Abstract
Homozygosity is a commonly used summary of allele-frequency distributions at polymorphic loci. Because high-frequency alleles contribute disproportionately to the homozygosity of a locus, it often occurs that most homozygotes are homozygous for the most frequent allele. To assess the relationship between homozygosity and the highest allele frequency at a locus, for a given homozygosity value, we determine the lower and upper bounds on the frequency of the most frequent allele. These bounds suggest tight constraints on the frequency of the most frequent allele as a function of homozygosity, differing by at most  and having an average difference of
and having an average difference of  − π2/18 ≈ 0.1184. The close connection between homozygosity and the frequency of the most frequent allele—which we illustrate using allele frequencies from human populations—has the consequence that when one of these two quantities is known, considerable information is available about the other quantity. This relationship also explains the similar performance of statistical tests of population-genetic models that rely on homozygosity and those that rely on the frequency of the most frequent allele, and it provides a basis for understanding the utility of extended homozygosity statistics in identifying haplotypes that have been elevated to high frequency as a result of positive selection.
− π2/18 ≈ 0.1184. The close connection between homozygosity and the frequency of the most frequent allele—which we illustrate using allele frequencies from human populations—has the consequence that when one of these two quantities is known, considerable information is available about the other quantity. This relationship also explains the similar performance of statistical tests of population-genetic models that rely on homozygosity and those that rely on the frequency of the most frequent allele, and it provides a basis for understanding the utility of extended homozygosity statistics in identifying haplotypes that have been elevated to high frequency as a result of positive selection.
THE concept of homozygosity appears ubiquitously in population genetics, in the context of mathematical theory as well as in statistical methods for data analysis. Consider a locus with K ≥ 2 alleles, for which the frequency of allele i is pi > 0 and for which the alleles are placed in decreasing order of frequency so that pi ≥ pj if i < j. For diploids, the fraction of homozygotes expected under the assumption of Hardy–Weinberg proportions can be defined as
 |
(1) |
where
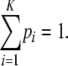 |
(2) |
In this article, we show that if all that is known about a locus is its expected homozygosity H, it is possible to localize the frequency p1 of its most frequent allele within a quite narrow range. Conversely, given p1, a narrow range can be specified for the value of H. Thus, we determine the upper and lower bounds on the frequency p1 of the most frequent allele as functions of homozygosity H. We also determine the bounds on H as functions of p1.
The connection between H and p1 provides a close relationship between two of the most basic quantities associated with a polymorphic locus. We use this relationship to explain a high correlation observed between H and p1 in human microsatellite data, as well as to provide a conceptual basis for the success of extended haplotype homozygosity methods in detecting positive selection. Note that expected heterozygosity under Hardy–Weinberg proportions is 1 − H; thus, by a simple transformation, our results can also be used to describe the relationship between heterozygosity and the frequency of the most frequent allele.
RESULTS
We consider a polymorphic locus with at least two alleles. We do not assume that the number of alleles with nonzero frequency is known; it is convenient to view the locus as having infinitely many alleles and to allow some of these alleles to have frequency 0. We refer to the frequency of the most frequent allele, p1, by M. Henceforth we use H and “homozygosity” to refer to expected homozygosity assuming Hardy–Weinberg proportions. Both M and H must lie in the interval (0, 1). The quantity ⌈x⌉ denotes the smallest integer larger than or equal to x. Our main results, which are proved in the appendix, are the bounds on M as functions of H (Theorem 1) and the bounds on H as functions of M (Theorem 2).
Theorem 1. Consider a sequence of the allele frequencies at a locus, 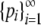 , with pi ∈ [0, 1),
, with pi ∈ [0, 1), 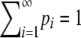 ,
, 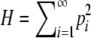 , M = p1, and i < j implies pi ≥ pj. Then
, M = p1, and i < j implies pi ≥ pj. Then
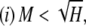 |
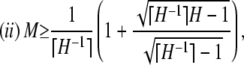 |
with equality if and only if pi = M for 1 ≤ i ≤ K − 1, pK = 1 − (K − 1)M, and pi = 0 for i > K, where K = ⌈H−1⌉ = ⌈M−1⌉.
Theorem 2. Consider a sequence of the allele frequencies at a locus, 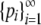 , with pi ∈ [0, 1),
, with pi ∈ [0, 1), 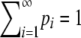 ,
, 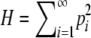 , M = p1, and i < j implies pi ≥ pj. Then (i) H > M2 and (ii) H ≤ 1 − M(⌈M−1⌉ − 1)(2 − ⌈M−1⌉M), with equality if and only if pi = M for 1 ≤ i ≤ K − 1, pK = 1 − (K − 1)M, and pi = 0 for i > K, where K = ⌈H−1⌉ = ⌈M−1⌉.
, M = p1, and i < j implies pi ≥ pj. Then (i) H > M2 and (ii) H ≤ 1 − M(⌈M−1⌉ − 1)(2 − ⌈M−1⌉M), with equality if and only if pi = M for 1 ≤ i ≤ K − 1, pK = 1 − (K − 1)M, and pi = 0 for i > K, where K = ⌈H−1⌉ = ⌈M−1⌉.
The bounds obtained in Theorems 1 and 2 are summarized in Table 1. Loosely speaking, Theorem 1 verifies that for a given homozygosity, the frequency of the most frequent allele is smallest when as many alleles as possible are tied as most frequent and greatest when there is one extremely frequent allele and many rare alleles. Theorem 2 shows that for a given frequency of the most frequent allele, homozygosity is smallest when many extremely rare alleles are present and greatest when as many alleles as possible are tied as most frequent. For each of the theorems, part i is straightforward to prove, and part ii follows from the fact that when considering all possible sets of nonnegative real numbers bounded above by a specified constant M and having a fixed sum C, the maximal sum of squares is obtained by greedily choosing as many of the numbers as possible to equal M and by assigning at most one additional number to be positive (Lemma 3 in the appendix).
TABLE 1.
Bounds on homozygosity and the frequency of the most frequent allele
| Quantity | Lower bound in terms of other quantity | Upper bound in terms of other quantity | Average difference between upper and lower bounds | Maximal difference between upper and lower bounds |
|---|---|---|---|---|
| M | 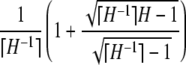 |
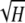 |
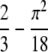 |
 at at 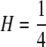
|
| H | M2 | 1 − M(⌈M−1⌉ − 1)(2 − ⌈M−1⌉M) | 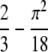 |
 at at 
|
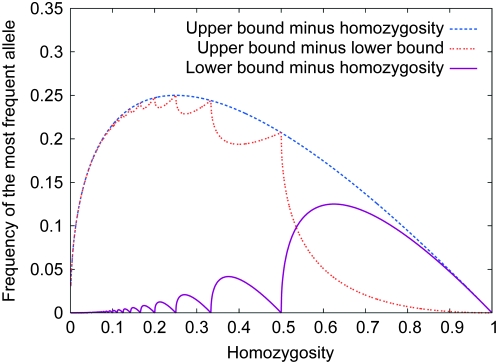
Theorems 1 and 2 can be visualized in Figures 1–5, and various properties of the bounds that can be observed in the figures are considered in the appendix. Figure 1 illustrates the upper and lower bounds on the frequency of the most frequent allele, as functions of homozygosity. The peculiar yet continuous and monotonic nature of the lower bound can be observed, as can the relatively confined range between the upper and lower bounds—with an average difference of 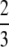 − π2/18 ≈ 0.1184—in which the frequency of the most frequent allele must lie. The stepped shape for the lower bound results from transitions at reciprocals of integers for the number of alleles contained in the collection of allele frequencies that achieves the lower bound.
