Abstract
This study investigated the possible association of the interleukin-1 beta (IL-1β) C-511T promoter polymorphism and the interleukin-1 receptor antagonist (IL-1Ra) (86bp)n variable number of tandem repeats (VNTR) polymorphism with antidepressant response to paroxetine and mirtazapine treatment. The study group consisted of 101 patients suffering from DSM-IV major depression participating in a randomized double-blind controlled clinical trial. Patients homozygous for the IL-1β-511T allele had a significantly faster and more pronounced response to paroxetine treatment than IL-1β-511C allele carriers. No association was found for the IL-1β C-511T polymorphism with mirtazapine treatment response. The IL-1Ra VNTR showed neither an association with paroxetine nor with mirtazapine treatment response. Our results provide further suggestive evidence that time course of response and antidepressant efficacy of paroxetine, but not of mirtazapine, is influenced in a clinically relevant manner by the IL-1β C-511T gene variant. Our data do not support the hypothesis that the IL-1Ra (86bp)n VNTR affects antidepressant treatment response to paroxetine or mirtazapine. An independent replication of our finding is needed. If replicated, the IL-1β C-511T promoter polymorphism could be considered useful for prospective confirmatory pharmacogenetic trials in patients with major depression.
Keywords: major depression, antidepressive agents, treatment outcome, interleukin-1 beta, interleukin-1 receptor antagonist, genetic polymorphisms
Introduction
A growing body of evidence supports the hypothesis that cytokine activity is involved in the etiology and pathophysiology of behavioral and somatic symptoms of major depression (MD) (Hayley et al 2005; Raison et al 2006). Among the cytokines, animal and clinical studies have demonstrated that interleukin 1beta (IL-1β) may have a key role in major depression and in the effects of antidepressant treatment (Licinio and Wong 1999; Kenis and Maes 2002). In MD patients, IL-1β levels have been investigated under several experimental conditions. Some of the results showed an increase in the blood or cerebrospinal fluid (CSF) of depressed patients (Levine et al 1999; Anisman et al 1999; Owen et al 2001; Thomas et al 2005). It was also reported that CSF IL-1β levels were correlated with depression subtype (Kaestner et al 2005), depression severity (Levine et al 1999), age of onset, and duration of illness (Anisman et al 1999). However, other studies have found that the IL-1β levels were unchanged (Brambilla and Maggioni 1998; Kagaya et al 2001), and not correlated with the severity of depression (Rothermund et al 2001). To elucidate the complex neurobiological effects of IL-1 in brain, several investigations have been performed in animal models. Systemic administration of IL-1 in rats increased dopamine (DA), norepinephrine (NE), and serotonin (5-HT) activities in hypothalamic nuclei, nucleus accumbens, and limbic regions including the hippocampus (Merali et al 1997; Anisman and Merali 1999; Dunn et al 1999; Song et al 1999). Increases in hippocampal extracellular 5-HT levels have also been found following intracerebroventricular (i.c.v.) administration of IL-1β in rats (Linthorst et al 1995). IL-1β has been shown to up-regulate hippocampal expression of the 5-HT transporter (Ramamoorthy et al 1995) and to activate neuronal 5-HT (Zhu et al 2006).
The action of IL-1β is regulated by a complex network of molecules including the anti-inflammmtory interleukin-1 receptor antagonist (IL-1Ra) (Anisman 2004; Licinio et al 2007), which represents a naturally occuring endogenous antagonist of IL-1 actions. Several studies support a neuroprotective function of IL-1Ra (Licinio and Wong 1999), which is constitutively expressed in brain (Licinio et al 2007). Additionally, i.c.v. administrated IL-1Ra prevented the development of behavioral deficits in the learned helplessness model of depression (Maier and Watkins 1995). In patients with MD, IL-1Ra serum levels were found to be elevated compared to normal controls (Kubera et al 2000).
Several studies addressed the effects of antidepressive treamtent on cytokine secretion and plasma levels in depressed patients, indicating a normalization of cytokine plasma levels and cytokine production after antidepressant treatment (Castanon et al 2002; Kenis and Maes 2002). However, in brain the chronic administration of antidepressants led to an increased expression of IL-1Ra mRNA and IL-1β mRNA (Suzuki et al 1996).
Taking into account the existing evidence for the close relation between IL-1β, IL-1Ra and MD, the genes encoding IL-1β and IL-1Ra can be considered candidate genes for antidepressant treatment response in MD. Both genes map on chromosome 2 in the region q13-q21 (Nicklin et al 1994). A single nucleotide polymorphism (C/T) has been identified in the 5′- regulatory region (position -511) of the IL-1β gene (di Giovine et al 1992). Pociot and colleagues (1992) demonstrated that individuals homozygous for the IL-1β-511T allele secreted significantly more IL-1β than heterozygous individuals, who secreted significantly more than IL-1β-511C homozygous individuals. Intron 2 of the IL-1Ra gene contains a penta-allelic variable number of tandem repeats (VNTR) of a 86bp sequence (five alleles comprising between two and six repeats). In the general population, the IL-1Ra allele 1 (86bp)4 is more common (0.74) than allele 2 (86bp)2 (0.21); the remaining alleles occur at a combined frequency of less than 5% (Tarlow et al 1993). Functionally, allele 2 was associated with an increased plasma level of IL-1Ra in Finnish healthy volunteers (Hurme and Santtila 1998).
