly smaller (∼10%) in diameter than those of wild-type IFM and N-terminally 11-mer–tagged Act88F transformants. Just as with wild-type flies, the thin and thick filaments of both N-terminally tagged transformants are arranged in a highly regular hexagonal array. However, the myofibrils from flies expressing the 11-mer–tagged Act88F exhibit occasional disturbances in the hexagonal packing of thin and thick filaments (Figure 5D, white arrows and inset). These disturbances are clearly distinct from the sporadic missing of a thick filament in an otherwise undisturbed hexogonal myofilament lattice of wild-type flies (Figure 5A, black arrowheads; Sparrow et al., 1991). Moreover, lattice discontinuities at the periphery (Figure 5D, black arrows) are consistent with the fraying of thick filaments along the edge of myofibrils observed in longitudinal sections (Figure 4D, arrows). In contrast, no thin filaments or hexagonal packing of thick filaments were observed in cross-sections of the C-terminally tagged Act88F transformants, just randomly distributed thick filaments with no clear delineation into distinct myofibrils (Figure 5B).
In flies expressing C-terminally tagged Act88F, the complete absence of myofibril morphology, from the absence of thin filaments to the absence of sarcomeric organization, accounts for the flightless phenotype observed for the transformants. In contrast, IFMs of N-terminally tagged transformants exhibit a largely normal muscle morphology with well-organized sarcomeres and highly ordered myofibrils comprising thin and thick filaments. As a result, these transformants are able to fly, albeit slightly less efficiently than wild-type flies. It is conceivable that the morphological imperfections observed in the IFMs of N-terminally tagged transformants (as described above) are responsible for the reduction in flight ability.
The Position of the Tag Influences the Amount of Act88F Present in the IFM
To test whether the failure to form proper myofibrils in the C-terminally tagged Act88F transformants was correlated with reduced amounts of recombinant actin in the IFM, we compared the amounts of tagged Act88F in the IFMs of transformants with the amounts of endogenous Act88F in wild-type flies. For this purpose, protein extracts of the null mutant KM88, wild-type, and transgenic flies were analyzed by immunoblotting with a monoclonal anti-actin antibody that recognizes most actin isoforms (see MATERIALS AND METHODS). As documented in Figure 6, the blot probed with this antibody, which equally recognized endogenous Act88F in wild-type flies as well as tagged Act88F in transformants, revealed C-terminally tagged transformants to accumulate significantly less actin in their IFM than wild-type flies. In addition, the ubiquitinated actin species that migrates as a 55-kDa band in IFM extracts from wild-type flies is not detectable in IFM extracts from C-terminally tagged transformants. Because only every seventh Act88F molecule is ubiquitinated in wild-type flies (Ball et al., 1987), the amount of tagged arthrin in IFM extracts from C-terminally tagged transformants may well be below the limits of detection. However, overloading gels with IFM extracts to yield amounts of tagged Act88F equivalent to the amount of wild-type Act88F where arthrin is detectable did not result in the detection of tagged arthrin in the transformants (our unpublished results). Alternatively, the tag at the C terminus might interfere with the ubiquitination process.
Figure 6.
The position of the tag affects the level of Act88F expression. Immunoblotting of IFM extracts from KM88 null mutants, from wild-type flies, and from tagged Act88F transformants with a monoclonal antibody that recognizes different actin isoforms reveals that the amounts of C-terminally tagged actin are significantly lower than the amounts of Act88F in wild-type (WT) flies. Act88F with an N-terminal tag is expressed at levels similar to endogenous Act88F in wild-type IFM. N-terminally tagged arthrin (∼55 kDa) is also detected by the anti-actin antibody. Consistent with the immunoblot shown in Figure 2, the antibody recognizes multiple 11-mer–tagged Act88F and arthrin bands. The band in the IFM extract of KM88 represents non-IFM actin isoforms.
In contrast, the expression levels of the N-terminally tagged Act88F are similar to wild-type Act88F levels, and ubiquitin conjugation occurs in 6xHis– as well as 11-mer–tagged Act88F transformants. In the latter, the monoclonal anti-actin antibody recognized multiple bands, which correspond to the three tagged actin and the two tagged arthrin bands detected by the P5D4 mAb to the 11-mer epitope (Figure 2B, right). This result confirms that the individual bands represent variants of 11-mer–tagged recombinant actin. Somewhat unexpected, small amounts of actin (∼5% of the wild-type amount) were also detected in the KM88 null mutant. It has been suggested that cytoplasmic actin is a minor actin species in the IFM (Fyrberg et al., 1983). Although the band detected in extracts from KM88 could represent cytoplasmic actin from IFM, we believe it rather represents other Drosophila actin isoforms from surrounding muscle or nonmuscle tissue, especially because in the absence of a discernible myofibrillar structure in KM88, it is extremely difficult to exclusively dissect IFM.