− π2/18 ≈ 0.1184—in which the frequency of the most frequent allele must lie. The stepped shape for the lower bound results from transitions at reciprocals of integers for the number of alleles contained in the collection of allele frequencies that achieves the lower bound.
Figure 1.—
Upper and lower bounds on the frequency of the most frequent allele, as functions of homozygosity.
Figure 2.—
The difference between the upper and lower bounds on the frequency of the most frequent allele, for a given homozygosity, and the difference between the bounds and homozygosity itself.
Figure 3.—
The lower bound on the fraction of homozygosity contributed by homozygotes for the most frequent allele. The upper bound is 1.
Figure 4.—
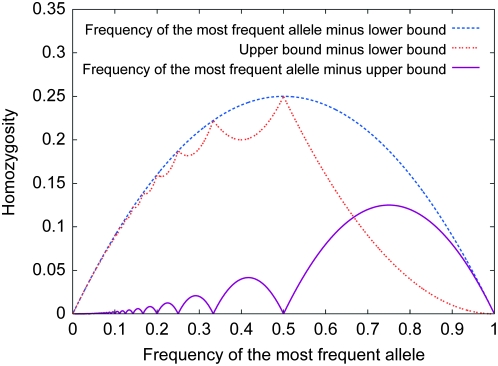
Upper and lower bounds on homozygosity, as functions of the frequency of the most frequent allele. These upper and lower bounds are the inverse functions of the lower and upper bounds on the frequency of the most frequent allele, given homozygosity.
Figure 5.—
The difference between the upper and lower bounds on homozygosity given the frequency of the most frequent allele, and the difference between the frequency of the most frequent allele and the bounds.
Figure 2 shows the pairwise differences among the upper bound, the lower bound, and the homozygosity itself. From this figure, it is possible to see that the lower bound on the frequency of the most frequent allele is greater than or equal to the homozygosity, with equality when homozygosity is the reciprocal of an integer. It can also be seen that the difference between the lower bound and the homozygosity has numerous local maxima, the highest point being at ( ,
,  ), and that the difference between the upper bound and the lower bound has local maxima at reciprocals of integers and local minima in the intervening intervals. The maximal difference between the upper and lower bounds occurs at (
), and that the difference between the upper bound and the lower bound has local maxima at reciprocals of integers and local minima in the intervening intervals. The maximal difference between the upper and lower bounds occurs at ( ,
,  ), and the highest of the local minima is nearby.
), and the highest of the local minima is nearby.
Figure 3 displays the minimal fraction of homozygosity contained in homozygotes for the most frequent allele. This function is monotonically increasing, so that for homozygosities substantially > , nearly all homozygotes are homozygous for the most frequent allele, regardless of the total number of alleles.
, nearly all homozygotes are homozygous for the most frequent allele, regardless of the total number of alleles.
The upper and lower bounds on homozygosity in terms of the frequency of the most frequent allele are the inverse functions of the lower and upper bounds on the frequency of the most frequent allele in terms of homozygosity. Thus, there is a close relationship between the bounds on H in terms of M shown in Figure 4 and the bounds on M in terms of H shown in Figure 1.
As functions of the frequency of the most frequent allele, Figure 5 depicts the pairwise differences among the upper bound on homozygosity, the lower bound, and the frequency of the most frequent allele itself. The frequency of the most frequent allele is greater than or equal to the upper bound, equaling the upper bound at reciprocals of integers. The difference between this frequency and the upper bound has a collection of local maxima, the highest being at (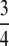 ,
,  ). The difference between the upper and lower bounds has local maxima at reciprocals of integers and local minima in the intervening intervals. The maximal difference between the upper and lower bounds occurs at (
). The difference between the upper and lower bounds has local maxima at reciprocals of integers and local minima in the intervening intervals. The maximal difference between the upper and lower bounds occurs at ( ,
,  ), near the highest of the local minima.
), near the highest of the local minima.
APPLICATION TO DATA
To demonstrate the bounds with actual allele frequencies, we consider the homozygosity and frequency of the most frequent allele for 783 multiallelic microsatellite loci studied in a sample of 1048 individuals drawn from worldwide human populations (Rosenberg et al. 2005). Although our theoretical results are useful for any collection of multiallelic loci, this data set provides a particularly illustrative example, as levels of variability of human microsatellites span quite a wide range. For each locus, we assume that the allele frequencies in the sample are parametric allele frequencies, and we obtain values for H and M in the full collection of 1048 individuals.
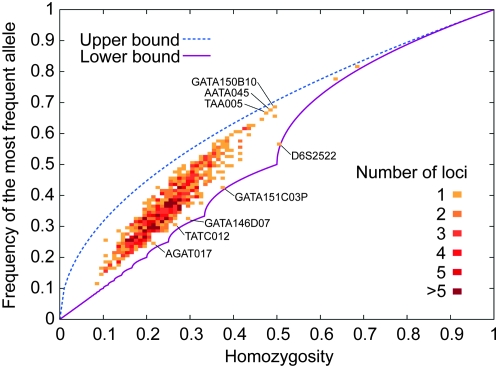
Figure 6 plots H and M for the 783 loci, illustrating a high degree of correlation between the two quantities. Homozygosity ranges from 0.0837 to 0.6872, and the frequency of the most frequent allele ranges from 0.1136 to 0.8146. Several loci have values of M quite close to the lower bound for their homozygosity values (Table 2). The lists of allele frequencies for these loci are fairly close to the lists that achieve the lower bound. For example, locus AGAT017 has homozygosity 0.2118, between 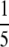 and
and  , and its four most frequent alleles have frequencies 0.2425, 0.2410, 0.2300, and 0.1979. At homozygosity 0.2118, the lower bound for the frequency of the most frequent allele is achieved when four alleles have frequency 0.2243 and a fifth allele has frequency 0.1027.
, and its four most frequent alleles have frequencies 0.2425, 0.2410, 0.2300, and 0.1979. At homozygosity 0.2118, the lower bound for the frequency of the most frequent allele is achieved when four alleles have frequency 0.2243 and a fifth allele has frequency 0.1027.
Figure 6.—
Homozygosity and frequency of the most frequent allele for 783 microsatellite loci. Each bin is 0.01 × 0.01, and the upper and lower bounds on the frequency of the most frequent allele are shown for comparison. The correlation coefficient of homozygosity and the frequency of the most frequent allele is 0.9439. Tables 2 and 3 give the frequencies of all alleles at the marked microsatellite loci.
TABLE 2.