Paroxetine is widely and successfully used in the treatment of MD. Like all SSRI, it exerts its antidepressant effects mainly through inhibition of the serotonin transporter (5-HTT). Yu and colleagues (2003) reported that depressed patients homozygous for the -511T allele had a more favorable fluoxetine treatment response than -511C allele carriers, so that one could hypothesize a similar effect in our sample of paroxetine-treated patients. Mirtazapine is an antidepressant that is believed to antagonize central α2-receptors located on both noradrenergic and serotonergic neurons. As a result, mirtazapine enhances both the neurotransmission of 5-HT and NE. Additionally, mirtazapine inhibits 5-HT2A, 5-HT2C and 5-HT3 receptors, thereby focusing the enhanced serotonergic neurotransmission on 5-HT1A receptors (Anttila and Leinonen 2001). To our knowledge, there is no previous report on an investigation of IL-1 gene complex variants and mirtazapine treatment response.
The aim of the present study, which is a part of a more comprehensive clinical genetics project on treatment response in depression (Szegedi et al 2005; Tadić et al 2007a, 2007b), was to investigate the possible effect of the IL-1β C-511T and IL-1Ra (86bp)n intron 2 polymorphism on the outcome of antidepressant response to paroxetine and mirtazapine treatment in patients with MD.
Methodology
Patients
A total of 269 outpatients were enrolled in a multicenter, randomized, double-blind comparison of paroxetine and mirtazapine conducted at 50 centers in Germany. Of these patients, 116 gave written informed consent for the collection of blood samples for DNA genotyping and the analyses of possible associations between genotypes and clinical data (eg, response to treatment). All study components have been approved by the local ethics committees.
A more detailed description of the recruitment procedure, treatment schedule, and clinical assessments has been reported in a previous paper (Benkert et al 2000). Briefly, females and males, aged from 18 to 70 years suffering from acute major depressive episode (MDE) with a sum score ≥18 points on the 17-item version of the Hamilton Rating Scale for Depression (HAMD-17) at the start and end of a placebo washout period were eligible for the study. Diagnosis of MDE according to DSM-IV criteria was assigned by eleven clinically experienced research assistants, who underwent several rater trainings prior to the start of the study. Exclusion criteria were the lack of response to at least two adequate antidepressant treatments during the current depressive episode, current episode’s duration more than twelve months, more than three previous episodes that did not respond to adequate treatment, a reduction of ≥25% in the HAMD-17 sum score during placebo washout period, suicide risk defined as a score offour to six on item ten of the Montgomery-Åsberg-Depression-Rating-Scale (MADRS), history of bipolar affective disorder, depressive disorder not otherwise specified, panic disorder (with or without agoraphobia), agoraphobia without a history of panic disorder, schizophrenia, organic mental disorder, eating disorder, specific phobia, social phobia, or generalized anxiety disorder.
After a 3- to 7-day washout period, patients were randomly assigned to either mirtazapine or paroxetine treatment for six weeks. Mirtazapine was first administered at 15 mg (days 1 and 2), from day 3 onward at 30 mg/day; after 2 weeks, an increased dose of 45 mg/day was given to nonresponders, defined by Clinical Global Impressions scale (CGI) ratings in the therapeutic effect item of “slight” or “unchanged/worsened” and no “outweighs therapeutic efficacy” ratings in the tolerability item. Paroxetine dose was 20 mg/day and could be increased to 40 mg/day after 2 weeks in nonresponders according to the same pre-specified CGI criterion. No concomitant psychotropic treatment was allowed during the study. All patients were assessed at baseline (day 0), and on days 7, 14, 21, 28, and 42 of active treatment using the HAMD-17.