DISCUSSION
In C-terminally Tagged Transformants Functional Thin Filaments Are Not Detectable
The low amount of C-terminally tagged Act88F present in the IFMs of corresponding transformants suggests that the thus modified actin is either not properly folded and/or cannot polmerize into actin-containing thin filaments, thereby becoming susceptible to degradation. The inability of actin to assemble into thin filaments, in turn, leads to the absence of sarcomeric organization and, ultimately, to the loss of flight ability. These results indicate that the actin conformation and/or polymerization is sensitive to certain modifications of its C terminus. A number of experiments involving deletions or mutations of the C-terminal sequence have demonstrated the importance of this region for proper F-actin polymerization and stability. For example, mutating the conserved cysteine at position 374 to a serine in the chicken β-cytoplasmic actin increased the critical concentration for polymerization by more than fivefold (Aspenström et al., 1993). Removal of either the two or three C-terminal residues of actin resulted in actin filaments with increased fragility and flexibility (O’Donoghue et al., 1992; Mossakowska et al., 1993).
Interestingly, other modifications of the C terminus such as labeling of Cys-374 with fluorescent probes (Kouyama and Mihashi, 1981; Cooper et al., 1983; for review see Miki et al., 1992) or undecagold (Milligan et al., 1990) do not significantly interfere with the polymerization characteristics of actin or the filament structure as predicted by the Holmes model (Holmes et al., 1990; Lorenz et al., 1993). Likewise, it has been reported that addition of the VSV-G 11-mer epitope at the C terminus of muscle as well as nonmuscle actin isoforms did not impede their in vitro copolymerization with rabbit skeletal muscle actin (von Arx et al., 1995; Mounier et al., 1997). After transfection, coassembly or association of recombinantly expressed tagged actin isoforms with the preexisting microfilament system was observed for a variety of cell types. It should be noted, however, that recombinant actin usually amounts to <10% of the total actin in transfected cells. Thus, it is conceivable that the excessive amounts of endogenous actin mask the effective properties of the less abundant recombinant actin.
The Heidelberg model of the F-actin filament, which has evolved from the atomic structure of the monomeric actin–DNaseI complex (Kabsch et al., 1990), in combination with x-ray fiber diffraction data (Holmes et al., 1990; Lorenz et al., 1993) appears largely consis-tent with the extensive biochemical data on actin at hand. However, a significantly different model has been proposed by Schutt et al. (1995a,b, 1997) underscoring that the ultimate structure of the actin filament at atomic scale has remained elusive. In particular, there are some uncertainties regarding the highly mobile N terminus and the C-terminal helix (residues 368–375). Because the definite structure in the filament of precisely those regions of the molecule that are modified by the tags is unknown, predictions on the structural consequences of the tag are subject to speculation. In fact, only in rare instances have the consequences on filament structure caused by the various mutations in actin been analyzed (Orlova et al., 1997). Nevertheless, based on the predicted location of the C terminus in the vicinity of the interface between the two long-pitch helical strands, it appears reasonable to assume that modifications in this region somehow affect subunit–subunit interactions. Both the 6xHis and the 11-mer tags are significantly larger than pyrene, N-iodoacetyl-N′-(5-sulfo-1-naphtyl)ethylene-diamine or even undecagold and, therefore, in contrast to these modifications, may interfere with the proper interaction of the two long-pitch helical strands of the F-actin filament so that no stable filaments are formed, and then, in an in vivo environment, the tagged actin is rapidly degraded. The low levels of C-terminally tagged Act88F observed in our transformants are consistent with this hypothesis. According to the Holmes model of the F-actin filament, there is sufficient space at the C terminus in the Holmes model to sterically accommodate the 6 histidines or the 11 amino acids of the 11-mer tag without causing a major structural change of the filament. However, it has been shown that conformational coupling of C-terminal modifications with more distant sites does exist for both the monomer and polymer forms of actin (Drummond et al., 1992; Crosbie et al., 1994; Moraczewska et al., 1996). Hence, addition of the tags to the C terminus of Act88F might affect the F-actin structure and/or conformation. Such structural changes might have an effect on the interaction of the C terminus with actin-binding proteins so that ultimately the function of the F-actin is modified.