Five microsatellite loci with frequency of the most frequent allele close to the lower bound
| Locus | Total data points | H | Lower bound on M | Upper bound on M | p1 (M) | p2 | p3 | p4 | p5 | p6 | p7 | p8 | p9 | p10 | p11 | p12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGAT017 | 1996 | 0.21 | 0.22 | 0.46 | 0.24 | 0.24 | 0.23 | 0.20 | 0.05 | 0.02 | <0.01 | <0.01 | <0.01 | <0.01 | ||
| TATC012 | 2046 | 0.26 | 0.28 | 0.51 | 0.31 | 0.30 | 0.27 | 0.09 | 0.02 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| GATA146D07 | 1940 | 0.30 | 0.31 | 0.54 | 0.32 | 0.31 | 0.31 | 0.03 | 0.02 | <0.01 | <0.01 | <0.01 | ||||
| GATA151C03P | 2076 | 0.37 | 0.41 | 0.61 | 0.43 | 0.42 | 0.10 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 | ||||
| D6S2522 | 1950 | 0.50 | 0.55 | 0.71 | 0.56 | 0.43 | <0.01 | <0.01 | <0.01 |
Two other loci whose most frequent alleles have frequency close to the lower bound—TATC012 and GATA146D07—have homozygosities between  and
and 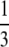 . For homozygosities in this interval, the lower bound is achieved when the three highest allele frequencies have the same value; indeed both loci have three high-frequency alleles with frequency near the lower bound. Similarly, locus GATA151C03P, with homozygosity between
. For homozygosities in this interval, the lower bound is achieved when the three highest allele frequencies have the same value; indeed both loci have three high-frequency alleles with frequency near the lower bound. Similarly, locus GATA151C03P, with homozygosity between 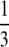 and
and  , has two high-frequency alleles with frequency near the lower bound.
, has two high-frequency alleles with frequency near the lower bound.
Table 3 displays the allele frequencies for three loci with values of M close to the upper bound. The upper bound is approximated when a locus has one allele with a particularly high frequency and many alleles with low frequencies. Consistent with their M values near the upper bound, each of the three loci has a single high-frequency allele and several low-frequency alleles.
TABLE 3.
Three microsatellite loci with frequency of the most frequent allele close to the upper bound
| Locus | Total data points | H | Lower bound on M | Upper bound on M | p1 (M) | p2 | p3 | p4 | p5 | p6 | p7 | p8 | p9 | p10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TAA005 | 2002 | 0.47 | 0.49 | 0.69 | 0.66 | 0.13 | 0.12 | 0.03 | 0.03 | 0.01 | <0.01 | <0.01 | <0.01 | |
| AATA045 | 2090 | 0.48 | 0.49 | 0.70 | 0.67 | 0.13 | 0.11 | 0.06 | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| GATA150B10 | 2068 | 0.50 | 0.50 | 0.71 | 0.69 | 0.12 | 0.10 | 0.04 | 0.03 | 0.02 | 0.01 | <0.01 | <0.01 | <0.01 |
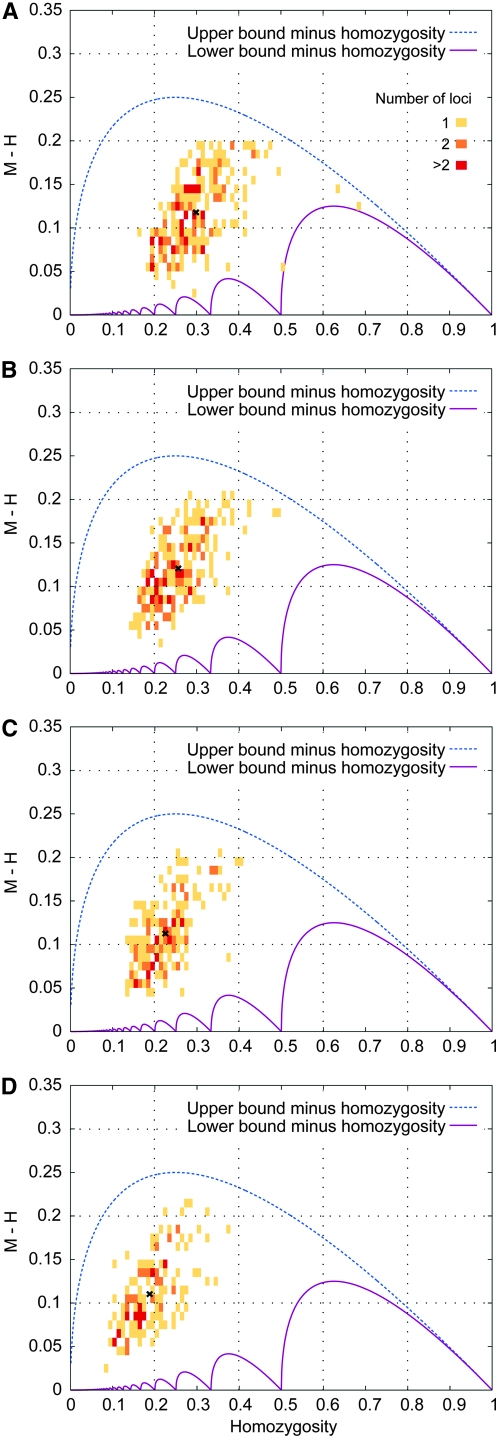
Subdividing loci on the basis of their numbers of alleles, Figure 7 illustrates a trend of decreasing H with an increasing number of alleles. Considering the four plots, the mean value of M − H is greatest in Figure 7B, in which the mean homozygosity is near 0.25. This observation is explained by the fact that the range between the upper and lower bounds on M is greatest for a homozygosity of  . As the mean homozygosity moves away from
. As the mean homozygosity moves away from  in Figure 7, A, C, and D, the mean value of M − H decreases.
in Figure 7, A, C, and D, the mean value of M − H decreases.
Figure 7.—
Frequency of the most frequent allele minus homozygosity for 783 microsatellite loci. Each bin is 0.01 × 0.01, and the upper and lower bounds on M − H are shown for comparison. For each plot, the mean 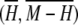 is marked by an x. (A) Loci with 4–9 distinct alleles (233): the mean is (0.2989, 0.1179). (B) Loci with 10–11 distinct alleles (222): the mean is (0.2570, 0.1208). (C) Loci with 12–14 distinct alleles (175): the mean is (0.2262, 0.1126). (D) Loci with 15–35 distinct alleles (153): the mean is (0.1890, 0.1102).
is marked by an x. (A) Loci with 4–9 distinct alleles (233): the mean is (0.2989, 0.1179). (B) Loci with 10–11 distinct alleles (222): the mean is (0.2570, 0.1208). (C) Loci with 12–14 distinct alleles (175): the mean is (0.2262, 0.1126). (D) Loci with 15–35 distinct alleles (153): the mean is (0.1890, 0.1102).
DISCUSSION
For a biallelic locus, an exact relationship exists between homozygosity (H) and the frequency of the most frequent allele (M), as H = 2M2 − 2M + 1 and 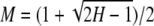 . Although in general the value of H or M is not uniquely specified from the value of the other quantity, we have found that a close connection between H and M does in fact exist. Our analysis verifies that measured values for homozygosity (and heterozygosity) consist largely of the contribution of the most common allele, and that the contribution made by rarer alleles is relatively small. Especially if homozygosity is very high or if the most frequent allele has a high frequency, each of the two summaries H and M greatly limits the possible values of the other quantity, so that both quantities provide similar information about an underlying allele-frequency distribution.
