Abstract
Many mutations cause obvious abnormalities only when combined with other mutations. Such synthetic interactions can be the result of redundant gene functions. In Caenorhabditis elegans, the synthetic multivulva (synMuv) genes have been grouped into multiple classes that redundantly inhibit vulval cell fates. Animals with one or more mutations of the same class undergo wild-type vulval development, whereas animals with mutations of any two classes have a multivulva phenotype. By varying temperature and genetic background, we determined that mutations in most synMuv genes within a single synMuv class enhance each other. However, in a few cases no enhancement was observed. For example, mutations that affect an Mi2 homolog and a histone methyltransferase are of the same class and do not show enhancement. We suggest that such sets of genes function together in vivo and in at least some cases encode proteins that interact physically. The approach of genetic enhancement can be applied more broadly to identify potential protein complexes as well as redundant processes or pathways. Many synMuv genes are evolutionarily conserved, and the genetic relationships we have identified might define the functions not only of synMuv genes in C. elegans but also of their homologs in other organisms.
GLOBAL analyses of loss-of-function mutants have revealed that many null mutations do not cause obvious phenotypic abnormalities (Park and Horvitz 1986; Winzeler et al. 1999; Fraser et al. 2000; Kamath et al. 2003). In some cases, the wild-type phenotypes of such mutants result from genetic redundancy, which has been suggested to confer phenotypic robustness to developmental and behavioral processes (Harrison et al. 2007b; Sieber et al. 2007). Genetic redundancy can be studied using synthetic interactions, in which an animal mutant in two genes displays a phenotype that is not seen in either single mutant. Genes that display such a synthetic interaction are typically thought to act in separate processes or pathways to mediate the same broad biological function. Numerous synthetic-lethal interactions have been identified in Saccharomyces cerevisiae (Tong et al. 2001, 2004; Ooi et al. 2003). In the nematode Caenorhabditis elegans, mutations in the synthetic multivulva (synMuv) genes cause synthetic defects in the development of the hermaphrodite vulva (Ferguson and Horvitz 1989).
In C. elegans, three of six equipotent progenitor cells are specified to adopt vulval cell fates by three cell-signaling cascades involving receptor tyrosine kinase (RTK)/Ras, Notch, and Wnt pathways (Sundaram 2005; Sternberg 2006). The three cells that adopt vulval fates divide several times and give rise to the vulva. The remaining three cells divide once, and their progeny adopt nonvulval fates and fuse with the underlying hypodermis (Sulston and Horvitz 1977). The synMuv genes prevent the adoption of ectopic vulval cell fates (Ferguson and Horvitz 1989).
The synMuv genes originally were grouped into two classes, A and B; animals mutant for one or more genes within the same synMuv class do not have vulval abnormalities, but a multivulva (Muv) phenotype is observed in class A–B double mutants (Ferguson and Horvitz 1989). This synthetic interaction indicates that the class A and class B synMuv genes act in two redundant pathways or processes to inhibit the adoption of vulval cell fates. The class A synMuv genes encode nuclear proteins proposed to regulate transcription (Clark et al. 1994; Huang et al. 1994; Davison et al. 2005). Many class B synMuv genes encode homologs of regulators of transcription and chromatin, including a nucleosome remodeling and deacetylase (NuRD) complex (Solari and Ahringer 2000; Unhavaithaya et al. 2002), an Rb/E2F4/DP complex (Lu and Horvitz 1998; Ceol and Horvitz 2001), two histone methyltransferases (Andersen and Horvitz 2007), heterochromatin protein 1 (HP1) (Couteau et al. 2002) and the DP/Rb/MuvB (DRM) complex (Harrison et al. 2006). Ceol and Horvitz (2004) proposed the existence of a third class of synMuv gene, called C. These genes encode proteins similar to members of the Tip60/NuA4 histone acetyltransferase complex. In this article, we show that these putative class C synMuv genes are not distinct from and should be considered class B synMuv genes. The gene lin-3, which encodes the EGF-like ligand of the RTK/Ras pathway required for the specification of vulval cell fates (Hill and Sternberg 1992), is an apparent direct or indirect transcriptional target of at least some synMuv genes (Cui et al. 2006). Given this observation and the molecular nature of many synMuv proteins, the synMuv proteins likely act as transcriptional repressors to prevent the ectopic expression of the RTK/Ras pathway ligand LIN-3 and in this way prevent ectopic vulval induction.
To date, there are at least 4 class A synMuv genes and 25 class B synMuv genes (Ferguson and Horvitz 1989; Lu and Horvitz 1998; Hsieh et al. 1999; von Zelewsky et al. 2000; Couteau et al. 2002; Dufourcq et al. 2002; Unhavaithaya et al. 2002; Thomas et al. 2003; Poulin et al. 2005; Ceol et al. 2006; Andersen and Horvitz 2007; Tseng et al. 2007). Two class B synMuv genes, met-1 and met-2, act redundantly with each other to inhibit vulval cell fates (Andersen and Horvitz 2007), and the Muv phenotypes caused by mutations in the class B synMuv genes let-418 and hpl-2 are enhanced by mutations in other class B synMuv genes (von Zelewsky et al. 2000; Couteau et al. 2002; Andersen and Horvitz 2007). These genetic interactions along with the molecular identities of the class B synMuv proteins led us to hypothesize that the class B genes do not act in a single pathway or process to inhibit vulval cell-fate specification.
By scoring the vulval phenotypes of strains sensitized using temperature and genetic background, we sought to determine whether genes within each of the two synMuv classes act together in the same pathway or process. In these assays, we compared the Muv phenotypes caused by single mutants with the Muv phenotypes caused by double mutants within the same synMuv class. We found that most but not all synMuv genes within a single class act in separate pathways or processes, as indicated by the more penetrant Muv phenotypes of double mutants within a class than those of the respective single mutants. The class C synMuv genes were originally postulated to define a separate class from the class B synMuv genes, because the putative class C mutations were weakly redundant with class B synMuv mutations. However, as our results show that nearly all class B synMuv genes are redundant with each other, the putative class C synMuv genes are not different from the class B synMuv genes. For the rest of this paper, we consider there to be only two synMuv classes, A and B.
In a few cases, we found that two genes within the same class did not show enhancement and hence might act in the same pathway or process. In several of these cases, the proteins encoded by these genes have been identified biochemically to be in a complex. We believe that such lack of genetic redundancy can identify sets of genes that mediate the same molecular function. Additionally, we showed that many synMuv proteins repress lin-3 transcription and that the redundancy observed in the vulval phenotype between class B synMuv genes likely results from an increase in lin-3 expression.
MATERIALS AND METHODS
Strains and genetics:
C. elegans was cultured on the bacterial strain OP50 as described (Brenner 1974). N2 was the wild-type strain. Mutant alleles used are listed below and in Table 1. Many alleles were described previously (Riddle et al. 1997) unless otherwise noted:
LGI: lin-35(n745) (Lu and Horvitz 1998), lin-53(n833) (Lu and Horvitz 1998), lin-61(n3447, n3809) (Harrison et al. 2007a), and met-1(n4337) (Andersen and Horvitz 2007).
LGII: lin-8(n2731) (Thomas et al. 2003), dpl-1(n3316) (Ceol and Horvitz 2001), trr-1(n3712) (Ceol and Horvitz 2004), lin-56(n2728) (Thomas et al. 2003), and lin-38(n751).
LGIII: lin-37(n4903) (Andersen et al. 2006), met-2(n4256) (Andersen and Horvitz 2007), lin-9(n112) (Beitel et al. 2000), lin-52(n771) (Thomas et al. 2003), and lin-36(n766) (Thomas and Horvitz 1999).