DNA analysis
Genotyping was performed blind to treatment and clinical course of the illness. Genomic DNA was prepared from 10 ml blood using the Quiagen Maxi DNA Extraction Kit (Quiagen, Hilden, Germany). Genotyping of the C-511T variant of the IL-1β gene (dbSNP: rs3087258) was performed using a polymerase chain reaction (PCR) – based method (Hurme and Santtila 1998). A 304-bp PCR fragment of the IL-1β promoter region was amplified using the primer pair 5′-TGGCATTGATCTGGTTCATC-3′ and 5′-GTTTAGGAATCTTCCCACTT-3′. Reaction was performed using 50 ng genomic DNA, 20 pmol of each primer (Invitrogen), 1 U Taq polymerase (Fermentas), 0.1 mM dNTP (Fermentas), 15 mM MgCl2, 60 mM Tris-HCl und 15 mM ammoniumsulfate. PCR conditions were as follows: 95 °C for 2 min, 55 °C for 1 min, 74 °C for 1 min, then 38 cycles of 95 °C for 1 min, 55 °C for 1 min, 74 °C for 1 min and finally 74 °C for 4 min. The products were digested with 6 U of AvaI at 37 °C for 3h. Fragments were analyzed by electrophoresis and stained with ethidium bromide. This gave products that either remained intact (IL-1β -511T allele; allele 2) or were cut into two fragments of 190 and 114 bp (IL1β -511C allele; allele 1). For the analysis of the pentaallelic VNTR within intron 2 of the IL-1Ra gene (Tarlow et al 1993), oligonucleotides 5′-TGGCATTGATCTGGTTCATC-3′ and 5′-TCCTGGTCTGCAGGTAA-3′ were used as primers in PCR. Reaction was performed using 50 ng genomic DNA, 20 pmol of each primer (Invitrogen), 1 U Taq polymerase (Fermentas), 0.1 mM dNTP (Fermentas), 3.5 mM MgCl2, 60 mM Tris-HCl und 15 mM ammoniumsulfate. PCR conditions were: initial denaturation at 96 °C for 1 min followed by 35 cycles at 94 °C for 1 min, 60 °C for 1 min, 70 °C for 1 min and finally 72 °C for 5 min. The PCR products were separated by electrophoresis on 2% agarose gel and visualized by ethidium bromide staining. Allele 1 (four repeats) was 410 bp long, allele 2 (two repeats) 240 bp, allele 3 (three repeats) 325 bp, and allele 4 (five repeats) 500 bp.
Data analysis
Data analyses were carried out using SPSS (version 12.0). In both treatment groups, clinical and demographic characteristics were compared between genotype groups with one-way ANOVA or t-test, when appropriate. Continuous data: Within treatment groups, HAMD-17 sum score differences between genotype groups across the treatment period were analyzed using a mixed-effects model repeated-measures methodology (MMRM), as MMRM has been shown to provide highly accurate modeling of treatment outcome while accounting for nonrandom missing data (Mallinckrodt et al 2001, 2003). In this model, we included HAMD-17 sum scores as dependent variables. The independent variables time and genotype as well as the interaction time × genotype were included as fixed effects, cases as random effects. Age and gender were included as potential covariates. For this analysis, patients with valid HAMD-17 scores for at least two weeks of treament (and baseline) were included (IL-1β*paroxetine/IL-1β*mirtazapine: n = 44/48 patients; IL-1Ra*paroxetine/IL-1Ra*mirtazapine: n = 39/46 patients). Proportions of missing data (IL-1β*paroxetine/IL-1β*mirtazapine: 3.4%/3.1%; IL-1Ra*paroxetine/IL-1Ra*mirtazapine: 3.0%/2.5%) were similar in all analyzed groups. Additionally, within the intention-to-treat (ITT) sample HAMD-17 sum score differences between genotype groups were analyzed for each assessment day and the individual endpoint with one-way ANOVA or t-test, if appropriate. Categorical data: Within the ITT sample, differences of time to remission (HAMD-17 sum score ≤7) between genotype groups were calculated applying Kaplan-Meier curve analyses (KM) with logrank tests in each treatment group separately. “Survival time” was defined as the time to first occurrence of remission or – in case of nonremission – as the time of the last observation. Again, age and gender were included as potential covariates by means of Cox regression analysis. Additionally, we determined remitters and nonremitters for each assessment day and the individual endpoint. The division by genotype led to clear 2 × 2 contingency tables which were analyzed by Chi-square test. The level of significance was set at 0.05 (two-tailed). A correction for multiple testing was not performed because of the exploratory nature of the study.
Results
One-hundred-and-sixteen patients gave written informed consent. Of the 116 patients, n = 12 patients had to be excluded because of missing HAMD-17 data at baseline. Considering the data for IL-1β C-511T analysis, n = 3 patients had to be excluded because offailed genotyping. In the case of the IL-1Ra (86bp)n polymorphism, n = 4 patients had to be excluded because offailed genotyping. Almost all patients were IL-1Ra*1 or IL-1Ra*2 allele carriers. In order to minimize the chances of spurious associations with very rare alleles, subjects bearing any of the other alleles (n = 6) were excluded from the analysis. Therefore, the total numbers of patients in the ITT samples differ between the analysis of the IL-1β (n = 101; females/males: n = 74/27) and the IL-1Ra polymorphism (n = 94; females/males: n = 73/21). With the exception of a lower proportion offemales in the group of patients without genotyping (sex ratio f/m = 1.4 vs. 2.9, p < 0.01), patients with or without genotyping were structurally similar with regard to age, depression severity at baseline and response to double-blind treatment in the trial (age, HAMD-17 sum score at baseline, day 42 and at endpoint, remitter rates at d42 and at endpoint: p > 0.1 for each analysis).
Genotype distribution, clinical and demographic data
The genotype distributions of IL-1β C-511T and IL-1Ra (86bp)n polymorphisms in our sample as well as clinical and demographic characteristics are shown in Table 1.