. Although in general the value of H or M is not uniquely specified from the value of the other quantity, we have found that a close connection between H and M does in fact exist. Our analysis verifies that measured values for homozygosity (and heterozygosity) consist largely of the contribution of the most common allele, and that the contribution made by rarer alleles is relatively small. Especially if homozygosity is very high or if the most frequent allele has a high frequency, each of the two summaries H and M greatly limits the possible values of the other quantity, so that both quantities provide similar information about an underlying allele-frequency distribution.
These results have implications for population-genetic methods that rely on H or M in analyses of multiallelic loci. Various neutrality tests have been developed that identify deviations from null population-genetic models on the basis of unusual values of homozygosity (Watterson 1977, 1978), heterozygosity (Depaulis and Veuille 1998; Depaulis et al. 2001; Markovtsova et al. 2001), or the frequency of the most frequent allele (Hudson et al. 1994). The close connection between homozygosity and the frequency of the most frequent allele suggests that tests using H and those using M detect similar features of the allele-frequency distribution. This observation potentially explains a high level of agreement seen in Table 7 of Innan et al. (2005) for the haplotype diversity test (Depaulis and Veuille 1998), based on haplotype heterozygosity, and the Hudson et al. (1994) haplotype test, based on the frequency of the most frequent haplotype.
Our results are also informative in relation to recently proposed methods that use “extended haplotype homozygosity”—pairwise identity of long haplotypes in the neighborhood of an index site—in detecting the signature of partial selective sweeps (Sabeti et al. 2002; Toomajian et al. 2006; Voight et al. 2006; Tang et al. 2007; Zeng et al. 2007). During such sweeps, a favored mutant allele rises to high frequency, carrying with it neighboring alleles that were near the selected site on the haplotype on which the mutation originally occurred. Thus, the detection of partial selective sweeps is a search for long high-frequency haplotypes that have not had sufficient time to be broken down by recombination. Because of the close connection between homozygosity and the frequency of the most frequent allele, genomic regions that have long high-frequency haplotypes will largely be coincident with regions that have long stretches of high haplotype homozygosity. Consequently, extended haplotype homozygosity methods provide an effective basis for accessing the signal of partial selective sweeps contained in extended high-frequency haplotypes.
Finally, the connection between homozygosity and the frequency of the most frequent allele may be useful for examining the properties of a variety of additional functions of allele frequencies that are based on homozygosity. Notably, the genetic differentiation measure FST and related quantities can be assembled from the homozygosities of various subgroups of a population—especially when viewed in the formulation of the GST measure of Nei (1987). From the connection between H and M, it follows that constraints on FST as functions of M undoubtedly exist; such constraints potentially provide the conceptual basis for understanding a frequency dependence observed for values of FST (Long and Kittles 2003; Hedrick 2005).
Acknowledgments
We thank S. Boca for numerous discussions of this work. Grant support was provided by a University of Michigan Center for Genetics in Health and Medicine postdoctoral fellowship, by National Institutes of Health grant R01 GM081441, by an Alfred P. Sloan Research Fellowship, and by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
APPENDIX
In addition to verifying Theorems 1 and 2, this appendix formalizes many of the features visible in Figures 1–5. We begin with the proofs of the theorems. We then obtain properties of the frequency of the most frequent allele in terms of homozygosity and properties of homozygosity in terms of the frequency of the most frequent allele. For convenience, we label the bounds as follows:
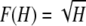 |
(A1) |
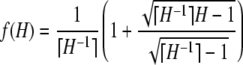 |
(A2) |
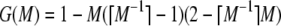 |
(A3) |
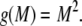 |
(A4) |
For integers K ≥ 2, we also denote the half-open interval [1/K, 1/(K − 1)) by IK.
The key result is Lemma 3, which considers sets of nonnegative numbers with a fixed positive sum C, in which the numbers in the set are bounded above by a positive constant M. The square of a positive number x is greater than or equal to the sum of squares for each collection of nonnegative numbers whose sum is x. As a result, considering all sets of nonnegative numbers with maximum M and with sum equal to C, we can show that the maximal sum of squares is obtained when as many of the numbers as possible are equal to M and when at most one remaining number is smaller than M. Lemma 3 makes it possible to obtain the maximal homozygosity as a function of M and, ultimately, to find the minimal M as a function of H.
Lemma 3. Suppose M > 0 and C > 0 and that ⌈C/M⌉ is denoted K. Considering all sequences  with pi ∈ [0, M],
with pi ∈ [0, M],  , and i < j implies pi ≥ pj,
, and i < j implies pi ≥ pj, 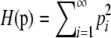 is maximal if and only if pi = M for 1 ≤ i ≤ K − 1, pK = C − (K − 1)M, and pi = 0 for i > K, and its maximum is K(K − 1)M2 − 2C(K − 1)M + C2.
is maximal if and only if pi = M for 1 ≤ i ≤ K − 1, pK = C − (K − 1)M, and pi = 0 for i > K, and its maximum is K(K − 1)M2 − 2C(K − 1)M + C2.
Proof. We use induction on K. Suppose K = 1, so that C ≤ M. Because 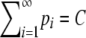 ,
,
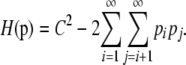 |
(A5) |
Because a nonnegative term is subtracted in Equation A5, the maximum of H(p) occurs when this term is zero. As a result, at the maximum, p1 = C ≤ M, pi = 0 for i > 1, and H(p) = C2. This establishes the base case.
Assume that the desired result is true for all C and M with ⌈C/M⌉ = K − 1. Now suppose ⌈C/M⌉ = K. The proposed value of p that maximizes H has pi = M for 1 ≤ i ≤ K − 1, pK = C − (K − 1)M, and pi = 0 for i > K. Label this sequence by p*. Then
 |
(A6) |
By assumption, ⌈C/M⌉ = K and M ≥ C/K. As Equation A6 describes a parabola in M with positive leading term, regardless of the value of M, H(p*) is greater than or equal to the value at the minimum of the parabola, or C2/K.
We now show that no other sequence p can achieve a value of H as high as H(p*). Suppose p1 < C/K. Because pi ≤ p1 for i > 1,
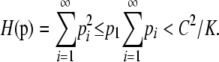 |
(A7) |
Because H(p*) ≥ C2/K, the sequence p that maximizes H cannot have p1 < C/K. This sequence must therefore have p1 ≥ C/K and ⌈C/p1⌉ ≤ K. However, because p1 ≤ M and ⌈C/M⌉ = K by assumption, ⌈C/p1⌉ ≥ ⌈C/M⌉ = K. Thus, ⌈C/p1⌉ = K and ⌈(C − p1)/p1⌉ = K − 1.