LGIV: ark-1(sy247) (Hopper et al. 2000).
LGV: let-418(n3536) (Ceol et al. 2006), hda-1(e1795) (Dufourcq et al. 2002), and mys-1(n3681) (Ceol and Horvitz 2004).
LGX: lin-15B(n744), lin-15A(n433, n767), and lin-15AB(e1763) (Ferguson and Horvitz 1985).
TABLE 1.
synMuv alleles used in this study
| Allele | Mutation | Null? | Complex | |
|---|---|---|---|---|
| Class A gene | ||||
| lin-8 | n2731 | Q113ochre | Yes | |
| lin-15A | n767 | Deletion | Yes | |
| lin-15A | n433 | A250Va | No | |
| lin-38 | n751 | R517Cb | Noc | |
| lin-56 | n2728 | Deletion | Yes | |
| Class B gene | ||||
| ark-1 | sy247 | Q528ochre | No | |
| dpl-1 | n3316 | Deletion | Yesd | DP/E2F/Rb, DRM |
| efl-1 | RNAie | NA | No | DP/E2F/Rb, DRM |
| hda-1 | e1795 | G182E | Noc | NuRD |
| let-418 | n3536 | P675L | Noc | NuRD |
| lin-9 | n112 | G341E | Noc | DRM |
| lin-15B | n744 | W485opal | Yes | |
| lin-35 | n745 | W151opal | Yes | DP/E2F/Rb, DRM |
| lin-36 | n766 | Y796ochre | No | |
| lin-37 | n4903 | Deletion | Yes | DRM |
| lin-52 | n771 | E36K | Noc | DRM |
| lin-53 | n833 | L292F | Noc | DRM, NuRD |
| lin-61 | n3809 | Q159ochre | Yes | |
| lin-61 | n3447 | S354N | No | |
| met-1 | n4337 | Deletion | Yes | |
| met-2 | n4256 | Deletion | Yes | |
| mys-1 | n3681 | G341R | Noc | NuA4 |
| trr-1 | n3712 | W2593amber | Yesd | NuA4 |
E. Davison and H. R. Horvitz, unpublished results.
A. M. Saffer, E. Davison and H. R. Horvitz, unpublished results.
This mutation is nonnull. Null alleles cause sterility or larval arrest.
This mutation causes a recessive sterile phenotype. Homozygotes descended from heterozygous mothers could possess some maternally provided wild-type gene activity.
For efl-1, we used RNAi to create a partial loss of gene function.
The following balancer chromosomes were used: mIn1 [mIs14 dpy-10(e128)] (Edgley and Riddle 2001), hT2 [qIs48], nT1 [qIs51] (Mathies et al. 2003), and qC1 [nIs189] (Andersen et al. 2006).
RNAi analyses:
RNAi of efl-1 was performed by injection using clone yk617e4 to synthesize dsRNA, as previously described (Andersen et al. 2006).
Scoring the vulval cell fate:
Using a dissecting microscope, we scored the vulval phenotypes of each strain from at least three and in many cases nine independently grown cultures. At least 100 animals were scored for each genotype. Animals were scored as Muv if one or more ectopic ventral protrusions were observed. We scored vulval induction of a few strains using Nomarski optics and found the frequencies of ectopic inductions of vulval cell fates to be equivalent to the frequencies of the Muv phenotypes observed using a dissecting microscope. The penetrance of the Muv phenotype was calculated as the fraction of total animals that were Muv multiplied by 100. The average percentage of Muv and standard deviation of the replicates were calculated. A strain was scored as enhanced when the Muv phenotype penetrance of the double mutant was more than one standard deviation greater than the Muv phenotype penetrances of the respective single mutants. In some cases, enhancement was observed only when comparing the Muv phenotypes of double mutants to the respective single mutants at one of a number of temperatures tested.
Quantitative PCR assay:
Synchronized wild-type and mutant animals were grown, and larvae were harvested at or near the L2-to-L3 larval transition, when vulval induction occurs. Total RNA was extracted using Trizol (Invitrogen). First-strand cDNA was prepared from 1 μg total RNA using the SuperScript III first-strand synthesis supermix for qRT–PCR (Invitrogen). Each real-time reverse transcriptase (RT) PCR mix contained 10 ng of RT products, SyBR Green PCR master mix (Applied Biosystems), and 0.4 μm of each primer. The real-time PCR was performed in triplicate on an Eppendorf Mastercycler ep realplex2. Two or three independent samples of each genotype were prepared, and levels of lin-3 and rpl-26 were quantified from each biological replicate. The ΔCT values for lin-3 were determined using rpl-26 as the internal reference, and the ΔΔCT values were calculated for each genotype by comparison with the wild type. All changes were normalized to the wild type. The error shown is the standard deviation of relative lin-3/rpl-26 ratios for the biological replicates.
RESULTS
To assess whether different synMuv genes function in the same process, we combined null or strong loss-of-function mutations (Table 1) and quantified the penetrance of the phenotype as compared to the phenotype of either single mutant. For simplicity below, we will use the word “process” instead of the phrase “pathway or process” to describe how groups of genes act together, but in each case such genes could act simultaneously (e.g., by encoding members of a protein complex) or sequentially (e.g., in a linear pathway). If a double synMuv mutant had a Muv phenotype equivalent in penetrance to that of either of the single mutants, we concluded that the two genes could act in the same process. By contrast, if a double synMuv mutant had a Muv phenotype that was higher in penetrance than that of either single mutant, we concluded that the two genes function in separate processes. The only way to score an increase in penetrance was to score strains in which the penetrance of the Muv phenotype is incomplete. We used both temperature and the addition of partial loss-of-function alleles of synMuv genes to sensitize the vulval phenotype to incomplete penetrances, increasing our ability to detect enhancement.
Temperature sensitizes the vulval phenotype:
As observed previously (Ferguson and Horvitz 1989; Clark et al. 1994; Lu and Horvitz 1998; Hsieh et al. 1999; Beitel et al. 2000; von Zelewsky et al. 2000; Ceol and Horvitz 2001; Unhavaithaya et al. 2002; Thomas et al. 2003; Ceol et al. 2006; Andersen and Horvitz 2007; Harrison et al. 2007a), single null mutations of synMuv genes did not cause a Muv phenotype at 20° or 25°, but class A mutations combined with class B mutations caused Muv phenotypes. The Muv phenotype caused by synMuv mutations is temperature sensitive: at higher temperatures the penetrance of the Muv phenotype increases for all tested synMuv double mutants (Ferguson and Horvitz 1989) and see below. For example, the penetrance of the lin-61(n3447); lin-56(n2728) Muv phenotype increased from 5% at 20° to 81% at 22.5°, and the penetrance of the lin-35(n745); lin-15A(n433) Muv phenotype increased from 11% at 15° to 100% at 20°. Both lin-56(n2728) and lin-35(n745) are presumptive null alleles, and some synMuv class A–B null double mutants have been shown to have temperature-sensitive Muv phenotypes (Ceol and Horvitz 2004; Ceol et al. 2006; Andersen and Horvitz 2007). Therefore, it is likely that it is the synMuv process and not the synMuv gene products that is temperature sensitive. Most synMuv double mutants have more penetrant Muv phenotypes at high temperatures and incompletely penetrant Muv phenotypes at low temperatures.