Table 1.
Clinical and demographic characteristics of our sample
| IL-1β C-511T |
IL-1Ra (86bp)n |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Paroxetine (n = 49) | Mirtazapine (n = 52) | Paroxetine (n = 44) | Mirtazapine (n = 50) | |||||||||
| Genotype | CC | CT | TT | CC | CT | TT | 11 | 12 | 22 | 11 | 12 | 22 |
| Number (%) | 15 (30.6) | 27 (55.1) | 7 (14.3) | 20 (38.5) | 27 (51.9) | 5 (9.6) | 18 (40.9) | 22 (50.0) | 4 (9.1) | 28 (56.0) | 17 (34.0) | 5 (10.0) |
| Gender (M/F; n°) | 7/8 | 5/22 | 2/5 | 8/12 | 3/24 | 2/3 | 6/12 | 4/18 | 1/3 | 4/24 | 4/13 | 2/3 |
| Age (years) | 48.4 ± 7.5 | 49.1 ± 12.6 | 46.7 ± 11.6 | 48.9 ± 10.9 | 48.9 ± 10.0 | 36.3 ± 10.5 | 49.9 ± 11.5 | 48.2 ± 10.6 | 45.3 ± 16.3 | 51.0 ± 11.4 | 48.5 ± 10.2 | 42.2 ± 6.8 |
| HAMD-17 at baseline | 22.5 ± 3.0 | 23.4 ± 4.1 | 23.1 ± 3.4 | 22.4 ± 2.4 | 22.4 ± 3.1 | 24.0 ± 3.8 | 22.6 ± 3.4 | 23.1 ± 4.0 | 23.5 ± 4.7 | 23.5 ± 3.0 | 21.9 ± 2.5 | 22.0 ± 3.5 |
| Mean daily drug dose (mg) | 23.8 ± 6.3 | 21.8 ± 4.4 | 22.2 ± 5.4 | 30.2 ± 2.8 | 30.8 ± 3.8 | 30.6 ± 5.1 | 21.6 ± 4.4 | 21.9 ± 4.8 | 28.9 ± 7.7 | 30.0 ± 2.9 | 31.2 ± 4.0 | 31.3 ± 4.5 |
Notes: Data are presented as mean (SD) unless otherwise indicated.
Abbreviations: HAMD-17, Hamilton Rating Scale for Depression (17 items); IL, interleukin.
Genotype frequencies of the two analyzed polymorphisms showed no deviation from Hardy-Weinberg equilibrium in both treatment groups. Genotype frequencies of both SNPs did not differ between treatment groups. In paroxetine-treated patients there were no significant differences among the three genotype groups of both polymorphisms in mean age, baseline mean HAMD-17 score or mean daily drug dose (p > 0.05 for each analysis). In mirtazapine-treated patients there were no significant differences of the same clinical and demographical data (p > 0.05 for each analysis), with the exception of younger IL-1β-511TT-carriers compared with -511CC- and -511CT- bearing patients (F(2) = 3.3; p = 0.045).
IL-1β C-511T polymorphism
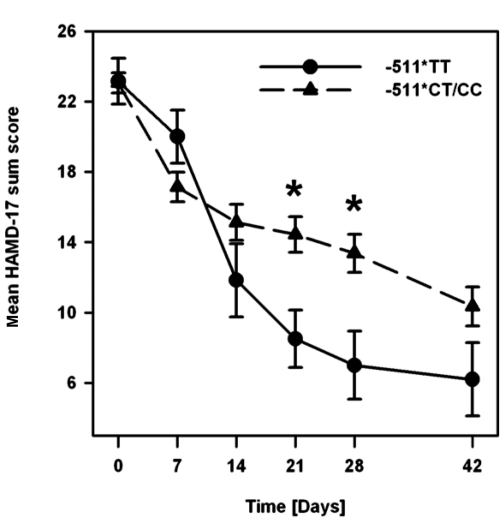
Paroxetine-treated patients homozygous for the IL-1β-511T allele displayed a more pronounced response during the course of treatment than IL-1β-511CT- or -511CC-carriers (data not shown). Therefore, IL-1β-511CT- and -511CC-carriers were pooled together for further analysis (-511CT/CC). The mean values of the HAMD-17 sum scores divided by genotype groups (-511TT and -511CT/CC) are plotted in Figure 1.
Figure 1.
Time course of changes in mean HAMD-17 sum score during treatment in paroxetine-treated patients stratified by IL-1β C-511T genotype groups. * = p < 0.05.
