Abstract
The molecular genetic changes underlying the transformation of wild plants into agricultural weeds are poorly understood. Here we use a sunflower cDNA microarray to detect variation in gene expression between two wild (non-weedy) Helianthus annuus populations from Utah and Kansas and four weedy H. annuus populations collected from agricultural fields in Utah, Kansas, Indiana, and California. When grown in a common growth chamber environment, populations differed substantially in their gene expression patterns, indicating extensive genetic differentiation. Overall, 165 uni-genes, representing ∼5% of total genes on the array, showed significant differential expression in one or more weedy populations when compared to both wild populations. This subset of genes is enriched for abiotic/biotic stimulus and stress response proteins, which may underlie niche transitions from the natural sites to agricultural fields for H. annuus. However, only a small proportion of the differentially expressed genes overlapped in multiple wild vs. weedy comparisons, indicating that most of the observed expression changes are due to local adaptation or neutral processes, as opposed to parallel genotypic adaptation to agricultural fields. These results are consistent with an earlier phylogeographic study suggesting that weedy sunflowers have evolved multiple times in different regions of the United States and further indicate that the evolution of weedy sunflowers has been accompanied by substantial gene expression divergence in different weedy populations.
AGRICULTURAL weeds have evolved repeatedly from their wild relatives to specialize in the unique environment of cultivated fields. These diverse weedy plants incur considerable economic and environmental damage. They lower crop yields, harbor insect pests and diseases, reduce the quality of rangelands, poison livestock and wildlife, destroy native plant and animal habitats, and lead to the expenditure of billions of dollars in chemical and biological control measures annually (Pimental et al. 2000). As a consequence, there is considerable incentive for understanding the molecular genetic changes that transform wild populations into weeds.
Although there is no unique set of traits found in all weedy plants, many weeds have adaptations to disturbed habitats such as cultivated fields. Traits often associated with agricultural weeds include short generation time, fast growth, substantial developmental plasticity, resistance to environmental stress, herbicide tolerance, predation and disease resistance, high reproductive output, efficient dispersal mechanisms, variable seed dormancy, and other traits (Baker 1974; Tilman 1994; Basu et al. 2004). Thus, in addition to strong economic incentives for understanding the evolution of weedy plants, weeds are excellent systems for studying the colonization of novel habitats and how this occurs at phenotypic and genetic levels.
We are just beginning to understand how plants become weeds. Although some plants may be preadapted to exploit anthropogenic disturbance, in other cases weed success appears to be a consequence of recent evolutionary changes (Abbott 1992; Ellstrand and Schierenbeck 2000; Lee 2002; Callaway and Maron 2006; Whitney et al. 2006; Rieseberg et al. 2007). Despite progress toward understanding the ecology of weedy plants, the genetic and molecular basis of weediness remains largely unexplored, owing to a lack of genetic and genomic resources for most weedy plants (Broz et al. 2007). A handful of studies have focused primarily on quantitative traits using QTL mapping (Paterson et al. 1995; F. Y. Hu et al. 2003; Gu et al. 2004), but limitations of QTL techniques have made it difficult to identify individual genes underlying weedy phenotypes.
Accumulating evidence from diverse organisms strengthens the hypothesis that the evolution and regulation of gene expression is often a major contributor to biological novelty and phenotypic variation (e.g., Doebley et al. 1995; Enard et al. 2002; Wray 2003; Carroll 2005; Clark et al. 2006; Gilad et al. 2006; Haerty and Singh 2006; Khaitovich et al. 2006; Prud'homme et al. 2006; McGregor et al. 2007). However, little is known about the role of gene expression changes in ecological divergence, in part because many large-scale gene expression studies are from model organisms (Brem et al. 2002; Rifkin et al. 2003; Nuzhdin et al. 2004) whose ecology is poorly understood (Whitehead and Crawford 2006b). Variation in gene expression between natural populations typically is highly heritable and has been suggested to be of adaptive importance (Gibson 2002; Whitehead and Crawford 2006b). Thus, examination of gene expression variation occurring in natural populations may provide significant insight into the origin of biological novelty, diversity, and adaptation (Barrier et al. 2001; Oleksiak et al. 2001, 2002; Bochdanovits et al. 2003; Derome and Bernatchez 2006; Derome et al. 2006; Juenger et al. 2006; Lai et al. 2006; Whitehead and Crawford 2006a). It is particularly interesting to evaluate gene expression changes in agricultural weeds because it enables us to test the prediction (Grime 1977; Blair and Wolfe 2005; Bossdorf et al. 2005; Rogers and Siemann 2005; Richards et al. 2006) that weedy populations have evolved faster growth rates partially by reducing investment in costly defenses against biotic and abiotic stresses. For example, if expression levels of genes involved in such stress tolerances were consistently reduced, this prediction would be supported.
The common sunflower Helianthus annuus, which is native to the central and western United States, is the progenitor of the domesticated sunflower (Harter et al. 2004) and is also a major weed in domesticated corn, soybean, sunflower, and wheat fields. Because of a wealth of genomic data (e.g., EST and BAC libraries, genetic maps, microarrays, etc.) developed by the Compositae Genome Project (compgenomics.ucdavis.edu), H. annuus represents an ideal example for studying weed evolution. Wild populations of H. annuus typically inhabit heavy, clay-based soils in open grasslands (Heiser et al. 1969), but some populations have evolved many typical traits of agricultural weeds (Whitney et al. 2006; Rieseberg et al. 2007). These populations show genetic differentiation from nearby wild populations, some of which is likely due to selection (Massinga et al. 2003), but show no evidence of recent genomewide bottlenecks in effective population size (Kane and Rieseberg 2008). Additionally, in this species, there is ample evidence for extensive local habitat adaptation (Kane and Rieseberg 2007). In this study, we collected seeds from two wild H. annuus populations from their ancestral range, as well as four weedy H. annuus populations from agricultural fields across the United States. We grew multiple individuals from each population in a common growth chamber environment and utilized a sunflower cDNA microarray (Lai et al. 2006) to examine variation in gene expression in these natural populations. Experiments described here address a number of questions relating to the evolution of weediness via changes in gene expression, including (1) How many genes are differentially expressed in wild vs. weedy populations of H. annuus?, (2) What are the functions of the differentially expressed genes?, (3) Are there similarities in gene expression changes involved in weediness in multiple populations of H. annuus?, and (4) What evolutionary forces are responsible for gene expression divergence among populations of H. annuus?