Partial loss-of-function mutations in class A or class B synMuv genes can sensitize the vulval phenotype:
In addition to temperature, genetic background can sensitize the vulval phenotype of synMuv mutant strains. To seek redundancy between class A genes, we partially inhibited a class B synMuv gene using the missense mutation lin-61(n3447) (Harrison et al. 2007a), which causes an incompletely penetrant synMuv defect in combination with strong or null class A mutations. This incompletely penetrant Muv phenotype might be enhanced by other null or strong class A mutations if the genes act in separate processes to inhibit vulval fates. To seek redundancy between class B genes, we partially inhibited a class A synMuv gene using the missense mutation lin-15A(n433), which causes a weak class A synMuv defect (Clark et al. 1994). The Muv phenotype caused by a partial loss of lin-15A function in combination with a null class B synMuv mutation might be enhanced by other null or strong class B mutations if these two class B synMuv genes act in separate processes to inhibit vulval fates.
Most class A–A double mutants have Muv phenotypes:
To look for enhancement, we scored strains at temperatures in which the Muv phenotypes were <100% penetrant and phenotypic enhancement could be quantified easily. We observed the vulval phenotypes of animals with null mutations in lin-8, lin-15A, and lin-56 (Table 1). We used a partial loss-of-function allele of lin-38, because a null mutation caused larval lethality, which precluded scoring of the vulval phenotype (A. M. Saffer and H. R. Horvitz, unpublished results). Whereas class A single mutants do not have Muv phenotypes at 20° (Ferguson and Horvitz 1989; Thomas et al. 2003; Davison et al. 2005), some class A synMuv mutations caused very weak Muv phenotypes at 25°, with <1% penetrance (Table 2). We also observed the vulval phenotypes of class A–A double mutants (Table 2). Most strains were not Muv at 20°, but many had Muv phenotypes at 25°, ranging from 1 to 33% penetrance. Only the class A–A double mutants lin-8 lin-56 and lin-56; lin-15A did not have a Muv phenotype greater than that of either single mutant at 25°.
TABLE 2.
Most class A synMuv double mutants have multivulva phenotypes
| % multivulva ± SDb
|
||
|---|---|---|
| Genotypea | 20° | 25° |
| Wild type | 0 ± 0 | 0 ± 0 |
| lin-8 | 0 ± 0 | 0 ± 0 |
| lin-56 | 0 ± 0 | 1 ± 0 |
| lin-38 | 0 ± 0 | 1 ± 1 |
| lin-15Ac | 0 ± 0 | 0 ± 0 |
| lin-8 lin-56 | 0 ± 0 | 0 ± 0 |
| lin-8 lin-38 | 1 ± 1 | 7 ± 2 |
| lin-8; lin-15Ac | 0 ± 0 | 13 ± 6 |
| lin-56 lin-38 | 0 ± 0 | 6 ± 4 |
| lin-56; lin-15Ac | 0 ± 0 | 1 ± 0 |
| lin-38; lin-15Ac | 0 ± 0 | 33 ± 10 |
The alleles used are described in Table 1.
Percentage of multivulva was determined as described in materials and methods. SD, standard deviation.
The null allele lin-15A(n767) was used for these analyses.
Therefore, not only do the class A and B synMuv genes act in separate processes, but, given these results, most class A synMuv genes can act in processes separate from those of other class A synMuv genes. Given that lin-8 lin-56 and lin-56; lin-15A double mutants were not enhanced when compared to their respective single mutants, the two genes in each of these gene pairs might act in the same process. Alternatively, they might act in different processes, with the decrease in synMuv gene function in these experiments being insufficient to reach the threshold necessary to cause a Muv phenotype, just as certain synMuv gene double mutants were Muv only if strong loss-of-function alleles were used, e.g., lin-8(n2741); lin-15B(n2245) was 1% Muv at 15°, while lin-8(n2376); lin-15B(n2245) was 98% Muv at 15° (Davison et al. 2005). Because the lin-8; lin-15A double mutant had an enhanced vulval phenotype, indicating that lin-8 and lin-15A act in separate processes, one or both of the class A–A synMuv gene pairs lin-8 lin-56 and lin-56; lin-15A must act in parallel.
lin-15A and lin-56 act in the same process to inhibit the specification of vulval cell fates:
For class A–A double mutants that contained at least one null mutation and for which no enhancement was observed, either the two synMuv genes act in the same process or they act in different processes and loss of function of both genes did not cause sufficient loss of inhibition of vulval cell fates to result in a Muv phenotype. Addition of a partial loss-of-function mutation in a synMuv gene from a different synMuv class could allow the detection of subtle redundancy by increasing the loss of inhibition of vulval fates to result in a Muv phenotype. We combined class A single or class A–A double mutations with the class B synMuv mutation lin-61(n3447).
We found that single class A synMuv mutations in combination with lin-61(n3447) caused Muv phenotypes at 20° and 22.5° (Table 3). In a lin-61(n3447) background, the Muv phenotypes caused by lin-15A or lin-38 mutations were more penetrant than those caused by lin-8 or lin-56, indicating that lin-15A and lin-38 cause a stronger inhibition of vulval cell fates. Because lin-8 and lin-56 mutations each cause a relatively weak inhibition of vulval cell fates, the lack of enhancement observed in lin-8 lin-56 and lin-56; lin-15A double mutants at 25° could be because neither class A–A double mutant had sufficient loss of vulval cell-fate inhibition to cause a Muv phenotype.
TABLE 3.
Mutations in most class A synMuv genes enhance the Muv phenotypes caused by mutations in other class A synMuv genes in a class B synMuv mutant background
| lin-8 | lin-15A | lin-38 | lin-56 | |
|---|---|---|---|---|
| 20° | ||||
| lin-8 | 8 ± 6 | 60 ± 7 | 69 ± 8 | 21 ± 11 |
| lin-15A | 21 ± 8 | 78 ± 10 | 14 ± 7 | |
| lin-38 | 32 ± 18 | 49 ± 13 | ||
| lin-56 | 5 ± 1 | |||
| 22.5° | ||||
| lin-8 | 69 ± 3 | 100 ± 1 | 100 ± 0 | 95 ± 2 |
| lin-15A | 97 ± 2 | 100 ± 0 | 97 ± 2 | |
| lin-38 | 99 ± 1 | 100 ± 0 | ||
| lin-56 | 81 ± 8 |
Each strain was grown at the temperature shown. All strains were homozygous for the class B mutation lin-61(n3447). The alleles used are described in Table 1. lin-15A(n767) was used in these analyses. Class A–A–B triple mutants with Muv phenotypes not enhanced when compared to the respective class A–B double mutants are in boldface type. The percentage of Muv was determined as described in materials and methods, and the average percentage of Muv and standard deviation are shown.
To address possible redundancy between the class A synMuv genes more sensitively, we constructed triple mutants with two class A synMuv genes and the class B synMuv gene lin-61 (Table 3). The group of class A–A mutants that with an added lin-61 mutation had Muv phenotypes greater than that of each respective class A; lin-61 double mutant included the group of class A–A double mutants that had Muv phenotypes at 25°; thus, the two class A synMuv genes in each of these double mutant combinations were shown to act in separate processes by two different observations. The Muv phenotype of the lin-61; lin-8 lin-56 triple mutant was stronger than the Muv phenotypes of either lin-61; lin-8 or lin-61; lin-56 animals at 20° and 22.5°. Therefore, lin-8 and lin-56 act in different processes. By contrast, the Muv phenotype of the lin-61; lin-56; lin-15A triple mutant was not stronger than lin-61; lin-56 or lin-61; lin-15A. Because all other class A; lin-15A; lin-61 and class A; lin-56; lin-61 triple mutants had Muv phenotypes enhanced as compared to the respective class A; lin-61 double mutants, the lack of enhancement observed in the lin-61; lin-56; lin-15A triple mutant suggests that lin-56 and lin-15A act in the same process.