The global comparison of HAMD-17 score changes across the course of treatment showed no clear genotype effect (MMRM: F (1,54.6) = 3.4, p = 0.072), but a significant interaction of time × genotype (MMRM: F (5,136.8) = 2.9, p = 0.017). Neither age (main effect: F (1,59.1) = 0.2, p = 0.68) nor gender (main effect: F (1,54.1) = 0.3, p = 0.62; all interactions p > 0.05) had a significant impact on these results and were hence omitted as factors in the final model. Mean HAMD-17 scores at particular time points were lower in -511TT- compared with -511CT/CC-bearing patients from day 14 onwards with statistically significant differences on day 21 (t (41) = 2.3, p = 0.028) and day 28 (t (39) = 2.3, p = 0.025) as well as nonsignificant differences on day 42 (6.2 ± 4.7 vs. 10.4 ± 6.4) and at the endpoint (5.5 ± 4.5 vs. 11.7 ± 7.2). Mean time to remission (± SE) was significantly shorter in the -511TT-group (29 ± 6 days) compared with the -511CT/CC-group (36 ± 2 days) (KM: logrank-test ; p = 0.043). In Cox regression analysis, age (main effect: p = 0.98) and gender (main effect: p = 0.81; all interactions p > 0.1) did not show a statistically significant effect on these results and were thus not included as potential confounders in the final model. Remitter rates were continuously higher in -511TT- compared with -511CT/CC- bearing patients from day 14 onwards (day 14: 33.3 vs. 13.2%) with significant differences on day 21 (50.0 vs. 10.8%; Pearsons’s , p = 0.016) and at the endpoint (83.3 vs. 32.5%; ; p = 0.017) as well as nonsignificant differences on day 28 (50.0 vs. 20.0%) and day 42 (80.0 vs. 35.3%).
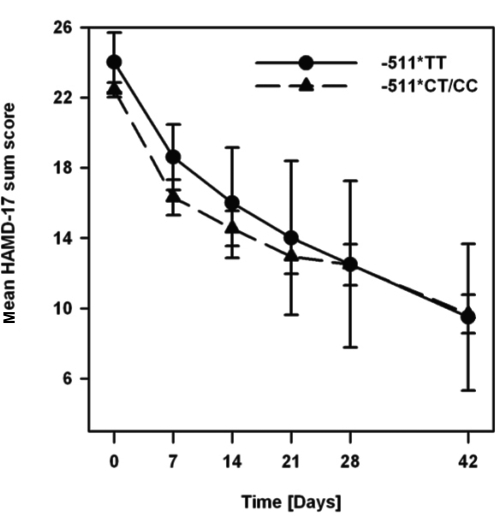
The course of mean HAMD-17 scores across the study period in mirtazapine-treated patients stratified by IL-1β C-511T genotype groups is depicted in Figure 2.
Figure 2.
Time course of changes in mean HAMD-17 sum score during treatment in mirtazapine-treated patients stratified by IL-1β C-511T genotype groups.
Between genotype groups, there were neither significant differences of HAMD-17 score changes over the treatment period (covariate age: F (1,54.2) = 0.3, p = 0.59; covariate gender: F (1,55.5) = 0.9, p = 0.36; all interactions p > 0.1) nor on particular assessment points (p > 0.05 for each analysis). Mean times to remission (± SE) were similar in both -511TT- (33 ± 6 days) and -511CT/CC-patients (32 ± 2 days; p > 0.05) without significant effects of age (p = 0.65) and gender (p = 0.67; all interactions p > 0.1) in Cox regression analysis. Proportions of remitters of both genotype groups did not differ significantly at any assessment day of the study (p > 0.05 for each analysis).
IL-1Ra (86bp)n polymorphism
A possible impact of the IL-1Ra (86bp)n polymorphism on different treatment response characteristics to paroxetine and mirtazapine was analyzed analogously to the analysis of the IL-1β C-511T SNP. There were neither clinically meaningful nor statistically significant differences between the three genotype groups IL-1Ra*11, IL-1Ra*12 and IL-1Ra*22 in any analyzed outcome measure (p > 0.05 for each analysis) in paroxetine as well as in mirtazapine-treated patients.
Discussion
Our results provide further suggestive evidence that the IL-1β C-511T gene variant affects the response to SSRI treatment in patients suffering from major depression. This conclusion is derived from the finding that paroxetine-treated patients with the IL-1β-511TT genotype showed a faster and more pronounced response to paroxetine than patients carrying the IL-1β-511CT or -511CC genotype. Our finding is in line with the report from Yu and colleagues (2003), who also reported an advantageous outcome in IL-1β-511TT-patients to fluoxetine treatment for 4 weeks. The mechanisms responsible for the observed differences in SSRI antidepressant response are not yet established. On the molecular genetic level the effects of the IL-1β-511 SNP may be mediated by strong linkage disequilibrium with a TATA box polymorphism at position -31 from the starting sequence of the IL-1β gene affecting protein (presumably transcription factors) binding to DNA (El-Omar et al 2000). Neurobiologically, one could have expected that subjects carrying the more active IL-1β -511TT genotype are more resistant to antidepressant treatment. However, this was neither the case in the sample studied by Yu et al (2003) nor in our sample. An explanation might be related to higher levels of IL-1β in -511TT carrying patients compared to IL-1β-511C carriers. If some of the effect of SSRIs is to diminish the production of central or peripheral proinflammatory cytokines such as IL-1 (Yirmiya et al 2001; Castanon et al 2004; Obuchowicz et al 2006) the response to SSRIs could indeed be greater in IL-1β-511TT carriers.