MATERIALS AND METHODS
Plant materials:
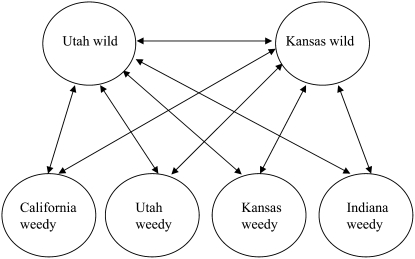
Two wild H. annuus populations from Utah and Kansas and four weedy H. annuus populations from Utah, Kansas, Indiana, and California were chosen for this study (Figure 1A). Wild populations were collected from permanently uncultivated fields, which are typical habitat for this species. Weedy populations were collected from within intensively cultivated corn fields. Populations from which seeds were collected comprised at least 500–10,000 individuals or more, depending on whether nearby populations are counted as well. One seed head per plant was collected along one or more transects in each population for at least 48 plants spaced at least 3 m apart. Seeds were germinated in the lab, and plants were grown in the growth chamber at 22°–25° with 16-hr light cycle. Experiments were conducted 4–6 weeks after germination when seedlings were 4–8 cm high and had four to eight true leaves. Seedlings including roots, shoots, and leaves were harvested, quick frozen in liquid nitrogen, and stored at −80° until RNA was extracted. To minimize the effect of individual differences within populations, two or three individuals were pooled as one biological sample and the microarray experiment was carried out following the design in Figure 2, which emphasizes comparisons of wild vs. weedy populations, with four biological replicates per comparison.
Figure 1.—
(A) Map showing geographic location of common sunflower populations employed in this study. Open circles, wild populations collected from natural sites; solid circles, weedy populations collected from cultivated cornfields. (B) Unrooted neighbor-joining tree showing the relationships among the six populations, based on microsatellite genetic distances [(δμ)2].
Figure 2.—
Experimental design employed for microarray study. The arrows represent the two samples labeled with Alexa Fluor 647, and two opposite samples were labeled with Alexa Fluor 555, with full dye swapping. Four biological replicates were done for each comparison.
Sunflower cDNA microarray and gene annotation:
An abiotic stress sunflower cDNA array (Lai et al. 2006) was employed in this study. Given that the primary difference between the wild and weedy form of sunflower is in the habitat that each occupy (wild in water/nutrient-limited natural sites and weedy in water/nutrient-rich cultivated fields), an abiotic stress array seemed ideal for identifying key expression differences between the two forms. The array was primarily developed from ESTs from environmentally stressed tissues, as well as salt and drought-subtracted libraries. An additional 384 ESTs with known map locations (Heesacker et al. 2008) were added to the sunflower cDNA microarray (Lai et al. 2006). In all, 4127 cDNAs, representing ∼3198 uni-genes, were spotted on the array. Gene annotation, as well as printing, post-processing, and storage of the array, followed our previous study (Lai et al. 2006).
RNA extraction and preparation of probes:
For each population, total RNA was extracted from four pools of two to three seedlings per pool. Each pooled extraction was considered a biological replicate and used for probe synthesis. RNA extractions followed Lai et al. (2006), except that on-column DNase I digestion (QIAGEN) was employed to eliminate possible genomic DNA contamination. Probes were made with the dendrimer labeling method (array 50 kit, Genisphere) following the manufacturer's instructions. Approximately 10–15 μg of total RNA was reverse transcribed to first-strand cDNA in the presence of deoxynucleotide triphosphate mix and special RT dT primer. The fluorescent 3DNA reagent hybridizes to the cDNA because it includes a “capture sequence” that is complementary to a sequence on the 5′-end of the RT primer. In our experiment, four biological replicates were carried out for each pairwise comparison, with full dye swapping (two of four cDNA samples from each population were synthesized using dT primer with sequence tag complementary to Alexa Fluor 555 and another two were synthesized using dT primer with sequence tag complementary to Alexa Fluor 647).
Hybridization, washing, and scanning:
After reverse transcription and RNA hydrolysis, one whole volume of cDNA (12.7 μl) from each replicate was mixed with sequence tag complementary to Alexa Fluor 555 or Alexa Fluor 647. For each slide used, one population was mixed with each type of tag (e.g., Utah wild population primed with Alexa Fluor 555 complementary sequence tag and Indiana weedy population primed with Alexa Fluor 647 complementary sequence tag, etc.).
The slides were incubated in prehybridization buffer containing 5× SSC, 0.1% SDS, and 1% I-Block (w/v) at 55° for 1.5 hr during the probe preparation. Following the prehybridization treatment, the slides were thoroughly washed three times with Milli-Q water for 1 min each and dried by centrifugation. The above cDNA probe was applied to the prehybridized microarray slides, covered with a cover slip (LifterSlips, Erie Scientific, Portsmouth, NH), and placed into hybridization chambers (Corning, NY). Hybridization chambers were submerged in the water bath at 55° overnight. After incubation, slides were washed in a large volume (400 ml) of 2× SSC/0.2% SDS to remove the cover slip. Then, slides were washed with 2× SSC/0.2% SDS at 55° for 10 min, 2× SSC at room temperature for 10 min, and 0.2× SSC at room temperature for 10 min, respectively.