Note that 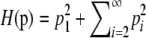 . We can therefore apply the inductive hypothesis to
. We can therefore apply the inductive hypothesis to 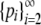 with C − p1 in place of C and p1 in place of M. By the inductive hypothesis, the maximum of
with C − p1 in place of C and p1 in place of M. By the inductive hypothesis, the maximum of  occurs if and only if pi = p1 for 2 ≤ i ≤ K − 1, pK = (C − p1) − (K − 2)p1, and pi = 0 for i > K. As a result,
occurs if and only if pi = p1 for 2 ≤ i ≤ K − 1, pK = (C − p1) − (K − 2)p1, and pi = 0 for i > K. As a result,
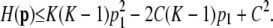 |
(A8) |
This function is monotonically increasing in p1 for p1 ≥ C/K and therefore achieves its maximum when p1 is as large as possible—that is, when p1 = M. ▪
Proof of Theorem 2. (i) This result follows from the definition of H and from the fact that p2 > 0; (ii) this follows from Lemma 3, taking C = 1 so that K = ⌈M−1⌉. ▪
Lemma 4. (i) 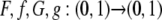 are monotonically increasing, continuous, and bijective; (ii) f and G are differentiable on (1/K, 1/(K − 1)) for each integer K ≥ 2.
are monotonically increasing, continuous, and bijective; (ii) f and G are differentiable on (1/K, 1/(K − 1)) for each integer K ≥ 2.
Proof. The result is trivial for F and g. For integers K ≥ 2, G(1/K) = 1/K, and G is monotonically increasing on each interval IK, where ⌈M−1⌉ has the fixed value K. Thus, G is monotonically increasing on (0, 1). From the form of G it is clear that on (1/K, 1/(K − 1)), G is continuous and differentiable. For each K, as M = 1/K is approached from either direction, G(M) approaches 1/K. Thus, G is continuous on (0, 1). Given H ∈ (0, 1), there is a unique M for which G(M) = H, so that G is bijective. Similar reasoning holds for f. ▪
As a consequence of this lemma, since G(1/K) = 1/K for integers K ≥ 2, if M ∈ (0, 1) and ⌈M−1⌉ = K, then G(M) lies in the interval IK. Similarly, if ⌈H−1⌉ = K for H ∈ (0, 1), then f(H) also lies in IK.
Lemma 5. F and g are inverse functions on (0, 1), as are f and G.
Proof. The result is trivial for F and g. As bijections, both f and G are invertible. Noting that M ∈ IK implies G(M) ∈ IK, ⌈M−1⌉ = ⌈G(M)−1⌉, from which we can solve for M in terms of G(M) on each interval IK to find that on each interval the inverse of G is f. ▪
Proof of Theorem 1.
This result follows from the definition of H and from the fact that p2 > 0.
By Theorem 2, for a given value of M and a given sequence
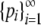 with
with 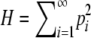 , G(M) ≥ H with equality if and only if pi = M for 1 ≤ i ≤ K − 1, pK = 1 − (K − 1)M, and pi = 0 for i ≥ K, where K = ⌈M−1⌉. Applying the monotonically increasing function f to the inequality G(M) ≥ H, f(G(M)) ≥ f(H) with the same equality condition. Because f is the inverse of G, M ≥ f(H) with the same equality condition. ▪
, G(M) ≥ H with equality if and only if pi = M for 1 ≤ i ≤ K − 1, pK = 1 − (K − 1)M, and pi = 0 for i ≥ K, where K = ⌈M−1⌉. Applying the monotonically increasing function f to the inequality G(M) ≥ H, f(G(M)) ≥ f(H) with the same equality condition. Because f is the inverse of G, M ≥ f(H) with the same equality condition. ▪
Note that for a given value of H, we can find a set of allele frequencies for which the value of M comes arbitrarily close to its upper bound of  . This can be accomplished by supposing that a locus has one common allele with frequency M and N rare alleles each with frequency ɛ. If all other alleles have zero frequency, such a locus must have M2 + Nɛ2 = H and M + Nɛ = 1. Solving this pair of equations for M in terms of N (taking the larger root) and letting
. This can be accomplished by supposing that a locus has one common allele with frequency M and N rare alleles each with frequency ɛ. If all other alleles have zero frequency, such a locus must have M2 + Nɛ2 = H and M + Nɛ = 1. Solving this pair of equations for M in terms of N (taking the larger root) and letting 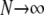 ,
, 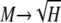 . Similar reasoning yields sets of allele frequencies that for a given value of M have values of H arbitrarily close to the lower bound of M2.
. Similar reasoning yields sets of allele frequencies that for a given value of M have values of H arbitrarily close to the lower bound of M2.
Frequency of the most frequent allele in terms of homozygosity:
We now derive the properties of the upper and lower bounds on the frequency of the most frequent allele, as functions of homozygosity. Most of the results that follow are relatively straightforward to prove, and they are included for completeness.
Proposition 6 determines the mean values of the bounds, finding that the difference between them has a rather small mean of  − π2/18 ≈ 0.1184. Lemma 7 then shows that the lower bound on the frequency of the most frequent allele is greater than or equal to the homozygosity itself; the mean values of the differences of the upper and lower bounds from the homozygosity are then obtained in Proposition 8. Results 9–15 concern additional properties of the differences among the upper and lower bounds and the homozygosity and properties of various maxima and minima associated with the upper and lower bounds. The section concludes with Proposition 16, which determines a lower bound on the fraction of homozygosity that is due to the most frequent allele.
− π2/18 ≈ 0.1184. Lemma 7 then shows that the lower bound on the frequency of the most frequent allele is greater than or equal to the homozygosity itself; the mean values of the differences of the upper and lower bounds from the homozygosity are then obtained in Proposition 8. Results 9–15 concern additional properties of the differences among the upper and lower bounds and the homozygosity and properties of various maxima and minima associated with the upper and lower bounds. The section concludes with Proposition 16, which determines a lower bound on the fraction of homozygosity that is due to the most frequent allele.
Proposition 6. Averaging across values of H ∈ (0, 1), (i) the mean of F(H) is 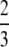 ; (ii) the mean of f(H) is π2/18; (iii) the mean of F(H) − f(H) is
; (ii) the mean of f(H) is π2/18; (iii) the mean of F(H) − f(H) is 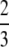 − π2/18.
− π2/18.
Proof.
The mean of F(H) is
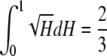 .
.- Because (0, 1) =
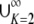 IK, and because ⌈H−1⌉ = K for H ∈ IK, the mean of f(H) can be written
IK, and because ⌈H−1⌉ = K for H ∈ IK, the mean of f(H) can be written
Because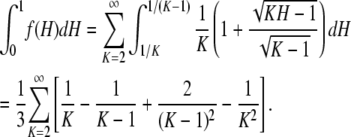
(A9) 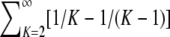 reduces to −1 and because
reduces to −1 and because 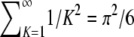 , Equation A9 simplifies to π2/18.
, Equation A9 simplifies to π2/18. That the mean of F(H) − f(H) is
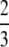 − π2/18 follows directly from i and ii together with the fact that F(H) > f(H) for H ∈ (0, 1). ▪
− π2/18 follows directly from i and ii together with the fact that F(H) > f(H) for H ∈ (0, 1). ▪
Lemma 7. For H ∈ (0, 1), f(H) ≥ H, with equality if and only if H = K−1 for some integer K.