Most class B–B double mutants do not have Muv phenotypes:
We also assayed redundancy among the class B synMuv genes using temperature to sensitize the vulval cell-fate decision. We used null mutations of the class B synMuv genes dpl-1, lin-35, lin-61, lin-37, met-1, met-2, and trr-1 (Table 1). We used partial loss-of-function alleles of let-418, lin-52, and mys-1, because null alleles cause larval lethality or sterility. We studied the pairwise interactions among this set of 10 class B synMuv genes comprehensively. We did not test all 378 pairwise interactions involving all 28 class B genes. Unlike the class A genes, most class B single and class B–B double synMuv mutants did not have Muv phenotypes at 20° or 25° (Table 4). The exceptions were let-418(n3536) and trr-1(n3712), each of which caused very low penetrance Muv defects as single mutants. Many class B–B double mutants had larval-lethal phenotypes at 25°, precluding scoring vulval phenotypes at high temperature. These larval-lethal phenotypes indicate that in the control of viability the class B synMuv genes act redundantly, but this redundancy might not be indicative of functions in vulval development. Also, the class B–B double null mutants lin-61(n3809) lin-35(n745) and lin-61(n3809); lin-37(n4903) had very low penetrance Muv phenotypes. Because most class B–B double mutants were not appreciably Muv at high temperature, the class B synMuv genes might all act in the same process. Given that there are a number of examples of redundancy within the class B synMuv genes (von Zelewsky et al. 2000; Couteau et al. 2002; Andersen and Horvitz 2007), it is more likely that many of these genes act in separate processes, and the loss of inhibition of vulval cell fates caused by the class B synMuv mutations was insufficient to reach the threshold necessary to cause a Muv phenotype.
TABLE 4.
Class B synMuv single and class B–B double mutants do not have appreciable Muv phenotypes
| % multivulva ± SDb
|
||
|---|---|---|
| Genotypea | 20° | 25° |
| Wild type | 0 ± 0 | 0 ± 0 |
| lin-35 | 0 ± 0 | 0 ± 0 |
| lin-61c | 0 ± 0 | 0 ± 0 |
| dpl-1d | 0 ± 0 | 0 ± 0 |
| trr-1d | 2 ± 1 | 3 ± 2 |
| lin-37 | 0 ± 0 | 0 ± 0 |
| lin-52 | 0 ± 0 | 0 ± 0 |
| let-418 | 0 ± 0 | 1 ± 0 |
| mys-1 | 0 ± 0 | 0 ± 0 |
| efl-1 | 0 ± 0 | 0 ± 0 |
| lin-35 lin-61c | 0 ± 0 | 1 ± 0 |
| lin-35; lin-37 | 0 ± 0 | 0 ± 0 |
| lin-35; lin-52 | 0 ± 0 | 0 ± 1 |
| lin-35; let-418 | 0 ± 0 | Lvl |
| lin-35; mys-1 | 0 ± 0 | Lvl |
| lin-35; efl-1 | 0 ± 0 | 0 ± 0 |
| lin-61c; lin-37 | 0 ± 0 | 1 ± 1 |
| lin-61c; lin-52 | 0 ± 0 | 0 ± 0 |
| lin-61c; let-418 | 2 ± 2 | Lvl |
| lin-61c; mys-1 | 0 ± 0 | Lvl |
| dpl-1; efl-1d | 0 ± 0 | 0 ± 0 |
| trr-1; mys-1d | 4 ± 3 | Lvl |
| lin-37; let-418 | 0 ± 0 | 0 ± 0 |
| lin-37; mys-1 | 0 ± 0 | Lvl |
| lin-37; efl-1 | 0 ± 0 | 0 ± 0 |
| lin-52; let-418 | 0 ± 0 | 0 ± 0 |
| lin-52; mys-1 | 0 ± 0 | Lvl |
| let-418 mys-1 | 0 ± 0 | Lvl |
Lvl, larval lethality.
The alleles used are described in Table 1.
Percentage of multivulva was determined as described in materials and methods. SD, standard deviation.
The null allele lin-61(n3809) was used for these analyses.
These animals were descended from synMuv mutant heterozygotes, because the mutations cause recessive sterility, as described in Table 1.
Most class B synMuv genes act redundantly with genes of the same class to inhibit vulval cell fates:
To observe subtle redundancies among the class B synMuv genes and to assess the relative input of each of the class B genes into vulval cell-fate inhibition, we combined class B single and class B–B double mutants with the weak class A mutation lin-15A(n433) (Tables 5 and 6). We tested many class B genes with viable null phenotypes. dpl-1, lin-35, lin-37, and met-2 caused the strongest inhibition of vulval cell fates, as mutants in each of these genes had highly penetrant Muv phenotypes at 17.5° and 20° in combination with lin-15A(n433). Mutants in lin-61, lin-52, let-418, met-1, mys-1, and trr-1 did not have appreciably Muv phenotypes at 17.5° or 20° in combination with lin-15A(n433), suggesting that these genes caused a weaker inhibition of vulval cell fates.
TABLE 5.
Mutations of most class B and C genes enhance the Muv phenotypes caused by mutations in other class B genes in a class A synMuv mutant background
| lin-35 | met-1 | dpl-1a | trr-1a | lin-37 | met-2 | lin-52 | lin-61 | let-418 | mys-1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 15° | ||||||||||
| lin-35 | 11 ± 3 | 93 ± 5 | 13 ± 3 | Lvl | 72 ± 6 | 98 ± 3 | 23 ± 3 | 97 ± 3 | 50 ± 5 | 67 ± 9 |
| met-1 | 0 ± 0 | 33 ± 15 | 65 ± 6 | 100 ± 0 | 62 ± 10 | 2 ± 1 | 4 ± 3 | 1 ± 1 | 1 ± 1 | |
| dpl-1 | 0 ± 0 | 55 ± 13 | 69 ± 5 | 90 ± 11 | 0 ± 0 | 46 ± 21 | 0 ± 0 | 0 ± 0 | ||
| trr-1 | 1 ± 1 | Lvl | 61 ± 6 | 11 ± 12 | 18 ± 10 | 6 ± 9 | 1 ± 2 | |||
| lin-37 | 4 ± 3 | 100 ± 0 | 22 ± 16b | 95 ± 3 | 33 ± 5 | 29 ± 5 | ||||
| met-2 | 7 ± 7 | 79 ± 7 | 67 ± 20 | 39 ± 4 | 20 ± 10 | |||||