After washing, the slides were dried by centrifugation. Then, the denatured fluorescent 3DNA reagent with Alexa Fluor 555 and Alexa Fluor 647 dye was applied to the slides in succession. As above, the slides were covered with a cover slip and placed into hybridization chambers, and hybridization chambers were submerged in the water bath at 55° for 5 hr. After hybridization, slides were washed in a large volume (400 ml) of 2× SSC/0.2% SDS to remove the cover slip. The slides were then washed for 10 min under each of the following conditions: 2× SSC/0.2% SDS at 65°, 2× SSC at room temperature, and 0.2× SSC at room temperature. After drying, the slides were scanned as described before (Lai et al. 2006).
Data collection, normalization, and cluster analysis:
Data collection, normalization, and analysis followed our previous study (Lai et al. 2006). Statistically significant expression differences were identified by setting the threshold false discovery rate (FDR) at <0.05. The false discovery rate, which measures the fraction of identified “significant” results that are spurious, is not the same as the more widely used false positive rate (P-value), which measures the fraction of truly nonsignificant results that are falsely identified as significant. The false discovery rate uses a more sophisticated approach, enabling researchers to balance the competing goals of maximizing the discovery of true positives while minimizing false positives and is thus ideal for use in genomewide studies with multiple tests (Storey and Tibshirani 2003). Cluster analysis of differentially expressed genes employed Euclidean distance and average linkage clustering, as implemented by The Institute for Genomic Research MultiExperiment Viewer (MeV) software. Branch lengths indicate the degree of similarity between genes in their patterns of expression (Eisen et al. 1998).
Quantitative PCR:
RNAs extracted for microarray were also used for quantitative PCR (Q-PCR) to verify expression patterns. First-strand cDNA was synthesized as described before (Lai et al. 2006). Primers for Q-PCR (supplemental Table 2) were designed using primer3, available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi. The lengths of PCR products were between 100 and 250 bp (supplemental Table 2).
Q-PCR was carried out using Stratagene MX3000p (Stratagene, La Jolla, CA) in 15 μl of total volume with 7.5 μl platinum SYBR green qPCR SuperMix-UDG, 0.3 μl SYBR green dye, 0.3 μl ROX reference dye, and 1.5 μl fourfold diluted first-strand cDNA and in the presence of 200 nm gene-specific forward and 200 nm reverse primer. Each reaction was run in triplicate. The cycling program was a two-step cycling method: 50° for 2 min hold, 95° for 2 min hold, 40 cycles of 95° for 15 sec and 60° for 30 sec. Dissociation curve analysis followed after PCR amplification. Standard curves were generated by the Stratagene QPCR software with a cDNA mix from all six populations as a template and corresponding gene-specific forward and reverse primers. Actin or 60S ribosomal protein served as a control to normalize the actual cDNA amount of each sample. The data were analyzed using the MxPro QPCR software version 3.00.
RESULTS
Validation of microarray results:
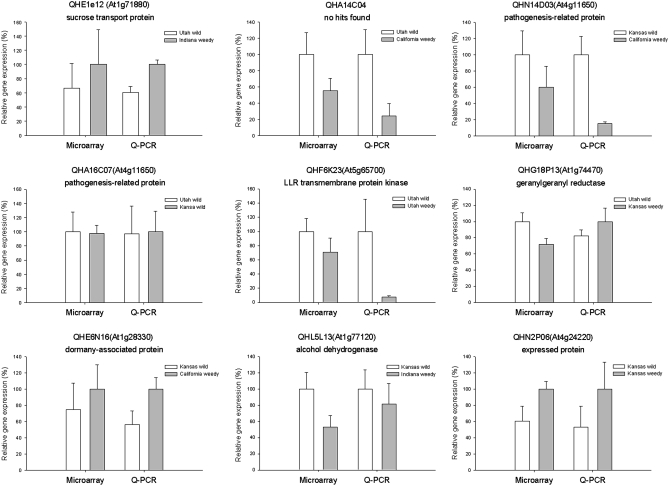
Nine ESTs were arbitrarily selected for validation of both identity and expression pattern (Figure 3). The identity of the PCR products was confirmed by resequencing, and expression pattern was validated with Q-PCR. As in our previous study (Lai et al. 2006), Q-PCR revealed similar patterns or tendencies of expression when compared with the microarray results (Figure 3). For example, a dormancy-associated protein (At1g28330) displayed the same expression pattern in both Q-PCR and microarray assays: elevated expression in Indiana weedy and California weedy populations compared with Utah wild and Kansas wild populations. Likewise, a pathogenesis-related protein in H. annuus (At4g11650) showed elevated expression in Utah wild and Kansas wild populations when compared with the California weedy population in both Q-PCR and microarray analyses. Overall, there was ∼85% agreement between Q-PCR and microarray data observed in this study (data not shown).
Figure 3.—
Gene expression differences as measured by microarray vs. Q-PCR. The x-axis represents assay method; the y-axis displays the relative gene expression assessed by Q-PCR and microarray. For both the microarray and Q-PCR, the population with the highest level of expression was set at 100%, and the expression level of another population was calculated relative to this standard. Error bars are based on the standard deviation from the mean. For a detailed calculation, see Lai et al. (2006).
Number of differentially expressed genes among populations:
To identify differences in gene expression between wild and weedy populations of the common sunflower, we compared expression patterns of 3198 uni-genes between the two wild H. annuus populations and each of the four weedy H. annuus populations. For comparative purposes, we also measured gene expression differences between the two wild populations.