In contrast, we found no associaton between the IL-1β C-511T gene variant and mirtazapine treatment response. This might be related to different immuno-modulatory effects of mirtazapine treatment on IL-1β. Given that, in contrast to SSRI, mirtazapine treatment was associated with the upregulation of the pro-inflammatory tumor necrosis factor-α (TNF-α) (Kraus et al 2002; Kast 2003), different effects on IL-1β seem possible. However, the multiplicity of possible mechanisms of immune modulation by antidepressants (Schiepers et al 2005) suggests that several other reasons might be involved.
Additionally, we did not find an association of the IL-1Ra (86bp)n intron 2 polymorphism with antidepressant response to paroxetine and mirtazapine. This result does not support the hypothesis that the IL-1Ra intron 2 VNTR is involved in a mechanism affecting treatment outcome to paroxetine and mirtazapine. This finding is in line with the results from Kubera and colleagues (2000), who reported that IL-1Ra was elevated in depressed patients and did not normalize after response following antidepressant treatment. Thus, based on the available data, it can not be suggested that IL-1Ra is involved in neurobiological mechanisms underlying antidepressant treatment response. However, if the IL-1Ra gene is an uncommon locus affecting antidepressant treatment response or one with relatively modest effects, our power to detect its effects could have been too small.
The herein reported results are preliminary due to some limitations. There were some differences in gender distribution between the groups of non-genotyped and genotyped patients. However, a selective sample selection is improbable because our sample of genotyped patients with more females than males is comparable with many other patient samples in similar investigations and we did not found gender effects on treatment outcome. The small sample size did not allow us to control for unknown population stratification (eg, center effects) and possible influences of comorbid personality disorders. The exclusion criteria may further limit the generalizability of our findings. We did no correction for multiple testing for the obtained p-values. A Bonferroni correction for multiple testing (15 tests in each group) would lead to a corrected threshold of about 0.003 at the 5% significance level. If we perform Bonferroni correction for multiple testing in this exploratory study, all differences are nonsignificant, and thus the results must be interpreted with caution. The sample size is small for a genetic study examining the relationship between six genotypes per group, two polymorphisms and the treatment response. Feasible sample sizes in clinical studies are often too small to identify genetic variations that would withstand correction for multiple testing, unless they conferred major effects. However, this is very unlikely to be the case in phenotypes of complex inheritance where several interacting genes of small effect are predicted to contribute to antidepressant response. To circumvent this dilemma, replications ideally in larger samples will be needed to confirm the association.
In conclusion, our results provide further suggestive evidence that the IL-1β-511TT genotype is associated with an advantageous SSRI treatment response compared with the IL-1β-511CT/CC genotype, which is in line with previous yet hitherto unreplicated findings (Yu et al 2003). Differences in the mechanisms of action of paroxetine and mirtazapine may account for the divergent effects of the IL-1β C-511T gene variant on treatment outcome with these substances. Our results do not support the hypothesis that the IL-1Ra (86bp)n intron 2 VNTR is involved in treatment response to paroxetine or mirtazapine in major depression. Replications in large scale studies, which address the limitations of our study, are needed to further clarify the role of the IL-1β C-511T variant on SSRI treatment response. If replicated, the IL-1β C-511T promoter polymorphism could be considered useful for prospective confirmatory pharmacogenetic trials in patients with major depression.
References
- Anisman H. Considering cytokine panels. Brain Behav Immun. 2004;18:221–2. doi: 10.1016/j.bbi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Anisman H, Merali Z. Anhedonic and anxiogenic effects of cytokine exposure. Adv Exp Med Biol. 1999;461:199–233. doi: 10.1007/978-0-585-37970-8_12. [DOI] [PubMed] [Google Scholar]
- Anisman H, Ravindran A, Griffiths J, et al. Endocrine and cytokine correlates of major depression and dysthymia with typical or atypical features. Mol Psychiatry. 1999;4:182–8. doi: 10.1038/sj.mp.4000436. [DOI] [PubMed] [Google Scholar]