| lin-52 | 0 ± 1 | 17 ± 7 | 0 ± 0 | 1 ± 1 | ||||||
| lin-61 | 0 ± 0 | 1 ± 1 | 1 ± 1 | |||||||
| let-418 | 0 ± 0 | 1 ± 1 | ||||||||
| mys-1 | 0 ± 0 | |||||||||
| 17.5° | ||||||||||
| lin-35 | 65 ± 10 | 96 ± 4 | 60 ± 3 | Lvl | 100 ± 0 | 100 ± 0 | 93 ± 10 | 100 ± 1 | 100 ± 0 | 99 ± 1 |
| met-1 | 0 ± 0 | 81 ± 5 | 90 ± 8 | 100 ± 0 | 95 ± 3 | 26 ± 18 | 16 ± 3 | 2 ± 2 | 15 ± 16 | |
| dpl-1 | 2 ± 2 | 90 ± 2 | 100 ± 0 | 99 ± 2 | 13 ± 3 | 91 ± 2 | 3 ± 1 | 0 ± 1 | ||
| trr-1 | 4 ± 5 | Lvl | 79 ± 4 | 64 ± 2 | 88 ± 3 | 3 ± 4 | 5 ± 7 | |||
| lin-37 | 59 ± 11 | 100 ± 1 | 92 ± 5b | 100 ± 0 | 96 ± 6 | 92 ± 7 | ||||
| met-2 | 59 ± 19 | 86 ± 14 | 98 ± 1 | 89 ± 7 | 95 ± 6 | |||||
| lin-52 | 0 ± 0 | 58 ± 18 | 1 ± 2 | 6 ± 6 | ||||||
| lin-61 | 1 ± 1 | 9 ± 2 | 9 ± 6 | |||||||
| let-418 | 0 ± 0 | 9 ± 2 | ||||||||
| mys-1 | 0 ± 0 | |||||||||
| 20° | ||||||||||
| lin-35 | 100 ± 0 | 100 ± 0 | 100 ± 0 | Lvl | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 |
| met-1 | 8 ± 4 | 99 ± 1 | 100 ± 1 | 100 ± 0 | 100 ± 1 | 86 ± 10 | 96 ± 6 | 6 ± 1 | 88 ± 6 | |
| dpl-1 | 70 ± 7 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 87 ± 2 | 99 ± 1 | 97 ± 1 | 84 ± 5 | ||
| trr-1 | 12 ± 5 | Lvl | 100 ± 0 | 98 ± 2 | 100 ± 0 | 53 ± 11 | 6 ± 6 | |||
| lin-37 | 100 ± 0 | 100 ± 0 | 100 ± 0b | 100 ± 0 | 100 ± 0 | 100 ± 0 | ||||
| met-2 | 99 ± 1 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | |||||
| lin-52 | 15 ± 6 | 100 ± 0 | 89 ± 10 | 97 ± 1 | ||||||
| lin-61 | 19 ± 17 | 94 ± 5 | 93 ± 6 | |||||||
| let-418 | 0 ± 0 | 31 ± 7 | ||||||||
| mys-1 | 2 ± 1 |
All strains were homozygous for the class A mutation lin-15A(n433). Each strain was grown at the temperature shown. The alleles used are described in Table 1. lin-61(n3809) was used in these analyses. Class A–B–B triple mutants with Muv phenotypes not enhanced when compared to the respective class A–B double mutants are in boldface type. The percentage of Muv was determined as described in materials and methods, and the average percentage of Muv and standard deviation are shown.
These animals were descended from synMuv mutant heterozygotes, because the mutations cause recessive sterility, as described in Table 1.
RNAi of lin-52 was used in the lin-37(n4903); lin-15A(n433) strain for this experiment.
TABLE 6.
Mutations in most class B synMuv genes enhance the Muv phenotypes caused by mutations of other class B synMuv genes in a class A synMuv mutant background
| % multivulva ± SDb
|
||||
|---|---|---|---|---|
| Genotypea | 15° | 17.5° | 18.5° | 20° |
| Wild type | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| ark-1 | 0 ± 0 | 0 ± 1 | 4 ± 2 | |
| dpl-1c | 0 ± 0 | 2 ± 2 | 70 ± 7 | |
| efl-1 | 0 ± 0 | 8 ± 12 | 92 ± 7 | |
| hda-1/+ | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| let-418 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| lin-9 | 4 ± 3 | 63 ± 11 | 81 ± 6 | 100 ± 0 |
| lin-35 | 10 ± 3 | 65 ± 10 | 86 ± 5 | 100 ± 0 |
| lin-36 | 0 ± 0 | 0 ± 0 | 5 ± 2 | |
| lin-37 | 4 ± 3 | 59 ± 11 | 32 ± 9 | 100 ± 0 |
| lin-52 | 0 ± 1 | 0 ± 0 | 0 ± 0 | 15 ± 6 |
| lin-53 | 0 ± 0 | 1 ± 1 | 7 ± 3 | 100 ± 1 |
| lin-61d | 0 ± 0 | 1 ± 1 | 19 ± 17 | |
| mys-1 | 0 ± 0 | 0 ± 0 | 2 ± 1 | |
| trr-1c | 1 ± 1 | 4 ± 5 | 12 ± 5 | |
| lin-35; ark-1 | 37 ± 23 | 92 ± 3 | 100 ± 0 | |
| lin-35; hda-1/+ | 36 ± 2 | 97 ± 1 | 100 ± 0 | |
| lin-36; let-418 | 1 ± 2 | 1 ± 0 | 99 ± 1 | |
| lin-36; mys-1 | 1 ± 1 | 15 ± 15 | 99 ± 1 | |
| lin-53; let-418 | 0 ± 1 | 4 ± 2 | 46 ± 13 | 99 ± 1 |
| lin-53; lin-9 | 13 ± 4 | 67 ± 9 | 93 ± 5 | 100 ± 0 |
| lin-53; lin-37 | 13 ± 1 | 42 ± 14 | 100 ± 0 | 100 ± 0 |
| lin-53; lin-52 | 0 ± 0 | 3 ± 2 | 16 ± 5 | 99 ± 1 |
| lin-53; mys-1 | 8 ± 13 | 47 ± 11 | 100 ± 1 | |
| lin-61d; ark-1 | 7 ± 8 | 34 ± 16 | 84 ± 26 | |
| lin-61d; hda-1/+ | 1 ± 1 | 7 ± 5 | 100 ± 0 | |
| lin-61d; lin-9 | 86 ± 12 | 100 ± 0 | 100 ± 0 | |
| lin-61d; lin-36 | 1 ± 1 | 21 ± 16 | 99 ± 1 | |
These strains were homozygous for the class A synMuv mutation lin-15A(n433). The alleles used are described in Table 1.
Percentage of multivulva was determined as described in materials and methods. SD, standard deviation.
These animals were descended from synMuv mutant heterozygotes, because the mutations cause recessive sterility, as described in Table 1.
lin-61(n3809) was used in these analyses.
We found using a sensitized genetic background that most class B genes act in separate processes to inhibit vulval cell fates (Tables 5 and 6). In combination with the weak class A mutation lin-15A(n433), most combinations of class B–B double mutants had more highly penetrant Muv phenotypes than those found in the respective single mutants. This difference was observed readily in strong double mutant combinations at 15°, intermediate combinations at 17.5°, and weak combinations at 20°. For example three weak synMuv genes, let-418, lin-52, and met-1, show insignificant enhancement at 17.5°, but at 20° enhancement is observed between let-418 and lin-52 but not between let-418 and met-1.