We first identified genes that were differentially expressed between populations using a threshold false discovery rate of <0.05 (Table 1). There were 24 genes differentially expressed between the two wild populations, whereas 22, 71, 276, and 315 genes were differentially expressed in comparisons between the wild population from Utah and the Utah weedy, Kansas weedy, Indiana weedy, and California weedy populations, respectively. Likewise, 28, 69, 188, and 96 genes exhibited significant expression differences between the wild population from Kansas and the Kansas weedy, Utah weedy, Indiana weedy, and California weedy populations, respectively.
TABLE 1.
Differentially expressed genes in populations of the common sunflower
| Comparison | Differentially expressed uni-genes (FDR < 0.05) |
|---|---|
| Utah wild and Kansas wild | 24 |
| Utah wild and Utah weedy | 22 |
| Utah wild and Kansas weedy | 71 |
| Utah wild and Indiana weedy | 276 |
| Utah wild and California weedy | 315 |
| Kansas wild and Kansas weedy | 28 |
| Kansas wild and Utah weedy | 69 |
| Kansas wild and Indiana weedy | 188 |
| Kansas wild and California weedy | 96 |
The numbers of differentially expressed genes between populations were strongly correlated with microsatellite genetic distances (Figure 1B; correlation coefficient = 0.86) reported in a parallel study (Kane and Rieseberg 2008) and weakly correlated with geographic distances (correlation coefficient = 0.48). For example, more differentially expressed genes were identified in comparisons between the two wild populations and Indiana weedy or California weedy populations, which are most distant genetically and geographically, than in comparisons between the two wild populations and nearby weedy populations. On the other hand, the Utah wild population was more strongly differentiated from the Indiana and California weedy populations than was the Kansas wild population, indicating that the number of significant differentially expressed genes does not always increase with geographic distance.
Ecological factors and the functions of differentially expressed genes:
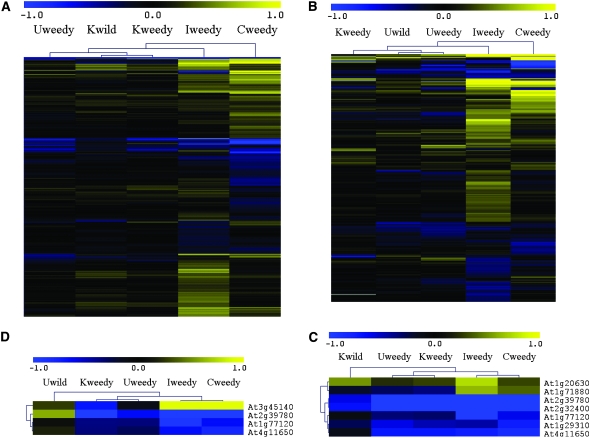
A total of 506 genes showed differential expression patterns in at least one comparison between Utah wild and other weedy populations. In contrast, only 276 genes were differentially expressed in at least one comparison between Kansas wild and other weedy populations (Figure 4). Taken together, 604 genes representing ∼19% of total genes on the array were differentially expressed in at least one comparison between wild and weedy populations.
Figure 4.—
Hierarchical clustering analysis of the following. (A) Five hundred and six genes were differentially expressed in at least one comparison between Utah wild and all other weedy populations. (B) Two hundred and seventy-six genes were differentially expressed in a pattern in at least one comparison between Kansas wild and all other weedy populations. (C) Seven genes were differentially expressed in Utah wild vs. all other weedy population comparisons. (D) Four genes were differentially expressed in Kansas wild vs. all other weedy population comparisons. Uwild, Utah wild population; Kwild, Kansas wild population; Uweedy, Utah weedy population; Kweedy, Kansas weedy population; Iweedy, Indiana weedy population; Cweedy, California weedy population. Color intensity is directly relative to the magnitude of differential expression ratios, which were transformed to log base 2 and subject to Euclidean distance and average linkage clustering.
Of these differentially expressed genes, 165 (representing ∼5% of the genes on the array) were differentially expressed in both wild populations relative to one or more weedy populations (supplemental Table 1). Gene annotation according to biological process revealed that 12% encode proteins that function in response to abiotic or biotic stimulus and 13% encode proteins that function in response to stress. These two functional groups are significantly overrepresented, with only 4% and 3% of total uni-genes on the array having the same functions, respectively (χ2 = 44.44, P = 2.616e-11 and χ2 = 82.93, P < 2.2e-16, respectively; supplemental Figure 1).
Surprisingly few genes exhibited differential expression in multiple weedy populations (Table 2; supplemental Figure 2). When compared against both wild populations, 6, 13, 71, and 89 differentially expressed genes were identified in the Utah, Kansas, California, and Indiana weedy populations, respectively (supplemental Figure 2). Of these, 1 was unique to the Utah weedy population, 10 to Kansas weedy, 66 to Indiana weedy, and 48 to California weedy (supplemental Figure 2). Only six and three genes, respectively, showed consistent expression differences between the Utah wild and all weedy populations and between the Kansas wild and all weedy populations (Table 2, Figure 4). Interestingly, these candidate “weedy” genes, most of which contribute to biotic or abiotic stress tolerance, were predominantly downregulated in the weedy compared with the wild populations (Table 2, Figure 4).
TABLE 2.