Proof. This result follows from the fact that for H ∈ (0, 1), ⌈H−1⌉ > 1, and ⌈H−1⌉ − 1 < H−1 ≤ ⌈H−1⌉, with the equality occurring if and only if H = K−1 for an integer K. ▪
Proposition 8. Averaging across values of H ∈ (0, 1), (i) the mean of F(H) − H is  ; (ii) the mean of f(H) − H is π2/18 −
; (ii) the mean of f(H) − H is π2/18 −  .
.
Proof. By Theorem 1 and Lemma 7, F(H) > f(H) ≥ H on the interval (0, 1). The mean of F(H) − H [or f(H) − H] equals the mean of F(H) [or f(H)] minus the mean of H, or  . Consequently, using Proposition 6, (i) the mean of F(H) − H is
. Consequently, using Proposition 6, (i) the mean of F(H) − H is 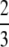 −
−  =
=  , and (ii) the mean of f(H) − H is π2/18 −
, and (ii) the mean of f(H) − H is π2/18 −  . ▪
. ▪
Proposition 9. On the interval [1/K, 1/(K − 1)), where K ≥ 2 is an integer, the maximal value of f(H) − H is 1/[4K(K − 1)], and it is achieved at H = (4K − 3)/[4K(K − 1)].
Proof. For H ∈ IK, ⌈H−1⌉ = K, and
 |
f(H) − H is continuous on the interval and differentiable except at the endpoints. Its only critical point on the interval is a maximum that occurs at ((4K − 3)/[4K(K − 1)], 1/[4K(K − 1)]). ▪
Corollary 10. On (0, 1), the maximal value of f(H) − H is  , and it is achieved at H =
, and it is achieved at H =  .
.
Proof. Because (0, 1) = 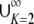 IK, f(H) − H has its maximum in IK for some K—in particular, for the K for which the maximal value of f(H) − H is greatest. By Proposition 9, the maximum of f(H) − H on IK is 1/[4K(K − 1)]. As 1/[4K(K − 1)] decreases for K ≥ 2, the maximum of f(H) − H on (0, 1) occurs in I2. Applying Proposition 9, this maximum is at (
IK, f(H) − H has its maximum in IK for some K—in particular, for the K for which the maximal value of f(H) − H is greatest. By Proposition 9, the maximum of f(H) − H on IK is 1/[4K(K − 1)]. As 1/[4K(K − 1)] decreases for K ≥ 2, the maximum of f(H) − H on (0, 1) occurs in I2. Applying Proposition 9, this maximum is at ( ,
,  ). ▪
). ▪
Proposition 11. On the interval [1/K, 1/(K − 1)], where K ≥ 2 is an integer,
i. For K ≥ 5, the maximal value of F(H) − f(H) is
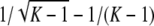 , and it is achieved at H = 1/(K − 1). For K = 2, 3, 4, the maximal value of F(H) − f(H) is
, and it is achieved at H = 1/(K − 1). For K = 2, 3, 4, the maximal value of F(H) − f(H) is 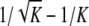 , and it is achieved at H = 1/K.
, and it is achieved at H = 1/K.- ii. The minimal value of F(H) − f(H) is
and it is achieved at H = (K − 1)/(K2 − K − 1).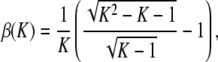
(A10)
Proof. Define χ(H) = F(H) − f(H). For H ∈ IK, ⌈H−1⌉ = K, and
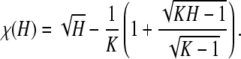 |
To verify ii, note that the only critical point of χ(H) on [1/K, 1/(K − 1)] is a minimum that occurs at ((K − 1)/(K2 − K − 1), β(K)).
To obtain i, note that because there is no maximum in the interior of [1/(K − 1), 1/K], the maximum of χ(H) occurs at the endpoint of the interval that produces the larger value of χ(H). At H = 1/K, 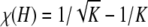 , and at H = 1/(K − 1),
, and at H = 1/(K − 1), 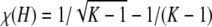 . Define
. Define 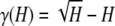 , and note that at points H = 1/K for integers K ≥ 1, γ(H) = χ(H). At the endpoints of [0, 1], γ(H) = 0, and on [0, 1], γ(H) has its maximum and only critical point at (
, and note that at points H = 1/K for integers K ≥ 1, γ(H) = χ(H). At the endpoints of [0, 1], γ(H) = 0, and on [0, 1], γ(H) has its maximum and only critical point at ( ,
,  ). Consequently, for H, H′ ∈ [0, 1], if H > H′ ≥
). Consequently, for H, H′ ∈ [0, 1], if H > H′ ≥  , then γ(H) < γ(H′), whereas if
, then γ(H) < γ(H′), whereas if  ≥ H > H′, then γ(H) > γ(H′). Thus, for K = 2, 3, 4, γ(1/K) = χ(1/K) > χ(1/(K − 1)) = γ(1/(K − 1)), whereas for integers K ≥ 5, γ(1/(K − 1)) = χ(1/(K − 1)) > χ(1/K) = γ(1/K). ▪
≥ H > H′, then γ(H) > γ(H′). Thus, for K = 2, 3, 4, γ(1/K) = χ(1/K) > χ(1/(K − 1)) = γ(1/(K − 1)), whereas for integers K ≥ 5, γ(1/(K − 1)) = χ(1/(K − 1)) > χ(1/K) = γ(1/K). ▪
Proposition 12. On (0, 1), the highest local minimum of F(H) − f(H) is 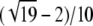 , and it occurs at H =
, and it occurs at H = 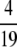 .
.
Proof. By Proposition 11, the minimal difference for a given interval [1/K, 1/(K − 1)] is achieved at H = (K − 1)/(K2 − K − 1) and is β(K). To find the integer K ≥ 2 where β(K) is greatest, we show that β(K) > β(K + 1) for K ≥ 6. It then follows that the largest value of β(K) occurs at the integer K ∈ [2, 6] that produces the highest value of β(K). This maximum occurs at K = 5, so that H = 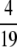 and
and 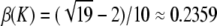 .
.
The following chain of inequalities yields the result:
 |
Corollary 13. The maximal value of F(H) − H is  , and it is achieved at H =
, and it is achieved at H =  .
.
Proof. This result was shown in the proof of Proposition 11 when it was found that 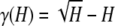 has its maximum on [0, 1] at H =
has its maximum on [0, 1] at H =  . ▪
. ▪
Corollary 14. The maximal value of F(H) − f(H) is  , and it is achieved at H =
, and it is achieved at H =  .
.
Proof. Because f(H) ≥ H, F(H) − f(H) ≤ F(H) − H. By Corollary 13, the maximum of F(H) − H occurs at  and is
and is  . Evaluating at H =
. Evaluating at H =  , F(H) − f(H) achieves this same upper bound. ▪
, F(H) − f(H) achieves this same upper bound. ▪
Proposition 15. The difference F(H) − f(H) is (i) greater than f(H) − H if 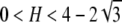 , (ii) equal to f(H) − H if
, (ii) equal to f(H) − H if 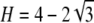 , and (iii) less than f(H) − H if
, and (iii) less than f(H) − H if  .
.