Some synMuv genes within the same class function nonredundantly to inhibit vulval cell fates:
Several class B–B double mutants had Muv phenotypes that were not enhanced as compared to the single class B mutants in a lin-15A(n433) background, indicating that these gene pairs likely act in the same process to inhibit vulval cell fates (Tables 5 and 6). Specifically, three pairs of genes—lin-35 and dpl-1; let-418 and met-1; and mys-1 and trr-1—did not show synthetic enhancement in a lin-15A(n433) background. This lack of genetic enhancement was detected even though these combinations involve alleles that in other combinations are sufficiently strong to cause Muv phenotypes. We suggest that our findings identify sets of genes that act nonredundantly, i.e., in the same processes.
lin-35 and dpl-1 encode the C. elegans homologs of human Rb and DP, respectively. Rb mediates transcriptional repression of the genes bound by the heterodimeric transcription factor E2F, which is composed of the proteins DP and E2F (Krek et al. 1993). In C. elegans, DPL-1 DP and EFL-1 E2F interact in vitro (Ceol and Horvitz 2001), and their homologs interact both in vitro and in vivo in other organisms (Bandara et al. 1993; Helin et al. 1993; Krek et al. 1993). We tested whether RNAi of the C. elegans homolog of E2F, efl-1 (Ceol and Horvitz 2001), would enhance the Muv phenotypes of lin-35; lin-15A and dpl-1; lin-15A mutants. RNAi of efl-1 in a lin-15A(n433) background caused a Muv phenotype that was 8 ± 12% penetrant at 17.5° (average ± standard deviation, see materials and methods). At 17.5°, dpl-1(n3316); lin-15A(n433) and lin-35(n745); lin-15A(n433) animals had 2 ± 2% and 65 ± 10% penetrant Muv phenotypes, respectively. RNAi of efl-1 in dpl-1(n3316); lin-15A(n433) or lin-35(n745); lin-15A(n433) mutants did not enhance the Muv phenotypes at 17.5° (7 ± 10% Muv and 72 ± 11% Muv, respectively). These data indicate that efl-1 E2F and dpl-1 DP, which together encode a heterodimeric transcription factor, act to inhibit vulval fates in the same molecular process both with each other and with lin-35 Rb.
We note that because null mutations of dpl-1 and trr-1 cause sterility, the dpl-1 and trr-1 homozygous animals we scored were descended from heterozygous mothers. Therefore, these animals could have maternal rescue of dpl-1 or trr-1. Additionally, RNAi of efl-1 and mys-1(n3681) likely caused partial loss of gene function, as null alleles of each gene cause sterility. We believe that the loss of gene function of each of these four genes was sufficient to allow us to detect enhancement of the Muv phenotype by our assays, because each allele or RNAi enhanced the Muv phenotypes of other synMuv combinations. For example, RNAi of efl-1 enhanced the Muv phenotype of lin-37(n4903); lin-15A(n433) mutants from 59 to 100% at 17.5°, and mys-1(n3681) enhanced other Muv phenotypes (Tables 5 and 6).
We chose several other class B synMuv genes to test in a limited number of class B–B double mutant combinations in a lin-15A(n433) background. Null or strong mutations in ark-1, lin-9, or lin-36 enhanced the Muv phenotypes caused by other class B genes (Table 6). The mutation hda-1(e1795) causes a recessive Muv phenotype (Dufourcq et al. 2002), and the Muv phenotype is enhanced by both class A and class B synMuv mutations (E. C. Andersen and H. R. Horvitz, unpublished results). Heterozygous hda-1(e1795) in three different mutant class B synMuv backgrounds caused enhancement of those Muv phenotypes, indicating that lin-35, lin-37, and lin-61 function in separate processes from hda-1. Using a strong partial loss-of-function allele of the class B synMuv gene lin-53 RbAp48 (Lu and Horvitz 1998), we found that in a lin-15A(n433) background lin-53(n833) failed to enhance the Muv phenotypes caused by lin-9(n112), lin-37(n4903), lin-52(n771), and let-418(n3536) at 17.5° or 20° but did enhance the Muv phenotype caused by mys-1(n3681). Because four class B; lin-53; lin-15A triple mutants were weakly Muv at 17.5° and completely Muv at 20°, we scored each of these strains at the intermediate temperature 18.5°. At this temperature, weak to moderate enhancement of the Muv phenotype was observed (Table 6), indicating that these genes act in separate processes.
In short, we identified four sets of synMuv genes—lin-15A and lin-56; lin-35, dpl-1 and efl-1; let-418 and met-1; and mys-1 and trr-1—for which the genes within each set likely act together to inhibit vulval fates.
The penetrances of synMuv vulval phenotypes correlate with the level of lin-3 mRNA expression:
Cui et al. (2006) reported that the lin-3 gene, which encodes the EGF ligand of the Ras pathway is a transcriptional target of some synMuv proteins. Three double mutants carrying a class A and a class B synMuv mutation had increased lin-3 EGF mRNA levels. We determined whether each of the class A genes and the class B genes we studied repress lin-3 transcription and whether the redundancy we found to control the vulval phenotype also controls lin-3 transcription.
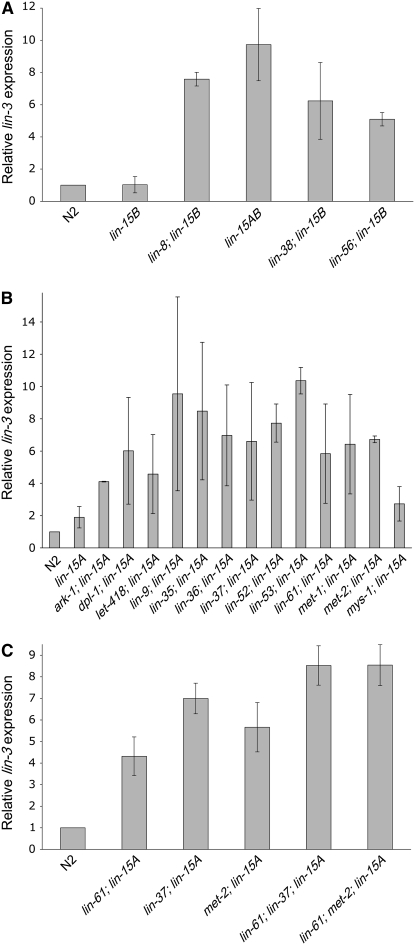
We found that all class A–B double mutant strains between mutations in each of the four class A genes and lin-15B(n744) had increased lin-3 mRNA levels as compared to the wild type (Figure 1A). These results suggest that all four class A synMuv proteins repress the transcription of lin-3 EGF. All class A–B double mutant strains between mutations in each of the 13 class B genes used in this study and lin-15A(n767) also had increased lin-3 mRNA levels as compared to the wild type (Figure 1B). This analysis greatly expands the number of genes implicated in the transcriptional repression of lin-3.
Figure 1.—
Regulation of lin-3 transcription by the synMuv genes. Real-time RT–PCR experiments were performed on the strains shown. Mean ΔΔCT values were used to calculate relative changes in lin-3 expression normalized to levels of rpl-26. Mean values and standard deviations of relative lin-3/rpl-26 ratios for the trials are shown. (A) Mutation of each class A synMuv gene in a class B synMuv mutant background has increased lin-3 transcription. (B) Mutation of each class B synMuv gene in a class A synMuv mutant background has increased lin-3 transcription. lin-15A(n767) and lin-61(n3809) were used for this experiment. (C) Two class B–B–A synMuv triple mutants have increased levels of lin-3 transcription compared to their respective class B–A double mutants. lin-15A(n433) and lin-61(n3809) were used for this experiment.
The level of increased lin-3 transcription in the class A–B synMuv double mutants roughly correlated with the phenotypic strength of the respective Muv phenotypes (supplemental Figure S1). For example, double mutants between lin-9 or lin-35 and a class A gene had very strong Muv phenotypes and high levels of lin-3 transcription. By contrast, mys-1; lin-15A animals had a weak Muv phenotype and a lower increase in lin-3 transcription.
Notably, ark-1(sy247); lin-15A(n767) had an increased level of lin-3 transcription, even though ark-1 encodes a protein proposed to directly downregulate Ras pathway activity through SEM-5 (Hopper et al. 2000). One possibility is that there is positive feedback through the Ras pathway during vulval induction, so that Ras pathway activity elevates levels of lin-3 mRNA. We measured the level of lin-3 transcription in mutants carrying the gain-of-function let-60 Ras allele n1046 and found lin-3 transcription was slightly increased (2.8-fold ± 0.9-fold) as compared to the wild type. Therefore, it is possible that at least some of the increase in lin-3 mRNA levels in the ark-1(sy247); lin-15A(n767) mutant is caused by positive feedback.