Genes with consistent pattern of differential expression between Utah wild Uw or Kansas wild Kw and all weedy populations
| Comparison | EST ID | Expression pattern | Ara_hits | Description |
|---|---|---|---|---|
| Uw vs. all weedy | QHB34L10 | Weedy upregulated | At1g20630 | Catalase 1 |
| Uw vs. all weedy | QHL5L13 | Weedy downregulated | At1g77120 | Alcohol dehydrogenase (EC 1.1.1.1) |
| Uw vs. all weedy | QHN19H20 | Weedy downregulated | At1g29310 | Protein transport protein sec61, putative |
| Uw vs. all weedy | QHN19J12 | Weedy downregulated | At2g39780 | S-like ribonuclease RNS2 |
| Uw vs. all weedy | QHN7A10 | Weedy downregulated | At2g32400 | Glutamate receptor family |
| Uw vs. all weedy | QHN8G11 | Weedy downregulated | At4g11650 | Pathogenesis-related protein in H. annuus |
| Kw vs. all weedy | QHL5L13 | Weedy downregulated | At1g77120 | Alcohol dehydrogenase (EC 1.1.1.1) |
| Kw vs. all weedy | QHN19J12 | Weedy downregulated | At2g39780 | S-like ribonuclease RNS2 |
| Kw vs. all weedy | QHN14D03 | Weedy downregulated | At4g11650 | Pathogenesis-related protein in H. annuus |
To elucidate the importance of local and regional adaptations (vs. weediness per se), we examined the genes that were differentially expressed in all Utah populations or in all Kansas populations. For this analysis, genes were identified as candidates for local or regional adaptations to Kansas if expression was significantly different in Kansas wild vs. all other non-Kansas populations (both weedy and wild) and also was significantly different in Kansas weedy vs. Utah wild, and not significantly different between Kansas wild and weedy populations. Similarly, to examine regional adaptations common to both Utah populations, we selected genes with significant differences in expression with wild Utah to all non-Utah comparisons, as well as weedy Utah to wild Kansas but not in comparison between wild Utah and weedy Utah. A single gene with strong homology to geranylgeranyl reductase (At1g74470) was found to be significantly expressed in both Kansas populations. This pattern may be of interest because differences in expression patterns of geranylgeranyl reductase are known to affect levels and quality of chlorophyll, tocopherol, and carotenoids, leading to differences in growth rates and other phenotypic effects in some environmental conditions (Tanaka et al. 1999). Two genes (At1g20630 and At4g33950) were differentially expressed in the two Utah populations and may be involved in adaptation to desert environments. One of these (At4g33950) was homologous to calcium-independent ABA-activated protein kinase, which responds to osmotic stress, salt stress, abscisic acid stimulus, pathogen, and regulation of seedling growth in Arabidopsis (Assmann 2003). The other (At1g20630) had strong homology to catalase proteins, which are thought to be involved in response to pathogens (Takahashi et al. 1997) through the breakdown of H2O2 in the peroxisome.
Hierarchical clustering:
To further explore the relationship between gene expression variation and genetic distance, geographic distance, and habitat, four sets of differentially expressed genes were analyzed by hierarchical clustering: 506 genes differentially expressed between Utah wild and one or more weedy populations, 276 genes differentially expressed between Kansas wild and one or more weedy populations, seven genes differentially expressed between Utah wild and all weedy populations, and four genes differentially expressed between Kansas wild and all weedy populations (Figure 4). Of the 11 genes that were differentially expressed in the weedy populations, 2 were polymorphic with respect to the direction of the expression difference (Figure 4). Cluster analyses revealed that expression patterns of the two large gene sets tracked population distance, with geographically and genetically closer populations exhibiting similar expression patterns. In contrast, expression profiles of the smaller gene sets united populations according to habitat. Thus, these genes may have evolved in response to selective forces common to agricultural habitats and are consistent with parallel adaptation (Figure 4).
DISCUSSION
Genetic relationships among wild and weedy populations:
The wild and weedy sunflower populations employed for expression analyses were part of a broader microsatellite study of phylogeography and selective sweeps within the common sunflower (Kane and Rieseberg 2008). Both phylogenetic (neighbor-joining) and Bayesian (Structure) clustering algorithms implied that the weedy populations were independently derived from nearby wild populations, although the possibility of a single origin of weed populations followed by gene flow with local wild populations could not be ruled out (Kane and Rieseberg 2008). Reanalysis of the microsatellite data for the subset of populations employed in the microarray analyses confirms this earlier result. Each weedy population, while genetically distinct, is most closely related to nearby wild populations (Figure 1B).
Analysis of differentially expressed genes in wild vs.
weedy sunflower populations: As noted in the Introduction, variation in gene expression between natural populations has been reported to be genetically determined and can underlie adaptation (Gibson 2002; Whitehead and Crawford 2006b). Therefore, examination of gene expression variation in natural wild populations and their closely related weedy populations provides insight into both the kind and percentage of genes whose expression may be related to the habitat transition from wild to agricultural field, at least in H. annuus.
We identified 165 genes (5%) with significant expression differences in one or more weedy populations when compared with both wild populations. Gene annotation revealed that abiotic or biotic stimulus-related proteins and stress-related proteins were significantly overrepresented among the weedy genes, implying possible ecological functions of genes that underlie important weedy traits. The overrepresentation of abiotic/biotic stimulus and stress-related proteins makes sense, given the contrast between the harsh, dry habitat of the wild populations and the nutrient-rich, irrigated corn fields where the weedy populations occur.