- Anttila SA, Leinonen EV. A review of the pharmacological and clinical profile of mirtazapine. CNS Drug Rev. 2001;7:249–64. doi: 10.1111/j.1527-3458.2001.tb00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert O, Szegedi A, Kohnen R. Mirtazapine compared with paroxetine in major depression. J Clin Psychiatry. 2000;61:656–63. doi: 10.4088/jcp.v61n0911. [DOI] [PubMed] [Google Scholar]
- Brambilla F, Maggioni M. Blood levels of cytokines in elderly patients with major depressive disorder. Acta Psychiatr Scand. 1998;97:309–13. doi: 10.1111/j.1600-0447.1998.tb10005.x. [DOI] [PubMed] [Google Scholar]
- Castanon N, Medina C, Mormede C, et al. Chronic administration of tianeptine balances lipopolysaccharide-induced expression of cytokines in the spleen and hypothalamus of rats. Psychoneuroendocrinology. 2004;29:778–90. doi: 10.1016/S0306-4530(03)00142-2. [DOI] [PubMed] [Google Scholar]
- Castanon N, Leonard BE, Neveu PJ, et al. Effects of antidepressants on cytokine production and actions. Brain Behav Immun. 2002;16:569–74. doi: 10.1016/s0889-1591(02)00008-9. [DOI] [PubMed] [Google Scholar]
- di Giovine FS, Takhsh E, Blakemore AI, et al. Single base polymorphism at -511 in the human interleukin-1 beta gene (IL-1 beta) Hum Mol Genet. 1992;1:450. doi: 10.1093/hmg/1.6.450. [DOI] [PubMed] [Google Scholar]
- Dunn AJ, Wang J, Ando T. Effects of cytokines on cerebral neurotransmission. Comparison with the effects of stress. Adv Exp Med Biol. 1999;461:117–27. doi: 10.1007/978-0-585-37970-8_8. [DOI] [PubMed] [Google Scholar]
- El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- Hayley S, Poulter MO, Merali Z, et al. The pathogenesis of clinical depression: stressor- and cytokine-induced alterations of neuroplasticity. Neuroscience. 2005;135:659–78. doi: 10.1016/j.neuroscience.2005.03.051. [DOI] [PubMed] [Google Scholar]
- Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1β genes. Eur J Immunol. 1998;28:2598–602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Kaestner F, Hettich M, Peters M, et al. Different activation patterns of proinflammatory cytokines in melancholic and non-melancholic major depression are associated with HPA axis activity. J Affect Disord. 2005;87:305–11. doi: 10.1016/j.jad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Kagaya A, Kugaya A, Takebayashi M, et al. Plasma concentrations of interleukin-1beta, interleukin-6, soluble interleukin-2 receptor and tumor necrosis factor alpha of depressed patients in Japan. Neuropsychobiology. 2001;43:59–62. doi: 10.1159/000054867. [DOI] [PubMed] [Google Scholar]
- Kast RE. Anti- and pro-inflammatory considerations in antidepressant use during medical illness: bupropion lowers and mirtazapine increases circulating tumor necrosis factor-alpha levels. Gen Hosp Psychiatry. 2003;25:495–6. doi: 10.1016/s0163-8343(03)00093-8. [DOI] [PubMed] [Google Scholar]
- Kenis G, Maes M. Effects of antidepressants on the production of cytokines. Int J Neuropsychopharmacol. 2002;5:401–12. doi: 10.1017/S1461145702003164. [DOI] [PubMed] [Google Scholar]
- Kraus T, Haack M, Schuld A, et al. Body weight, the tumor necrosis factor system, and leptin production during treatment with mirtazapine or venlafaxine. Pharmacopsychiatry. 2002;35:220–5. doi: 10.1055/s-2002-36390. [DOI] [PubMed] [Google Scholar]
- Kubera M, Kenis G, Bosmans E, et al. Plasma levels of interleukin-6, interleukin-10, and interleukin-1 receptor antagonist in depression: comparison between the acute state and after remission. Pol J Pharmacol. 2000;52:237–41. [PubMed] [Google Scholar]
- Levine J, Barak Y, Chengappa KN, et al. Cerebrospinal cytokine levels in patients with acute depression. Neuropsychobiology. 1999;40:171–6. doi: 10.1159/000026615. [DOI] [PubMed] [Google Scholar]
- Licinio J, Mastronardi C, Wong ML. Pharmacogenetics of neuroimmune interactions in human psychiatric disorders. Exp Physiol. 2007;92:807–11. doi: 10.1113/expphysiol.2007.038471. [DOI] [PubMed] [Google Scholar]
- Licinio J, Wong ML. The role of inflammatory mediators in the biology of major depression: central nervous system cytokines modulate the biological substrate of depressive symptoms, regulate stress-responsive systems, and contribute to neurotoxicity and neuroprotection. Mol Psychiatry. 1999;4:317–27. doi: 10.1038/sj.mp.4000586. [DOI] [PubMed] [Google Scholar]
- Linthorst AC, Flachskamm C, Müller-Preuss P, et al. Effect of bacterial endotoxin and interleukin-1 beta on hippocampal serotonergic neurotransmission, behavioral activity, and free corticosterone levels: an in vivo microdialysis study. J Neurosci. 1995;15:2920–34. doi: 10.1523/JNEUROSCI.15-04-02920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier SF, Watkins LR. Intracerebroventricular interleukin-1 receptor antagonist blocks the enhancement offear conditioning and interference with escape produced by inescapable shock. Brain Res. 1995;695:279–82. doi: 10.1016/0006-8993(95)00930-o. [DOI] [PubMed] [Google Scholar]