Ubiquitination of C-terminally Tagged Act88F Actin Does Not Occur in Homozygous Transformants
The apparent absence of ubiquitinated actin in C-terminally tagged homozygotes (see Figures 2A and 6) suggested that the modification of the C terminus interfered with IFM-specific ubiquitination. However, in heterozygotes with one copy of the C-terminally tagged Act88F gene and one wild-type Act88F gene, we have observed thin filaments in the IFM. In extracts of these IFMs, a ubiquitin conjugate was detected by antibodies recognizing the tag (our unpublished data), indicating that interference of the C-terminal tag with ubiquitination is an unlikely explanation for the absence of tagged arthrin in homozygous transformants. Ubiquitination lags several hours behind Act88F expression and thus parallels myofibril formation (Ball et al., 1987). It is conceivable that actin ubiquitination might require or be regulated by thin filament formation. Our findings support this notion, because in the IFM of homozygous transformants, C-terminally tagged Act88F is unable to form thin filaments, and hence, ubiquitination does not occur.
Tagging the N Terminus of Act88F Does Not Significantly Interfere with Myofibrillar Structure and Function
Evidently, the amino acid sequence at the N terminus of actin is crucial for its structural and functional integrity (Cook et al., 1991, 1992; Reedy et al., 1991; Aspenström et al., 1992; Miller et al., 1996). Thus, it was surprising that the N-terminal fusion of the relatively large green fluorescent protein to Dictyostelium actin did not prevent the hybrid protein from copolymerizing with endogenous actin (Westphal et al., 1997). Likewise, transformants exclusively expressing N-terminally tagged Act88F in their IFMs exhibit a virtually normal sarcomeric organization with alternating thin and thick filaments and are able to fly, although not as proficiently as wild-type flies. These results demonstrate that N-terminally tagged actin is by itself polymerization competent, and that the resulting thin filaments are at least partially functional in a flight test.
Consistent with the compromised function of the corresponding IFM, subtle effects on the morphological phenotype became apparent at the ultrastructural level. In IFMs of 11-mer–tagged Act88F flies, occasional disruptions of the hexagonal myofibril array were observed. A similar phenotype has been described in transformants with the single point mutation E316K in Act88F (Drummond et al., 1990). Unlike the N-terminal tags, this mutation involving a glutamic acid to lysine change at position 316 abolished flight ability. More specifically, it was found to alter cross-bridge kinetics, although it is distant from the nearest known myosin or tropomyosin contact. As discussed above in the context of the C-terminal modifications, this mutation may be affecting the interaction of actin with myosin through a long-range conformational change within the F-actin polymer. Moreover, in Drosophila expressing mutated myosin, defects in the actomyosin cross-bridges have been described to account for disruptions of the hexagonal packing of thin and thick filaments in the IFM (Mogami and Hotta, 1981; Warmke et al., 1992).
Tags at the N Terminus of Actin May Influence Actin–Myosin Interactions
Over the past few years, significant progress has been made on mapping the binding sites on the F-actin filament of a number of actin-binding proteins through the combination of maps obtained by electron microscopy with atomic structures determined by x-ray diffraction and structural nuclear magnetic resonance (for review see McGough, 1998, and references therein). These studies emphasized the importance of the filament geometry (i.e., the packing of actin subunits in the helix) and of conformational flexibility in defining the molecular interactions between actin and actin-binding proteins (Chik et al., 1996; McGough et al., 1997). In most cases, actin subdomain 1, where both the N- and C termini reside, contributed to the respective binding site. At the C terminus, residues from ∼340 to 355 appear to be involved in a number of binding sites (Rayment et al., 1993; McGough et al., 1994, 1997; Schmid et al., 1994; Hanein et al., 1997). In light of these findings, the absence of thin filaments in the IFMs of C-terminally tagged transformants may be explained by conformational changes brought about by the tags such that interactions with actin-binding proteins, which are relevant for myofibril assembly in the IFM, are altered or can no longer occur. Alternatively, the tags may reduce the mobility of the C terminus, which is required to substantially move during filament formation (Milligan, 1996).