Proof. Consider H ∈ [1/K, 1/(K − 1)], for K ≥ 3. On this interval, by Proposition 11, the minimum of F(H) − f(H) is β(K), and by Proposition 9, the maximum of f(H) − H is μ(K) = 1/[4K(K − 1)]. The following inequalities yield F(H) − f(H) > f(H) − H for H <  :
:
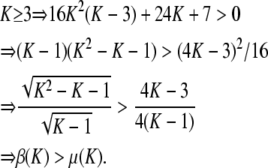 |
(A11) |
For H ∈ [ , 1), [F(H) − f(H)] − [f(H) − H] =
, 1), [F(H) − f(H)] − [f(H) − H] = 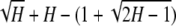 , which for H ∈ [
, which for H ∈ [ , 1) can be shown to fall on the same side of zero as H2 − 8H + 4. The only root of H2 − 8H + 4 = 0 for H ∈ [
, 1) can be shown to fall on the same side of zero as H2 − 8H + 4. The only root of H2 − 8H + 4 = 0 for H ∈ [ , 1) is
, 1) is  , at which the sign of H2 − 8H + 4 switches from positive to negative. ▪
, at which the sign of H2 − 8H + 4 switches from positive to negative. ▪
Proposition 16.
- i. The fraction of homozygosity due to homozygotes for the most frequent allele is greater than or equal to
with equality if and only if K = ⌈H−1⌉ = ⌈M−1⌉, p1 = p2 = … = pK−1 = M, and pK = 1 − (K − 1)M.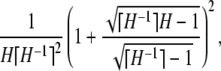
ii. The fraction of homozygosity due to homozygotes for the most frequent allele is greater than or equal to H, equality requiring H = K−1 for some integer K ≥ 2, and p1 = p2 = … = pK = H.
iii. The lower bound on the fraction of homozygosity due to homozygotes for the most frequent allele lies in [1/K, 1/(K − 1)), where K = ⌈H−1⌉.
iv. The lower bound on the fraction of homozygosity due to homozygotes for the most frequent allele is monotonically increasing with H on the interval (0, 1).
Proof. The fraction of homozygosity due to homozygotes for the most frequent allele is M2/H, so that i follows directly from Theorem 1ii.
ii. That M2/H ≥ f(H)2/H ≥ H2/H follows directly from Theorem 1ii and Lemma 7, with equality under the same conditions as specified by these results.
iii. That M2/H ≥ K−1 for H ∈ IK follows trivially from ii. Note that f(H)2/H < 1/(K − 1) is equivalent to
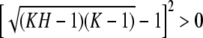 , which is true except if H = 1/(K − 1).
, which is true except if H = 1/(K − 1).- iv. Denote the lower bound in i by σ(H). The function σ is continuous on (0, 1), and at H = K−1 for integers K ≥ 2, σ(H) = K−1. To show that σ is monotonic on (0, 1) all that must be shown is that it is monotonic for H ∈ IK. On this interval, ⌈H−1⌉ = K, and the derivative of σ is
To show that the term inside the brackets is positive for H ∈ IK, we can begin with the inequality (K − 1)H2 − KH + 1 > 0, which holds for H ∈ IK, as the leading term is positive and the roots are located at 1/(K − 1) and 1. Multiplying by K2 and adding identical terms to both sides, we have (K − 1)(K2H2 − 4KH + 4) > (K2 − 4K + 4)(KH − 1). Noting that K ≥ 2 and for H ∈ IK, 2 − KH > 0, the square root of both sides can be taken to obtain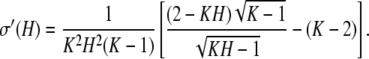
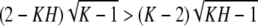 . ▪
. ▪
Homozygosity in terms of the frequency of the most frequent allele:
Many of the results in this section follow from those in the previous section, using the fact that the lower and upper bounds g and G for homozygosity are the respective inverse functions of the upper and lower bounds F and f for the frequency of the most frequent allele.
Proposition 17. Averaging across values of M ∈ (0, 1), (i) the mean of G(M) is 1 − π2/18; (ii) the mean of g(M) is 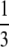 ; (iii) the mean of G(M) − g(M) is
; (iii) the mean of G(M) − g(M) is 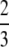 − π2/18.
− π2/18.
Proof.
iii. Because G and g are the inverse functions of f and F by Lemma 5, and because on (0, 1), G > g and F > f, the area between G and g equals the area between F and f. By Proposition 6, this area is
 − π2/18.
− π2/18.ii. The mean of g(M) is
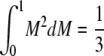 .
.i. That the mean of G(M) is 1 − π2/18 follows directly from ii and iii. ▪
Lemma 18. For M ∈ (0, 1), G(M) ≤ M, with equality if and only if M = K−1 for some integer K.
Proof. This result follows directly from Lemma 7 and the inverse relationship of G and f in Lemma 5. ▪
Proposition 19. Averaging across values of M ∈ (0, 1), (i) the mean of M − G(M) is π2/18 −  ; (ii) the mean of M − g(M) is
; (ii) the mean of M − g(M) is  .
.
Proof. From the inverse relationship between G and f (Lemma 5), the area between M and G(M) equals the area between f(H) and H, or π2/18 −  (Proposition 8ii), and from the inverse relationship between g and F, the area between M and g(M) equals the area between F(H) and H, or
(Proposition 8ii), and from the inverse relationship between g and F, the area between M and g(M) equals the area between F(H) and H, or  (Proposition 8i). ▪
(Proposition 8i). ▪
Proposition 20. On the interval [1/K, 1/(K − 1)), where K ≥ 2 is an integer, the maximal value of M − G(M) is 1/[4K(K − 1)], and it is achieved at H = (2K − 1)/[2K(K − 1)].
Proof. For M ∈ IK, ⌈M−1⌉ = K, and M − G(M) = M − [K(K − 1)M2 − 2(K − 1)M + 1]. M − G(M) is continuous on the interval and differentiable except at the endpoints. Its only critical point on the interval is a maximum that occurs at ((2K − 1)/[2K(K − 1)], 1/[4K(K – 1)]). ▪
Corollary 21. On (0, 1), the maximal value of M − G(M) is  , and it is achieved at M =
, and it is achieved at M = 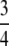 .
.
Proof. Because (0, 1) = 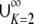 IK, M − G(M) has its maximum in IK for some K—in particular, for the K for which the maximal value of M − G(M) is greatest. By Proposition 20, the maximum of M − G(M) on IK is 1/[4K(K − 1)]. As 1/[4K(K − 1)] decreases for K ≥ 2, the maximum of M − G(M) on (0, 1) occurs in I2. Applying Proposition 20, this maximum is at (
IK, M − G(M) has its maximum in IK for some K—in particular, for the K for which the maximal value of M − G(M) is greatest. By Proposition 20, the maximum of M − G(M) on IK is 1/[4K(K − 1)]. As 1/[4K(K − 1)] decreases for K ≥ 2, the maximum of M − G(M) on (0, 1) occurs in I2. Applying Proposition 20, this maximum is at (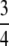 ,
,  ). ▪
). ▪
Proposition 22. On the interval [1/K, 1/(K − 1)], where K ≥ 2 is an integer,
i. For K ≥ 3, the maximal value of G(M) − g(M) is (K − 2)/(K − 1)2, and it is achieved at M = 1/(K − 1). For K = 2, the maximal value of G(M) − g(M) is
 , and it is achieved at M =
, and it is achieved at M =  .