We found that the Muv phenotypes of many class A–B–B synMuv triple mutants were enhanced as compared to the respective class A–B double mutants. We wondered if this redundancy extends to the control of lin-3 transcriptional repression. We compared the lin-3 transcription levels of lin-61(n3809); lin-15A(n433), lin-37(n4903); lin-15A(n433), and met-2(n4256); lin-15A(n433) double mutants to the lin-3 expression of lin-61(n3809); lin-37(n4903); lin-15A(n433) and lin-61(n3809); met-2(n4256); lin-15A(n433) triple mutants. We found that in each case there was an increase in lin-3 transcription in the triple mutants as compared to the respective double mutants (Figure 1C), indicating that for these synMuv genes there is redundant control of lin-3 transcriptional repression.
DISCUSSION
Genetic enhancement tests can distinguish processes that act in series or in parallel:
Genetic enhancement experiments are most simply interpreted when one or both of the mutations used in the study are null. If a null mutation is combined with another mutation, the phenotype can be more severe than the phenotypes of the respective single mutants if the genes act in parallel (redundant) processes but cannot be more severe if the genes act in the same process. At least one of the two mutations must be null for such a test, because enhancement of the mutant phenotype of a double mutant between two partial loss-of-function mutations could result even if the genes act in the same process. Additionally, the penetrance of the mutant phenotype must be incomplete, so that differences can be observed.
This type of genetic pathway analysis of biological systems defines logical relationships equivalent to “and” vs. “or” statements in mathematics and to “series” vs. “parallel” circuits in electrical engineering. If a double mutant has a phenotype more severe than that of either single mutant, the two genes must provide separate functions, e.g., act in parallel as a logical “or”; if either gene is active, some function is provided. By contrast, if a double mutant has a phenotype equivalent to that observed in either single mutant, the two genes might provide the same function, e.g., act together in series as a logical “and”; if either gene is inactive, no function is provided.
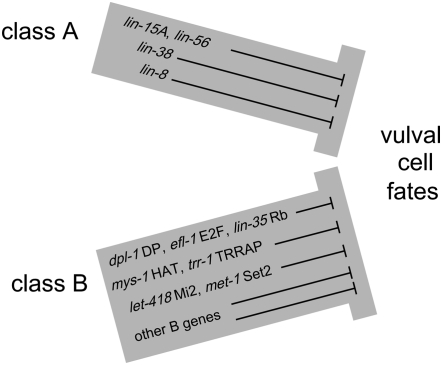
By these criteria, the class A and B synMuv genes act in parallel processes, as class A–B double mutants have almost completely penetrant Muv phenotypes at 20° (Ferguson and Horvitz 1989). It has been previously thought that within a synMuv gene class there is no enhancement in double mutants, in which case all of the synMuv genes within each synMuv class could act in a single process. However, we found that there are multiple levels of redundant processes involving the functions of both the class A and class B synMuv gene classes in inhibiting vulval cell fates (Figure 2). Most class A–A or class B–B double mutants either had Muv phenotypes or showed enhancement in a sensitized genetic background or at higher temperatures. By contrast, a few sets of genes within a single synMuv class failed to show enhancement and hence might act within the same process.
Figure 2.—
Model: The synMuv classes function in two redundant pathways or processes, each composed of separate molecular pathways or processes, to mediate the inhibition of vulval cell fates. The class A and B synMuv genes act in two distinct pathways to inhibit the expression of vulval cell fates. Additionally, each class is composed of many separate pathways as defined by genetic redundancy tests. A few synMuv genes act nonredundantly with each other: lin-15A and lin-56 act together within the class A genes, while within the class B genes dpl-1 DP, efl-1 E2F, and lin-35 Rb; mys-1 HAT and trr-1 TRRAP; and let-418 Mi2 and met-1 Set2 act together to inhibit vulval fates.
The synMuv genes define two distinct classes:
The class B synMuv genes met-1, met-2, hda-1, hpl-2, and let-418 cause a Muv phenotype in combination with either class A or class B synMuv mutations (von Zelewsky et al. 2000; Couteau et al. 2002; Dufourcq et al. 2002; Ceol and Horvitz 2004; Andersen and Horvitz 2007). Mutations in each of these genes cause stronger Muv phenotypes when combined with class A synMuv mutations than with class B synMuv mutations. The class C genes were considered a separate class because of their redundancy with both class A and class B genes. However, our findings indicate that the class C synMuv genes are not distinctive in this respect, as most class B synMuv genes are similarly weakly redundant with each other. Furthermore, many class A–A double mutants had weak Muv defects comparable in strength to or stronger than those of class B–C double mutants.
We propose that the synMuv genes should be classified on the basis of the strength of the Muv phenotype observed in a double mutant with a class A synMuv mutation as compared to a double mutant with a class B synMuv mutation. The double mutant combination with the more penetrant Muv phenotype distinguishes the class. For example, if a synMuv mutation when combined with a class A synMuv mutation caused an 80% penetrant Muv phenotype and when combined with a class B synMuv mutations caused a 15% penetrant Muv phenotype, this synMuv gene would be assigned to class B, because it is more Muv in combination with class A mutations than with class B mutations. Because mutations in met-1, met-2, hda-1, hpl-2, lin-13, let-418 or any of the class C genes all exhibit strong Muv phenotypes when combined with mutations in class A genes and weak Muv phenotypes when combined with mutations in class B genes, we suggest that each of these genes should be assigned as class B synMuv genes.
Lack of genetic enhancement can identify proteins that function in a complex or a process:
A few sets of synMuv genes in the same class did not display synthetic interactions. While all synMuv genes act redundantly with some other synMuv genes to inhibit vulval cell fates, a set of synMuv genes that acts nonredundantly might act together in a more specific molecular process, as discussed below.
Specifically, some sets of synMuv genes that did not show synthetic enhancement encode proteins expected or known to function together in vivo. For example, we found that dpl-1 DP, efl-1 E2F, and lin-35 Rb define a single process in the inhibition of vulval cell fates. The corresponding C. elegans proteins likely form a complex (Ceol and Horvitz 2001), and their homologs function together in a complex (Bandara et al. 1993; Helin et al. 1993; Krek et al. 1993). Similarly as shown previously (Ceol and Horvitz 2004), mys-1 HAT and trr-1 TRRAP define a single process, and their homologs have been identified together in a histone acetyltransferase complex (Ikura et al. 2000). Our data also indicate that the class A genes lin-15A and lin-56 act together in the same process. This observation is consistent with recent molecular data that indicate LIN-56 and LIN-15A are each required for the stability of the other in vivo and bind each other in vitro, supporting the hypothesis that LIN-15A and LIN-56 function in a protein complex (E. M. Davison and H. R. Horvitz, unpublished results). We suggest that our genetic enhancement tests have identified groups of genes that function together in protein complexes or that act sequentially in a pathway.