Proper actin–myosin interactions are thought to be important for the correct registration of thick and thin filaments within the sarcomere (Reedy and Beall, 1993). If so, alterations in cross-bridge formation attributable to actin or myosin mutations would be expected to cause aberrant myofibrillar assembly. At the N terminus of rabbit skeletal actin, the six negatively charged amino acids, Asp1, Glu2, Asp3, Glu4, Asp24, and Asp25, are getting in close contact with the myosin loop Tyr626 to Gln647 during the cross-bridge cycle and are believed to be predominantly involved in ionic interactions with myosin beside the primary myosin binding site involving the helix-loop-helix near the C terminus of actin (Ile341 to Gln354), a loop between Ala144 and Thr148 on the same actin monomer, and part of the DNase I binding loop (His40-Gly42) on an adjacent actin monomer (for review, see Milligan, 1996). Modification of the charge environment at the N terminus by either tag could affect these ionic interactions and interfere with the normal cross-bridge formation. Moreover, it has been shown that mutations in the N-terminal region of actin, which yielded a change of charge, affect myosin S1 binding to actin and thereby reduce in vitro motility (Aspenström and Karlsson, 1991; Aspenström et al., 1992; Sutoh et al., 1991; Miller et al., 1996). However, N-terminal mutations that do not induce charge changes may also affect actomyosin interactions. For example, in the Act88FG6AA7T double mutant, peripheral thin and thick filaments are out of register. Moreover, the unincorporated thin filaments at the periphery point in the opposite direction, as indicated by the reversed orientation of myosin chevrons (Reedy et al., 1989). Like the N-terminally tagged Act88F transformants, these mutants are flightless despite a core of precisely interdigitated thin and thick filaments.
It is conceivable that the extra charge introduced by the six histidines and/or the structural changes arising from the additional residues in the N-terminally tagged transformants also affect the actomyosin interactions. The unincorporated thick filaments at the periphery of the myofibrils support this hypothesis. However, these rather minor ultrastructural defects and the flight ability in particular argue against a severe effect of these two N-terminal tags on the actomyosin interaction. Future experiments should provide insight into the structural details of the actomyosin rigor complex.
Alternatively, the thin–thick filament lattice disturbances observed in the IFM (see Figures 4 and 5) could result from an imbalance of the actin-to-myosin stoichiometry. Consistent with an imbalance between thin and thick filaments (Beall et al., 1989; Cripps et al., 1994), we have observed fraying of their hexagonal packing with unintegrated thick filaments only residing at the periphery of myofibrils.
The accumulation of N- and C-terminally tagged Act88F in the IFM differs drastically. Comparison of the respective phenotypes strongly suggests that the amount of actin plays an important role in myofibril assembly and/or organization. Studies on Act88F mutations that yield a reduced amount of actin over myosin support the notion that reduced accumulation of actin in the IFM might produce structural and functional myofibrillar defects. For example, in the point mutant V339I (Drummond et al., 1991), the monomeric actin conformation appears largely unaltered; nevertheless, the mutant flies display a very disrupted IFM structure and functionally are flightless, a phenotype that has been related to reduced amounts of the mutant actin relative to myosin. A number of experiments provide evidence that an imbalance of the ratio between thin and thick filaments rather than the absolute deficit of one of these component appears to be responsible for the myofibrillar defects observed (Beall et al., 1989). Hence, it is conceivable that the subtle morphological defects observed in the N-terminally tagged Act88F transformants are possibly due to a small imbalance of tagged Act88F over myosin. Although immunoblot experiments indicate that expression of N-terminally tagged Act88F and wild-type Act88F is similar, small variations in expression that go undetected by this technique might nevertheless have definite structural and functional consequences.
Several myofibrillar protein heterozygous null mutants with only one copy of the normal gene exhibit out-of-register myofilaments at the periphery of the myofibrils, similar to those seen in the homozygous N-terminally tagged Act88F transformants. For example, heterozygotes for the KM88 null mutation, which are flightless, display myofibrils with a core of hexagonally packed thin and thick filaments surrounded by unintegrated thick filaments (Beall et al., 1989). We observed a corresponding phenotype in heterozygotes with one wild-type and one C-terminally tagged Act88F gene (our unpublished results). The core of hexagonally packed thin and thick filaments could be conceived as myofibril with a smaller diameter. For comparison, myofibrils of transformants homozygous for 6xHis-tagged Act88F also had a slightly smaller diameter than the IFM myofibrils of wild-type flies but were still able to fly.