- Mallinckrodt CH, Clark WS, David SR. Accounting for dropout bias using mixed-effects models. J Biopharm Stat. 2001;11:9–21. doi: 10.1081/BIP-100104194. [DOI] [PubMed] [Google Scholar]
- Mallinckrodt CH, Clark SW, Carroll RJ, et al. Assessing response profiles from incomplete longitudinal clinical trial data under regulatory considerations. J Biopharm Stat. 2003;13:179–90. doi: 10.1081/BIP-120019265. [DOI] [PubMed] [Google Scholar]
- Merali Z, Lacosta S, Anisman H. Effects of interleukin-1beta and mild stress on alterations of norepinephrine, dopamine and serotonin neurotransmission: a regional microdialysis study. Brain Res. 1997;761:225–35. doi: 10.1016/s0006-8993(97)00312-0. [DOI] [PubMed] [Google Scholar]
- Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382–4. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- Obuchowicz E, Kowalski J, Labuzek K, et al. Amitriptyline and nortriptyline inhibit interleukin-1 release by rat mixed glial and microglial cell cultures. Int J Neuropsychopharmacol. 2006;9:27–35. doi: 10.1017/S146114570500547X. [DOI] [PubMed] [Google Scholar]
- Owen BM, Eccleston D, Ferrier IN, et al. Raised levels of plasma interleukin-1beta in major and postviral depression. Acta Psychiatr Scand. 2001;103:226–8. doi: 10.1034/j.1600-0447.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- Pociot F, Molvig J, Wogensen L, et al. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends Immunol. 2006;27:24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S, Ramamoorthy JD, Prasad PD, et al. Regulation of the human serotonin transporter by interleukin-1 beta. Biochem Biophys Res Commun. 1995;216:560–7. doi: 10.1006/bbrc.1995.2659. [DOI] [PubMed] [Google Scholar]
- Rothermundt M, Arolt V, Peters M, et al. Inflammatory markers in major depression and melancholia. J Affect Disord. 2001;63:93–102. doi: 10.1016/s0165-0327(00)00157-9. [DOI] [PubMed] [Google Scholar]
- Schiepers OJ, Wichers MC, Maes M. Cytokines and major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:201–17. doi: 10.1016/j.pnpbp.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Song C, Merali Z, Anisman H. Variations of nucleus accumbens dopamine and serotonin following systemic interleukin-1, interleukin-2 or interleukin-6 treatment. Neuroscience. 1999;88:823–36. doi: 10.1016/s0306-4522(98)00271-1. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Shintani F, Kanba S, et al. Induction of interleukin-1 beta and interleukin-1 receptor antagonist mRNA by chronic treatment with various psychotropics in widespread area of rat brain. Neurosci Lett. 1996;215:201–4. doi: 10.1016/0304-3940(96)12985-2. [DOI] [PubMed] [Google Scholar]
- Szegedi A, Rujescu D, Tadić A, et al. The catechol-O-methyltransferase Val108/158Met polymorphism affects short-term treatment response to mirtazapine, but not to paroxetine in major depression. Pharmacogenomics J. 2005;5:49–53. doi: 10.1038/sj.tpj.6500289. [DOI] [PubMed] [Google Scholar]
- Tadić A, Rujescu D, Müller MJ, et al. A monoamine oxidase B gene variant and short-term antidepressant treatment response. Prog Neuropsychopharmacol Biol Psychiatry. 2007a;31:1370–7. doi: 10.1016/j.pnpbp.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Tadić A, Müller MJ, Rujescu D, et al. The MAOA T941G polymorphism and short-term treatment response to mirtazapine and paroxetine in major depression. Am J Med Genet B Neuropsychiatr Genet. 2007b;144:325–31. doi: 10.1002/ajmg.b.30462. [DOI] [PubMed] [Google Scholar]
- Tarlow JK, Blakemore AI, Lennard A, et al. Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet. 1993;91:403–4. doi: 10.1007/BF00217368. [DOI] [PubMed] [Google Scholar]
- Thomas AJ, Davis S, Morris C, et al. Increase in interleukin-1beta in late-life depression. Am J Psychiatry. 2005;162:175–7. doi: 10.1176/appi.ajp.162.1.175. [DOI] [PubMed] [Google Scholar]
- Yirmiya R, Pollak Y, Barak O, et al. Effects of antidepressant drugs on the behavioral and physiological responses to lipopolysaccharide (LPS) in rodents. Neuropsychopharmacology. 2001;24:531–44. doi: 10.1016/S0893-133X(00)00226-8. [DOI] [PubMed] [Google Scholar]
- Yu YW, Chen TJ, Hong CJ, et al. Association study of the interleukin-1 beta (C-511T) genetic polymorphism with major depressive disorder, associated symptomatology, and antidepressant response. Neuropsychopharmacology. 2003;28:1182–5. doi: 10.1038/sj.npp.1300172. [DOI] [PubMed] [Google Scholar]
- Zhu CB, Blakely RD, Hewlett WA. The proinflammatory cytokines interleukin-1beta and tumor necrosis factor-alpha activate serotonin transporters. Neuropsychopharmacology. 2006;31:2121–31. doi: 10.1038/sj.npp.1301029. [DOI] [PubMed] [Google Scholar]