Interestingly, the magnitude of expression differences between wild and weedy populations tended to be relatively small, ranging from a 1.08- to a 6.54-fold difference (data not shown), particularly in relation to an earlier comparison of different sunflower species (Lai et al. 2006). However, it is now well established that small relative changes in gene expression can be as functionally important as larger ones (Oleksiak et al. 2005; Whitehead and Crawford 2006a). Alternatively, it might be that adaptive genetic differences between the wild and weed populations are minor or that gene flow has prevented fixation of some alleles with large expression difference, thereby reducing the apparent magnitude of expression differences in pooled samples such as those employed here. A final explanation for the small expression differences is that they result from variation in the environment or among individual genotypes rather than from fixed genetic differences between these populations. However, this explanation seems unlikely since plants were grown in a common environment to minimize the physiological differences. Also, several individuals were pooled to reduce the within-population differences and to maximize the ability to detect between-population differences (Kendziorski et al. 2005). All plants appeared healthy and the entire seedling was used for RNA isolations. Thus, the majority of the variation detected in the experiment is not due to different physiological conditions or differences in tissue types. Instead, these differences should represent genetic differences between populations, although the possibility of maternal effects cannot be ruled out (Roach and Wulff 1987) since the maternal environment of these seeds necessarily varied with collection locality. Other factors that might be important in natural populations, such as population life history, population structure, and potential genotype–environment interactions are beyond the scope of study conducted here.
Candidate genes:
Our results indicate that variation in gene expression is strongly influenced by the geographical location of populations. Only a handful of the 165 genes (see above) exhibited differential expression in one or more weedy populations when compared with both wild populations, implying that most differentially expressed genes contribute to local adaptation or neutral processes rather than weediness per se. However, until a larger fraction of the genes in the genome can be analyzed, it remains unclear if the evolution of weediness in H. annuus mostly involves different sets of genes in different populations, as has been reported for adaptive melanism in mice (Hoekstra and Nachman 2003). Clearly, local adaptation has a key role in shaping the evolution of the weedy H. annuus transcriptome. Because these are natural populations, many characteristics other than wild vs. weedy habitat could influence the gene expression changes. Therefore, some of the observed variation in gene expression could have been due to different selective forces such as local climate, soil type, pathogens, and different methods of corn cultivation practiced in different regions.
Six genes showed consistent expression differences between all weedy populations and at least one wild population and three genes—alcohol dehydrogenase (ADH, At1g77120), S-like ribonuclease RNS2 (At2g39780), and pathogenesis-related protein of H. annuus (At4g11650)—showed consistent expression differences in all comparisons of wild and weedy populations (Table 2, Figure 4). ADH is of particular interest because earlier isozyme studies of H. annuus identified an E allele at Adh-1, so named because of the early cessation of activity in germinating seeds (Torres 1974; Torres and Diedenhofen 1979). Adh-1E was found to be associated with wet areas, such as agricultural fields where weedy sunflowers predominate. Torres and Diedenhofen (1979) speculated that Adh-1E facilitated germination under anaerobic conditions, such as those found in frequently flooded sites. In this study, the expression level of ADH was downregulated in the seedlings of weedy populations, which is consistent with the precocious cessation of Adh-1E activity observed in isozyme studies.
All of these three candidate weedy genes are known to be involved in stress responses and two have been shown to be upregulated in response to biotic or abiotic stress in other studies. For example, homologs of S-like ribonuclease RNS2 are known to be upregulated under drought stress, phosphate starvation and under senescence in rice, Antirrhinum, green alga, and Arabidopsis (Taylor et al. 1993; Liang et al. 2002; Salekdeh et al. 2002). Likewise, upregulation of pathogenesis-related protein of H. annuus (At4g11650) has been reported following chemical elicitation of pathogen resistance (X. Hu et al. 2003). In this study, both genes were found to be downregulated in weedy populations, which occur in less stressful environments.
Observations of constitutive downregulation of stress tolerance genes in seedlings from weedy populations (Table 2) are consistent with a possible general hypothesis for the evolution of weediness in sunflowers. Wild and weedy populations differ in seedling growth rate, with weedy seedlings growing almost twice as fast as their wild counterparts (L. H. Rieseberg, unpublished data). If there is a cost to abiotic and biotic tolerances, which seems likely, downregulation of costly genes and pathways might provide a competitive growth rate advantage to seedlings (Grime 1977; Blair and Wolfe 2005; Bossdorf et al. 2005; Rogers and Siemann 2005; Richards et al. 2006). Obviously, the number of common weedy genes identified here are too few to test such a hypothesis, but it does provide a prediction that can be tested in future studies of weedy sunflowers and other invasive plants.
Origin of expression variation in H. annuus:
As with other traits that are variable and heritable, gene expression variation is likely affected by both neutral drift and selection. If drift is the primary cause of variation in gene expression, then greater genetic divergence among populations should be correlated with greater divergence in gene expression (Whitehead and Crawford 2006a). However, this pattern could also be due to local or regional adaptations. In contrast, parallel shifts in gene expression are most likely a consequence of natural selection (Derome et al. 2006). In yeast, stabilizing selection appears to constrain the divergence of the gene expression transcriptomes (Denver et al. 2005), whereas a neutral model best accounts for expression variation in human populations (Khaitovich et al. 2004).
In our study, hierarchical clustering analysis of expression differences indicates that expression variation is strongly correlated with genetic differences among populations, a result consistent with either drift or local adaptation. However, analysis of the set of common weedy genes revealed parallel shifts in gene expression across different weedy populations and may represent an example of parallel adaptation to agricultural conditions.
Caveats:
Because only a small subset of the genome (∼10%) was analyzed, many of the key genes contributing to weediness likely were missed. Additionally, although spurious results were minimized by using FDR rather than P-values, we do expect that a small fraction (5%) of genes identified as different in any one comparison will be spurious. However, falsely identifying the same gene in more than one analysis is exceedingly unlikely using this approach, indicating that the majority of our important findings are on solid ground. Another caveat is that we focused exclusively on gene expression differences in whole seedlings and it might be that other ecological variables (e.g., mycorrhizal associations with roots) are most critical to the evolution of weediness. A comprehensive common garden study is underway to identify possible differences between wild and weedy sunflowers in abiotic and biotic tolerances, as well as in herbivory and mycorrhizal associations. This experiment will be employed to guide future microarray studies, which will employ a much more complete array currently under development (below).