The IFM provides a unique experimental system to assay both qualitatively and quantitatively the effects of modifying Act88F actin. In the absence of endogenous protein, specifically tagged Act88F actin can be tested at increasing levels of stringency ranging from specific protein accumulation to quantitative assessment of rescuing flight. Further analysis of the consequences of these tags on actin polymerization and filament structure will require physicochemical studies with purified proteins. Such experiments are now feasible with the use of affinity purification procedures based on the metal-binding properties of the polyhistidine tags.
ACKNOWLEDGMENTS
We thank Professor Jean-Claude Perriard (Swiss Federal Institute of Technology, Zürich, Switzerland) for providing the α-cardiac actin 11-mer construct. The P5D4 antibody was a kind gift from the late Professor Thomas Kreis (University of Geneva, Geneva, Switzerland). We are grateful to Jon Clayton and Belinda Bullard (European Molecular Biology Laboratory, Heidelberg, Germany) for teaching us how to dissect IFM for immunoblot analysis. We are also indebted to the members of Professor Walter J. Gehring’s laboratory (Biozentrum) for the use of their equipment and for their advice on all work involving flies. Last but not least, we thank Dr. Bernhard Heymann, Dr. Richard Kammerer (both Biozentrum), and Prof. Henry F. Epstein (Baylor College of Medicine, Houston, TX) for fruitful discussions and constructive comments on the manuscript. This work was supported by the Swiss National Science Foundation, the Canton Basel-Stadt, and the M.E. Müller Foundation of Switzerland.
Abbreviations used:
- F-actin
filamentous actin
- 6xHis
6x histidine
- IFM
indirect flight muscle
- UTR
untranslated region
- VSV-G
vesicular stomatitis virus G protein
REFERENCES
- Aspenström P, Karlsson R. Interference with myosin subfragment-1 binding by site directed mutagenesis. Eur J Biochem. 1991;200:35–41. doi: 10.1111/j.1432-1033.1991.tb21045.x. [DOI] [PubMed] [Google Scholar]
- Aspenström P, Lindberg U, Karlsson R. Site-specific amino-terminal mutants of yeast-expressed β-actin. FEBS Lett. 1992;303:59–63. doi: 10.1016/0014-5793(92)80477-x. [DOI] [PubMed] [Google Scholar]
- Aspenström P, Schutt C, Lindberg U, Karlsson R. Mutations in β-actin: influence on polymer formation and on interactions with myosin and profilin. FEBS Lett. 1993;1:163–170. doi: 10.1016/0014-5793(93)80215-g. [DOI] [PubMed] [Google Scholar]
- Ball E, Karlik CC, Beall CJ, Saville DL, Sparrow JC, Bullard B, Fyrberg EA. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell. 1987;51:221–228. doi: 10.1016/0092-8674(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Beall CJ, Sepanski MA, Fyrberg EA. Genetic dissection of Drosophila myofibril formation. Genes & Dev. 1989;3:131–140. doi: 10.1101/gad.3.2.131. [DOI] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE. The structure of an open state of β-actin at 2.65 Å resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Collucio LM, Tilney LG. Phalloidin enhances actin assembly by preventing monomer dissociation. J Cell Biol. 1984;99:529–535. doi: 10.1083/jcb.99.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Blake WT, Rubenstein PA. Removal of the amino-terminal acidic residues of yeast actin. J Biol Chem. 1992;267:9430–9436. [PubMed] [Google Scholar]
- Cook RK, Sheff DR, Rubenstein PA. Unusual metabolism of the yeast actin amino terminus. J Biol Chem. 1991;266:16825–16833. [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Becker KD, Mardahl M, Kronert WA, Hodges D, Bernstein SI. Transformation of Drosophila melanogaster with the wild-type myosin heavy-chain gene: rescue of mutant phenotypes and analysis of defects caused by overexpression. J Cell Biol. 1994;126:689–699. doi: 10.1083/jcb.126.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosbie RH, Miller C, Cheung P, Goodnight T, Muhlrad A, Reisler E. Structural connectivity in actin: effects of C-terminal modifications on the properties of actin. Biophys J. 1994;67:1957–1964. doi: 10.1016/S0006-3495(94)80678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. Characterization of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. The binding of mutant actins to profilin, ATP and DNase I. Eur J Biochem. 1992;209:171–179. doi: 10.1111/j.1432-1033.1992.tb17274.x. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Peckham M, Sparrow JC, White DCS. Alteration in cross-bridge kinetics caused by mutations in actin. Nature. 1990;348:440–442. doi: 10.1038/348440a0. [DOI] [PubMed] [Google Scholar]