.ii. The minimal value of G(M) − g(M) is ρ(K) = (K − 2)/(K2 − K − 1), and it is achieved at M = (K − 1)/(K2 − K − 1).
Proof. Define ξ(M) = G(M) − g(M). For M ∈ IK, ⌈M−1⌉ = K, and ξ(M) = (K2 − K − 1)M2 − 2(K − 1)M + 1. To verify ii, note that the only critical point of ξ(M) on [1/K, 1/(K − 1)] is a minimum that occurs at ((K − 1)/(K2 − K − 1), ρ(K)).
To obtain i, note that because there is no maximum in the interior of [1/(K − 1), 1/K], the maximum of ξ(M) occurs at the endpoint that produces the larger value of ξ(M). At M = 1/K, ξ(M) = 1/K − 1/K2, and at M = 1/(K − 1), ξ(M) = 1/(K − 1) − 1/(K − 1)2. Define δ(M) = M − M2, and note that at points M = 1/K for integers K ≥ 1, δ(M) = ξ(M). At the endpoints of [0, 1], δ(M) = 0, and on [0, 1], δ(M) has its maximum and only critical point at ( ,
,  ). Consequently, for M, M′ ∈ [0, 1], if M > M′ ≥
). Consequently, for M, M′ ∈ [0, 1], if M > M′ ≥  , then δ(M) < δ(M′), whereas if
, then δ(M) < δ(M′), whereas if  ≥ M > M′, then δ(M) > δ(M′). Thus, for K = 2, δ(1/K) = ξ(1/K) > ξ(1/(K − 1)) = δ(1/(K − 1)), whereas for integers K ≥ 3, δ(1/(K − 1)) = ξ(1/(K − 1)) > ξ(1/K) = δ(1/K). ▪
≥ M > M′, then δ(M) > δ(M′). Thus, for K = 2, δ(1/K) = ξ(1/K) > ξ(1/(K − 1)) = δ(1/(K − 1)), whereas for integers K ≥ 3, δ(1/(K − 1)) = ξ(1/(K − 1)) > ξ(1/K) = δ(1/K). ▪
Proposition 23. On (0, 1), the highest local minimum of G(M) − g(M) is  , and it occurs at M =
, and it occurs at M = 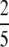 .
.
Proof. By Proposition 22, the minimal difference for a given interval [1/K, 1/(K − 1)] is achieved at M = (K − 1)/(K2 − K − 1) and is ρ(K). To find the integer K ≥ 2 where ρ(K) is greatest, note that
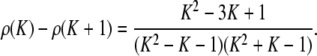 |
(A12) |
As a result, ρ(K) − ρ(K + 1) > 0 for K ≥ 3. It follows that ρ(K) is largest at the integer K ∈ [2, 3] that produces the highest value of ρ(K). This maximum occurs at K = 3, so that M =  and ρ(K) =
and ρ(K) = 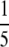 . ▪
. ▪
Corollary 24. The maximal value of M − g(M) is  , and it is achieved at M =
, and it is achieved at M =  .
.
Proof. This result was shown in the proof of Proposition 22 when it was found that δ(M) = M − M2 has its maximum on [0, 1] at M =  . ▪
. ▪
Corollary 25. The maximal value of G(M) − g(M) is  , and it is achieved at M =
, and it is achieved at M =  .
.
Proof. Because M ≥ G(M), G(M) − g(M) ≤ M − g(M). By Corollary 24, the maximum of M − g(M) occurs at  and is
and is  . Evaluating at M =
. Evaluating at M =  , G(M) − g(M) achieves this same upper bound. ▪
, G(M) − g(M) achieves this same upper bound. ▪
Proposition 26. The difference G(M) − g(M) is (i) greater than M − G(M) if 0 < M < 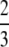 , (ii) equal to M − G(M) if M =
, (ii) equal to M − G(M) if M = 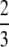 , and (iii) less than M − G(M) if
, and (iii) less than M − G(M) if 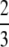 < M < 1.
< M < 1.
Proof. Consider M ∈ [1/K, 1/(K − 1)], for K ≥ 3. On this interval, by Proposition 22, the minimum of G(M) − g(M) is ρ(K), and by Proposition 20, the maximum of M − G(M) is μ(K) = 1/[4K(K − 1)]. The quantity ρ(K) − μ(K) can be simplified to
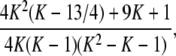 |
which is clearly positive for K > 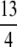 , and which is also positive for K = 3. As a result, G(M) − g(M) > M − G(M) for intervals IK with K ≥ 3, that is, for 0 < M <
, and which is also positive for K = 3. As a result, G(M) − g(M) > M − G(M) for intervals IK with K ≥ 3, that is, for 0 < M <  .
.
For M ∈ [ , 1), [G(M) − g(M)] − [M − G(M)] = 3M2 − 5M + 2. The only root of 3M2 − 5M + 2 = 0 for M ∈ [
, 1), [G(M) − g(M)] − [M − G(M)] = 3M2 − 5M + 2. The only root of 3M2 − 5M + 2 = 0 for M ∈ [ , 1) is M =
, 1) is M = 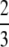 , at which the sign of 3M2 − 5M + 2 switches from positive to negative. ▪
, at which the sign of 3M2 − 5M + 2 switches from positive to negative. ▪
References
- Depaulis, F., and M. Veuille, 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15 1788–1790. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18 1136–1138. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 2005. A standardized genetic differentiation measure. Evolution 59 1633–1638. [PubMed] [Google Scholar]
- Hudson, R. R., K. Bailey, D. Skarecky, J. Kwiatowski and F. J. Ayala, 1994. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., K. Zhang, P. Marjoram, S. Tavaré and N. A. Rosenberg, 2005. Statistical tests of the coalescent model based on the haplotype frequency distribution and the number of segregating sites. Genetics 169 1763–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J. C., and R. A. Kittles, 2003. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 75 449–471. [DOI] [PubMed] [Google Scholar]
- Markovtsova, L., P. Marjoram and S. Tavaré, 2001. On a test of Depaulis and Veuille. Mol. Biol. Evol. 18 1132–1133. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Rosenberg, N. A., S. Mahajan, S. Ramachandran, C. Zhao, J. K. Pritchard et al., 2005. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1 660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. P. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419 832–837. [DOI] [PubMed] [Google Scholar]
- Tang, K., K. R. Thornton and M. Stoneking, 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5 1587–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian, C., T. T. Hu, M. J. Aranzana, C. Lister, C. Tang et al., 2006. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PLoS Biol. 4 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1977. Heterosis or neutrality? Genetics 85 789–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1978. The homozygosity test of neutrality. Genetics 88 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, K., S. Mano, S. Shi and C.-I. Wu, 2007. Comparisons of site- and haplotype-frequency methods for detecting positive selection. Mol. Biol. Evol. 24 1562–1574. [DOI] [PubMed] [Google Scholar]