Conclusions and future work:
This study interrogated ∼10% of the sunflower transcriptome in multiple wild and weedy populations and identified several candidate weedy genes that are the targets of both functional and molecular evolutionary study. For example, we have initiated studies of expression-control elements to determine that expression patterns are caused by selection in the vicinity of the genes themselves (cis) or by selection on interacting regulatory genes (trans). Additionally, sequencing studies are underway to determine if there is a demographic signature of selection at these loci. Finally, we are developing a 33,000 uni-gene array to test our hypothesis that downregulation of costly genes and pathways might provide a competitive growth rate advantage to weedy sunflowers.
Acknowledgments
We thank Brian Eads, Matthew Hahn, Andreas Rechtsteiner, and Xinguo Wang for helpful advice; Monica Sentmanat for assistance with microarray printing; and Amy Cash for assistance with microarray hybridizations. This work was supported by a grant from the National Science Foundation (DBI0421630) to L.H.R.
References
- Abbott, R., 1992. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 7 401–405. [DOI] [PubMed] [Google Scholar]
- Assmann, S. M., 2003. OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8 151–153. [DOI] [PubMed] [Google Scholar]
- Baker, H. G., 1974. The evolution of weeds. Annu. Rev. Ecol. Evol. S5 1–24. [Google Scholar]
- Barrier, M., R. H. Robichaux and M. D. Purugganan, 2001. Accelerated regulatory gene evolution in an adaptive radiation. Proc. Natl. Acad. Sci. USA 98 10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, C., M. D. Halfhill, T. C. Mueller and C. N. Stewart, Jr., 2004. Weed genomics: new tools to understand weed biology. Trends Plant Sci. 9 391–398. [DOI] [PubMed] [Google Scholar]
- Blair, A. C., and L. M. Wolfe, 2005. The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85 3035–3042. [Google Scholar]
- Bochdanovits, Z., H. van der Klis and G. de Jong, 2003. Covariation of larval gene expression and adult body size in natural populations of Drosophila melanogaster. Mol. Biol. Evol. 20 1760–1766. [DOI] [PubMed] [Google Scholar]
- Bossdorf, O., H. Auge, L. Lafuma, W. E. Rogers, E. Siemann et al., 2005. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144 1–11. [DOI] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296 752–755. [DOI] [PubMed] [Google Scholar]
- Broz, A. K., C. D. Broeckling, J. He, X. Dai, P. X. Zhao et al., 2007. A first step in understanding an invasive weed through its genes: an EST analysis of invasive Centaurea maculosa. BMC Plant Biol. 7 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, R. M., and J. L. Maron, 2006. What have exotic plant invasions taught us over the past 20 years? Trends Ecol. Evol. 21 369–374. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., 2005. Evolution at two levels: on genes and form. PLoS Biol. 3 e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. M., T. N. Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38 594–597. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, J. T. Streelman, S. K. Kim, M. Lynch et al., 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37 544–548. [DOI] [PubMed] [Google Scholar]
- Derome, N., and L. Bernatchez, 2006. The transcriptomics of ecological convergence between 2 limnetic coregonine fishes (Salmonidae). Mol. Biol. Evol. 23 2370–2378. [DOI] [PubMed] [Google Scholar]
- Derome, N., P. Duchesne and L. Bernatchez, 2006. Parallelism in gene transcription among sympatric lake whitefish (Coregonus clupeaformis Mitchill) ecotypes. Mol. Ecol. 15 1239–1249. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand, N. C., and K. A. Schierenbeck, 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl. Acad. Sci. USA 97 7043–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard, W., P. Khaitovich, J. Klose, S. Zollner, F. Heissig et al., 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296 340–343. [DOI] [PubMed] [Google Scholar]
- Gibson, G., 2002. Microarrays in ecology and evolution: a preview. Mol. Ecol. 11 17–24. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., A. Oshlack and S. A. Rifkin, 2006. Natural selection on gene expression. Trends Genet. 22 456–461. [DOI] [PubMed] [Google Scholar]
- Grime, J. P., 1977. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theroy. Am. Nat. 111 1169–1194. [Google Scholar]
- Gu, X. Y., S. F. Kianian and M. E. Foley, 2004. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty, W., and R. S. Singh, 2006. Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 23 1707–1714. [DOI] [PubMed] [Google Scholar]
- Harter, A. V., K. A. Gardner, D. Falush, D. L. Lentz, R. A. Bye et al., 2004. Origin of extant domesticated sunflowers in eastern North. Am. Nat. 430 201–205. [DOI] [PubMed] [Google Scholar]
- Heesacker, A. F., S. Tang, S. D. Gandhi, J. M. Kolkman, C. F. Barrios et al., 2008. Genetic mapping of 1,530 transcribed and non-transcribed loci in sunflower, a resource for comparative mapping, forward genetics, and molecular breeding. Theor. Appl. Genet. (in press).