- Estes JE, Selden LA, Gershman LC. Mechanism of action of phalloidin on the polymerization of muscle actin. Biochemistry. 1981;20:708–712. doi: 10.1021/bi00507a006. [DOI] [PubMed] [Google Scholar]
- Fyrberg E, Kelly M, Ball E, Fyrberg C, Reedy MC. Molecular genetics of Drosophila α-actinin: mutant alleles disrupt Z disk integrity and muscle insertions. J Cell Biol. 1990;110:1999–2011. doi: 10.1083/jcb.110.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg EA, Mahaffey JW, Bond J, Davidson N. Transcript of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 1983;33:115–123. doi: 10.1016/0092-8674(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Gimona M, Vandekerckhove J, Goethals M, Herzog M, Lando Z, Small JV. β-actin specific monoclonal antibody. Cell Motil Cytoskeleton. 1994;27:108–116. doi: 10.1002/cm.970270203. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matsudaira P, DeRosier DJ. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y, Hotta Y. Actin gene mutations in Drosophila; heat shock activation in the indirect flight muscles. EMBO J. 1985;4:1681–1687. doi: 10.1002/j.1460-2075.1985.tb03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y, Okamoto H, Gehring WJ, Hotta Y. Germline transformation with Drosophila mutant actin genes induces constitutive expression of heat shock genes. Cell. 1986;44:293–301. doi: 10.1016/0092-8674(86)90763-4. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of actin: DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kaech S, Fischer M, Doll T, Matus A. Isoform specificity in the relationship of actin to dendritic spines. J Neurosci. 1997;24:9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik CC, Saville DL, Fyrberg EA. Two missense alleles of the Drosophila melanogaster act88F actin gene are strongly antimorphic but only weakly induce synthesis of heat shock proteins. Mol Cell Biol. 1987;7:3084–3091. doi: 10.1128/mcb.7.9.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R, Weber U, Gehring WJ. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodo-acetamide-labeled F-actin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The Genome of Drosophila. San Diego: Academic Press; 1992. [Google Scholar]
- Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against x-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- Lubit BW, Schwartz JH. An antiactin antibody that distinguishes between cytoplasmic and skeletal muscle actin. J Cell Biol. 1980;86:891–897. doi: 10.1083/jcb.86.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A. F-actin-binding proteins. Curr Opion Struct Biol. 1998;8:166–176. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Way M, DeRosier D. Determination of the a-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki M, O’Donoghue SI, Dos Remedios CG. Structure of actin observed by fluorescence resonance energy transfer spectroscopy. J Muscle Res Cell Motil. 1992;13:132–145. doi: 10.1007/BF01874150. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Wong WW, Bobkova E, Rubenstein PA, Reisler E. Mutational analysis of the role of the N terminus of actin in actomyosin interactions. Comparison with other mutant actins and implications for the cross-bridge cycle. Biochemistry. 1996;35:16557–16565. doi: 10.1021/bi962388+. [DOI] [PubMed] [Google Scholar]
- Milligan RA. Protein-protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci USA. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan RA, Whittaker M, Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- Mogami K, Hotta Y. Isolation of Drosophila flightless mutants which affect myofibrillar proteins of indirect flight muscles. Mol Gen Genet. 1981;183:409–417. doi: 10.1007/BF00268758. [DOI] [PubMed] [Google Scholar]
- Moraczewska J, Strzelecka-Golaszewska H, Moens PDJ, Dos Remedios CG. Structural changes in subdomain 2 of G-actin observed by fluorescence spectroscopy. Biochem J. 1996;317:605–611. doi: 10.1042/bj3170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossakowska M, Moraczewska J, Khaitlina S, Strzelecka-Golaszewska H. Proteolitic removal of three C-terminal residues of actin alters the monomer-monomer interactions. Biochem J. 1993;289:897–902. doi: 10.1042/bj2890897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier N, Perriard J-C, Gabbiani G, Chaponnier C. Transfected muscle and nonmuscle actins are differentially sorted by cultured smooth muscle and nonmuscle cells. J Cell Sci. 1997;110:839–846. doi: 10.1242/jcs.110.7.839. [DOI] [PubMed] [Google Scholar]