From our genetic enhancement tests of class B synMuv genes, we identified one pair of genes that had not been found previously to act together, met-1 and let-418. The two histone methyltransferases met-1 and met-2 act redundantly to control vulval cell-fate inhibition; met-1 is similar to the yeast histone H3 lysine 36 methyltransferase Set2p (Andersen and Horvitz 2007). let-418 encodes the C. elegans homolog of human Mi2, an ATP-dependent chromatin-remodeling enzyme found in the NuRD complex (von Zelewsky et al. 2000). To date, neither MET-1 nor its homologs have been found in NuRD-like complexes. Perhaps the NuRD-like complex requires MET-1 to function in the inhibition of vulval fates. For example, MET-1 might methylate histone H3 lysine 36 at genes that normally promote vulval cell fates, leading to recruitment of the NuRD-like complex and transcriptional repression.
It is possible that members of a protein complex could display synthetic enhancement. However, genes that display synthetic enhancement cannot encode proteins that work as essential components of the same complex (i.e., if either component is absent, the complex has no function) or as members of a linear pathway in which each step requires the prior step for activity (i.e., if any component is absent, the pathway has no output). Many class B synMuv proteins, including LIN-9, LIN-35, LIN-37, LIN-52, LIN-53, LIN-54, DPL-1, and EFL-1, are components of the C. elegans DRM complex (Harrison et al. 2006). Nonetheless, most double mutants among lin-9, lin-35, lin-37, lin-52, and lin-53 showed weak redundancy, suggesting that most proteins in the DRM complex have redundant functions during vulval development. These results suggest that either the DRM complex is composed of members that can functionally substitute for each other or the complex (which was isolated from embryos) does not function in the inhibition of vulval cell fates. The synthetic interactions between many DRM complex members were much weaker than interactions between non-DRM complex members, suggesting that complex components might have subtle redundant functions.
Genetic enhancement tests can identify functionally related groups of evolutionarily conserved but uncharacterized genes:
The gene lin-3, which encodes the EGF ligand for the receptor tyrosine kinase RTK/Ras signal transduction cascade that drives vulval development, is a transcriptional target of some synMuv proteins (Cui et al. 2006). We found that all of the synMuv genes we examined repress lin-3 transcription. Each set of synMuv genes we hypothesize to act together in a single process to inhibit vulval cell fates might mediate a specific mechanism of lin-3 transcriptional repression. For example, two sets of class B synMuv genes, dpl-1, efl-1, and lin-35; and mys-1 and trr-1, probably act redundantly to control two different processes of transcriptional repression, namely the DNA binding of a transcriptional repressor and histone acetylation, respectively. We suggest that a third set of class B synMuv genes, let-418 and met-1, mediates a movement and methylation of histones important for transcriptional repression in a process distinct from those of the previous two examples.
An understanding of the distinct mechanisms used by different sets of synMuv genes in lin-3 repression could define how the homologs of these genes control repression of target genes in other organisms. We suggest that genetic enhancement studies of additional synMuv gene interactions might implicate the homologs of conserved but uncharacterized synMuv genes in well-characterized processes and in this way facilitate the understanding of how these genes act and of how they interact with other gene processes in both development and disease.
Acknowledgments
We thank Na An for strain management and Hillel Schwartz and David Harris for critical reading of this manuscript. Strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Heath (NIH) National Center for Research Resources. Yuji Kohara of the National Institute of Genetics in Japan kindly provided the efl-1 cDNA clone. E.C.A. is an Anna Fuller Cancer Research Fellow, and H.R.H. is the David H. Koch Professor of Biology at the Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant GM-24663.
References
- Andersen, E. C., and H. R. Horvitz, 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134 2991–2999. [DOI] [PubMed] [Google Scholar]
- Andersen, E. C., X. Lu and H. R. Horvitz, 2006. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133 2695–2704. [DOI] [PubMed] [Google Scholar]
- Bandara, L. R., V. M. Buck, M. Zamanian, L. H. Johnston and N. B. La Thangue, 1993. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 12 4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel, G. J., E. J. Lambie and H. R. Horvitz, 2000. The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene 254 253–263. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7 461–473. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6 563–576. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., F. Stegmeier, M. M. Harrison and H. R. Horvitz, 2006. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics 173 709–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. G., X. Lu and H. R. Horvitz, 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F., F. Guerry, F. Muller and F. Palladino, 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., J. Chen, T. R. Myers, B. J. Hwang, P. W. Sternberg et al., 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10 667–672. [DOI] [PubMed] [Google Scholar]
- Davison, E. M., M. M. Harrison, A. J. Walhout, M. Vidal and H. R. Horvitz, 2005. lin-8, which antagonizes Caenorhabditis elegans Ras-mediated vulval induction, encodes a novel nuclear protein that interacts with the LIN-35 Rb protein. Genetics 171 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq, P., M. Victor, F. Gay, D. Calvo, J. Hodgkin et al., 2002. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol. 22 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M. L., and D. L. Riddle, 2001. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266 385–395. [DOI] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 325–330. [DOI] [PubMed] [Google Scholar]
- Harrison, M. M., C. J. Ceol, X. Lu and H. R. Horvitz, 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA 103 16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, M. M., X. Lu and H. R. Horvitz, 2007. a LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics 176 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, R., B. Papp, C. Pal, S. G. Oliver and D. Delneri, 2007. b Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. USA 104 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht et al., 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7 1850–1861. [DOI] [PubMed] [Google Scholar]
- Hill, R. J., and P. W. Sternberg, 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358 470–476. [DOI] [PubMed] [Google Scholar]
- Hopper, N. A., J. Lee and P. W. Sternberg, 2000. ARK-1 inhibits EGFR signaling in C. elegans. Mol. Cell 6 65–75. [PubMed] [Google Scholar]
- Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al., 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang et al., 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102 463–473. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Krek, W., D. M. Livingston and S. Shirodkar, 1993. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262 1557–1560. [DOI] [PubMed] [Google Scholar]
- Lu, X., and H. R. Horvitz, 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95 981–991. [DOI] [PubMed] [Google Scholar]
- Mathies, L. D., S. T. Henderson and J. Kimble, 2003. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development 130 2881–2892. [DOI] [PubMed] [Google Scholar]
- Ooi, S. L., D. D. Shoemaker and J. D. Boeke, 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35 277–286. [DOI] [PubMed] [Google Scholar]
- Park, E. C., and H. R. Horvitz, 1986. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 113 821–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin, G., Y. Dong, A. G. Fraser, N. A. Hopper and J. Ahringer, 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., T. Blumenthal, B. J. Meyer and J. R. Priess, 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Sieber, P., F. Wellmer, J. Gheyselinck, J. L. Riechmann and E. M. Meyerowitz, 2007. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134 1051–1060. [DOI] [PubMed] [Google Scholar]
- Solari, F., and J. Ahringer, 2000. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr. Biol. 10 223–226. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., 2006. Pathway to RAS. Genetics 172 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 110–156. [DOI] [PubMed] [Google Scholar]
- Sundaram, M. V., 2005. The love-hate relationship between Ras and Notch. Genes Dev. 19 1825–1839. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., C. J. Ceol, H. T. Schwartz and H. R. Horvitz, 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H., and H. R. Horvitz, 1999. The C. elegans gene lin-36 acts cell autonomously in the lin-35 Rb pathway. Development 126 3449–3459. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Tseng, R. J., K. R. Armstrong, X. Wang and H. M. Chamberlin, 2007. The bromodomain protein LEX-1 acts with TAM-1 to modulate gene expression in C. elegans. Mol. Genet. Genomics 278 507–518. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya, Y., T. H. Shin, N. Miliaras, J. Lee, T. Oyama et al., 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111 991–1002. [DOI] [PubMed] [Google Scholar]
- von Zelewsky, T., F. Palladino, K. Brunschwig, H. Tobler, A. Hajnal et al., 2000. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127 5277–5284. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]