- Heiser, C. B., D. M. Smith, S. B. Clevenger and W. C. Martin, 1969. The North American sunflowers (Helianthus). Mem. Torrey Bot. Club 22 1–218. [Google Scholar]
- Hoekstra, H. E., and M. W. Nachman, 2003. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 12 1185–1194. [DOI] [PubMed] [Google Scholar]
- Hu, F. Y., D. Y. Tao, E. Sacks, B. Y. Fu, P. Xu et al., 2003. Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. USA 100 4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., D. L. Bidney, N. Yalpani, J. P. Duvick, O. Crasta et al., 2003. Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol. 133 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger, T. E., T. Wayne, S. Boles, V. V. Symonds, J. McKay et al., 2006. Natural genetic variation in whole-genome expression in Arabidopsis thaliana: the impact of physiological QTL introgression. Mol. Ecol. 15 1351–1365. [DOI] [PubMed] [Google Scholar]
- Kane, N. C., and L. H. Rieseberg, 2007. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics 175 1823–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, N. C., and L. H. Rieseberg, 2008. Genetics and evolution of weedy Helianthus annuus populations: adaptation of an agricultural weed. Mol. Ecol. 17 384–394. [DOI] [PubMed] [Google Scholar]
- Kendziorski, C., R. A. Irizarry, K. S. Chen, J. D. Haag and M. N. Gould, 2005. On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. USA 102 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., G. Weiss, M. Lachmann, I. Hellmann, W. Enard et al., 2004. A neutral model of transcriptome evolution. PLoS Biol. 2 E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich, P., W. Enard, M. Lachmann and S. Paabo, 2006. Evolution of primate gene expression. Nat. Rev. Genet. 7 693–702. [DOI] [PubMed] [Google Scholar]
- Lai, Z., B. L. Gross, Y. Zou, J. Andrews and L. H. Rieseberg, 2006. Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 15 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. E., 2002. Evolutionary genetics of invasive species. Trends Ecol. Evol. 17 386–391. [Google Scholar]
- Liang, L., Z. Lai, W. Ma, Y. Zhang and Y. Xue, 2002. AhSL28, a senescence- and phosphate starvation-induced S-like RNase gene in Antirrhinum. Biochim. Biophys. Acta 1579 64–71. [DOI] [PubMed] [Google Scholar]
- Massinga, R. A., K. Al-Khatib, P. S. Amand and J. F. Miller, 2003. Gene flow from imidazolinone-resistant domesticated sunflower to wild relatives. Weed Sci. 51 854–862. [Google Scholar]
- McGregor, A. P., V. Orgogozo, I. Delon, J. Zanet, D. G. Srinivasan et al., 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448 587–590. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., M. L. Wayne, K. L. Harmon and L. M. McIntyre, 2004. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol. Biol. Evol. 21 1308–1317. [DOI] [PubMed] [Google Scholar]
- Oleksiak, M. F., K. J. Kolell and D. L. Crawford, 2001. Utility of natural populations for microarray analyses: isolation of genes necessary for functional genomic studies. Mar. Biotechnol. 3 S203–S211. [DOI] [PubMed] [Google Scholar]
- Oleksiak, M. F., G. A. Churchill and D. L. Crawford, 2002. Variation in gene expression within and among natural populations. Nat. Genet. 32 261–266. [DOI] [PubMed] [Google Scholar]
- Oleksiak, M. F., J. L. Roach and D. L. Crawford, 2005. Natural variation in cardiac metabolism and gene expression in Fundulus heteroclitus. Nat. Genet. 37 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., K. F. Schertz, Y. R. Lin, S. C. Liu and Y. L. Chang, 1995. The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc. Natl. Acad. Sci. USA 92 6127–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimental, D., L. Lach, R. Zuniga and D. Morrison, 2000. Environmental and economic costs of non-indigenous species in the United States. Biosciences 50 53–65. [Google Scholar]
- Prud'homme, B., N. Gompel, A. Rokas, V. A. Kassner, T. M. Williams et al., 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440 1050–1053. [DOI] [PubMed] [Google Scholar]
- Richards, C. L., O. Bossdorf, N. Z. Muth, J. Gurevitch and M. Pigliucci, 2006. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9 981–993. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., S. C. Kim, R. A. Randell, K. D. Whitney, B. L. Gross et al., 2007. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129 149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin, S. A., J. Kim and K. P. White, 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat. Genet. 33 138–144. [DOI] [PubMed] [Google Scholar]
- Roach, D. A., and R. D. Wulff, 1987. Maternal effects in plants. Annu. Rev. Ecol. Syst. 18 209–235. [Google Scholar]
- Rogers, W. E., and E. Siemann, 2005. Herbivory tolerance and compensatory differences in native and invasive ecotypes of Chinese tallow tree (Sapium sebiferum). Plant Ecol. 181 57–68. [Google Scholar]
- Salekdeh, G. H., J. Siopongco, L. J. Wade, B. Ghareyazie and J. Bennett, 2002. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2 1131–1145. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Z. Chen, H. Du, Y. Liu and D. F. Klessig, 1997. Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11 993–1005. [DOI] [PubMed] [Google Scholar]
- Tanaka, R., U. Oster, E. Kruse, W. Rudiger and B. Grimm, 1999. Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C. B., P. A. Bariola, S. B. delCardayre, R. T. Raines and P. J. Green, 1993. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc. Natl. Acad. Sci. USA 90 5118–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D., 1994. Competition and biodiversity in spatially structured habitats. Ecology 75 2–16. [Google Scholar]
- Torres, A. M., 1974. Genetics of sunflower alcohol dehydrogenase: Adh2, nonlinkage to Adh1 and Adh1 early alleles. Biochem. Genet. 12 385–392. [DOI] [PubMed] [Google Scholar]
- Torres, A. M., and U. Diedenhofen, 1979. Baker sunflower populations revisited. J. Hered. 70 275–276. [Google Scholar]
- Whitehead, A., and D. L. Crawford, 2006. a Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. USA 103 5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, A., and D. L. Crawford, 2006. b Variation within and among species in gene expression: raw material for evolution. Mol. Ecol. 15 1197–1211. [DOI] [PubMed] [Google Scholar]
- Whitney, K. D., R. A. Randell and L. H. Rieseberg, 2006. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am. Nat. 167 794–807. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., 2003. Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 47 675–684. [PubMed] [Google Scholar]