- O’Donoghue SI, Miki M, dos Remedios CG. Removing the two C-terminal residues of actin affects the filament structure. Arch Biochem Biophys. 1992;293:110–116. doi: 10.1016/0003-9861(92)90372-4. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Hiromi Y, Ishikawa E, Yamada T, Isoda K, Maekawa H, Hotta Y. Molecular characterization of mutant actin genes which induce heat-shock proteins in Drosophila flight muscles. EMBO J. 1986;5:589–596. doi: 10.1002/j.1460-2075.1986.tb04251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A, Chen X, Rubenstein PA, Egelman ED. Modulation of yeast F-actin structure by a mutation in the nucleotide-binding cleft. J Mol Biol. 1997;271:235–243. doi: 10.1006/jmbi.1997.1163. [DOI] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn M, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C. Ultrastructure of developing flight muscle in Drosophila. I. Assembly of myofibrils. Dev Biol. 1993;160:443–465. doi: 10.1006/dbio.1993.1320. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C, Fyrberg E. Formation of reverse rigor chevrons by myosin heads. Nature. 1989;339:481–483. doi: 10.1038/339481a0. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C, Fyrberg E. Do variant residues among the six actin isoforms of Drosophila reflect functional specialization? Biophys J. 1991;59:187a. (abstract). [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sakai U, Okamoto H, Mogami K, Yamada T, Hotta Y. Actin with tumor-related mutation is antimorphic in Drosophila muscle: two distinct modes of myofibrillar disruption by antimorphic actins. J Biochem. 1990;107:499–505. doi: 10.1093/oxfordjournals.jbchem.a123074. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Mittal B, Sanger JM. Analysis of myofibrillar structure and assembly using fluorescently-labeled contractile proteins. J Cell Biol. 1984;98:825–833. doi: 10.1083/jcb.98.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Agris J, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-β motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt CE, Kreatsoulas C, Page R, Lindberg U. Plugging into actin’s architectonic socket. Nature Struct Biol. 1997;4:169–172. doi: 10.1038/nsb0397-169. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Rozycki MD, Chik J, Lindberg U. Structural studies on the ribbon-to-helix transition in profilin:actin crystals. Biophys J. 1995b;68:12s–18s. [PMC free article] [PubMed] [Google Scholar]
- Schutt CE, Rozycki MD, Myslik JC. A discourse on modelling F-actin. J Struct Biol. 1995a;115:186–198. doi: 10.1006/jsbi.1995.1043. [DOI] [PubMed] [Google Scholar]
- Skalli O, Ropraz P, Trzeciak A, Benzonana G, Gillessen D, Gabbiani G. A monoclonal antibody against α-smooth muscle actin: a new probe for smooth muscle differentiation. J Cell Biol. 1986;103:2787–2796. doi: 10.1083/jcb.103.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T, Perriard JC. Intracompartmental sorting of essential myosin light chains: molecular dissection and in vivo monitoring by epitope tagging. Cell. 1991;66:277–289. doi: 10.1016/0092-8674(91)90618-9. [DOI] [PubMed] [Google Scholar]
- Sparrow J, Reedy M, Ball E, Kyrtatas V, Molloy J, Durston J, Hennessey E, White D. Functional and ultrastructural effects of a missense mutation in the indirect flight muscle-specific actin gene of Drosophila melanogaster. J Mol Biol. 1991;222:963–982. doi: 10.1016/0022-2836(91)90588-w. [DOI] [PubMed] [Google Scholar]
- Sutoh K, Ando M, Toyoshima YY. Site-directed mutations of Dictyostelium actin: disruption of a negative charge cluster at the N-terminus. Proc Natl Acad Sci USA. 1991;88:7711–7714. doi: 10.1073/pnas.88.17.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arx P, Bantle S, Soldati T, Perriard JC. Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke J, Yamakawa M, Molloy J, Falkenthal S, Maughan D. Myosin light chain-2 mutation affects flight, wing beat frequency, and indirect flight muscle contraction kinetics in Drosophila. J Cell Biol. 1992;119(6):1523–1539. doi: 10.1083/jcb.119.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Mülbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Zentgraf H, Frey M, Schwinn S. Detection of histidine-tagged fusion proteins by using a high-specific mouse monoclonal antihistidine tag antibody. Nucleic Acids Res. 1995;16:3347–3348. doi: 10.1093/nar/23.16.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]