Abstract
The X chromosome plays a central role in the evolution of reproductive isolation, but few studies have examined the genetic basis of X-linked incompatibilities during the early stages of speciation. We report the results of a large experiment focused on the reciprocal introgression of the X chromosome between two species of house mice, Mus musculus and M. domesticus. Introgression of the M. musculus X chromosome into a wild-derived M. domesticus genetic background produced male-limited sterility, qualitatively consistent with previous experiments using classic inbred strains to represent M. domesticus. The genetic basis of sterility involved a minimum of four X-linked factors. The phenotypic effects of major sterility QTL were largely additive and resulted in complete sterility when combined. No sterility factors were uncovered on the M. domesticus X chromosome. Overall, these results revealed a complex and asymmetric genetic basis to X-linked hybrid male sterility during the early stages of speciation in mice. Combined with data from previous studies, we identify one relatively narrow interval on the M. musculus X chromosome involved in hybrid male sterility. Only a handful of spermatogenic genes are within this region, including one of the most rapidly evolving genes on the mouse X chromosome.
THE X chromosome often plays a central role in the evolution of reproductive isolation between animal species with sex chromosomes. For example, studies in Drosophila have shown that substitution of the X chromosome between species has a larger impact on hybrid male fertility than introgression of autosomes (Dobzhansky 1936; Orr 1987; Coyne and Orr 1989; Masly and Presgraves 2007). Likewise, levels of gene flow between naturally hybridizing species tend to be lower on the X chromosome relative to the autosomes (Tucker et al. 1992; Dod et al. 1993; Besansky et al. 2003), suggesting selection against X-linked incompatibilities. The contribution of the X chromosome seems particularly strong when considering reproductive isolation between closely related species. Hybrid incompatibilities between incipient species are often sex limited and manifest in the heterogametic sex first (i.e., Haldane's rule; Haldane 1922; Laurie 1997; Orr 1997) due, in part, to the exposure of hemizygous X-linked incompatibilities (Muller 1942; Turelli and Orr 1995; Turelli and Orr 2000). Nevertheless, we know relatively little about the specific details of the genetic basis and architecture of X-linked reproductive isolation during the early stages of speciation.
Hybrid sterility and other forms of intrinsic postzygotic isolation are thought to be a consequence of deleterious epistasis between interacting genes that have diverged between species [i.e., Dobzhansky–Muller incompatibilities (D–M incompatibilities); Bateson 1909; Dobzhansky 1937; Muller 1942]. Hybrid male sterility is especially relevant to the onset of intrinsic isolation between species with heterogametic males because it has been shown to evolve much more quickly than female sterility or inviability in either sex (Wu 1992; True et al. 1996; Coyne and Orr 1997; Tao et al. 2003; Wu and Ting 2004; Masly and Presgraves 2007). Multiple factors may contribute to the rapid evolution of hybrid male sterility, including the exposure of recessive X-linked D–M incompatibilities (Muller 1942; Turelli and Orr 1995; Turelli and Orr 2000), the rapid evolution of male-limited traits (Wu and Davis 1993; Wu et al. 1996; Presgraves and Orr 1998), and a higher rate of positive selection on the X chromosome (Charlesworth et al. 1987). These forces might quickly lead to a complex genetic architecture for hybrid male sterility, as seen in Drosophila by both the rapid accumulation of X-linked sterility factors (True et al. 1996; Tao et al. 2003; Masly and Presgraves 2007) and the rapid evolution of complex epistasis within sets of interacting male sterility loci (Davis and Wu 1996; Sawamura et al. 2004).
House mice are well suited to address the genetic architecture of hybrid male sterility during the early stages of speciation. House mice are a well-studied model organism (Dietrich et al. 1996; Mouse Genome Sequencing Consortium 2002; Su et al. 2004; Shifman et al. 2006) consisting of at least three closely related species: Mus domesticus, M. musculus, and M. castaneus (also referred to as subspecies of M. musculus: M. m. domesticus, M. m. musculus, and M. m. castaneus). Divergence of these lineages initiated ∼500,000 years ago (She et al. 1990; Boursot et al. 1993). M. castaneus is distributed throughout southeastern Asia, M. musculus occurs in eastern Europe and Asia, and M. domesticus occurs in western Europe, the Middle East, Africa, Australia, and the Americas (Boursot et al. 1993). House mice live in close association with humans, and their ranges have been extended through human-mediated dispersal. All three pairwise combinations of these taxa hybridize to some extent in nature along established hybrid zones (M. domesticus–M. musculus; Boursot et al. 1993) or in mosaic or admixed populations (M. castaneus–M. musculus, Boursot et al. 1993; M. domesticus–M. castaneus; Orth et al. 1998).
The X chromosome is thought to play a primary role in isolation between M. domesticus and M. musculus. In the European hybrid zone, the X chromosome shows lower levels of gene flow relative to the autosomes (Tucker et al. 1992; Dod et al. 1993, 2005; Munclinger et al. 2002; Macholán et al. 2007; Teeter et al. 2008). Payseur et al. (2004) surveyed 13 X-linked loci across a transect in southern Germany and found significantly reduced introgression at several adjacent markers located in the middle of the chromosome, suggesting that this genomic region harbors at least one hybrid incompatibility. Moreover, some laboratory crosses between wild-derived inbred strains of M. domesticus and M. musculus result in asymmetric F1 hybrid male sterility (Britton-Davidian et al. 2005; Good et al. 2008). In these asymmetric crosses, sterility only occurs when the maternal strain is of M. musculus origin. This pattern is consistent with hybrid sterility caused by epistatic interactions between the hemizygous M. musculus X-linked or mitochondrial mutations and dominant autosomal and/or hemizygous Y-linked mutations in M. domesticus (Britton-Davidian et al. 2005; Good et al. 2008).
Two recent studies offer further insights into the genetic basis of X-linked hybrid male sterility between M. musculus and M. domesticus. First, Storchová et al. (2004) substituted the X chromosome of M. musculus (wild-derived strain PWD/Ph) onto the genomic background of M. domesticus as represented by the classic laboratory strain C57BL/6J. The resulting X (heterosomic) consomic females were normal while consomic males were sterile with significantly smaller testes, lower sperm count, and abnormal sperm head morphology (Storchová et al. 2004). Second, Oka et al. (2004) observed male-limited sterility when they introgressed the X chromosome of M. m. molossinus (laboratory strain MSM/Ms) into C57BL/6J. M. m. molossinus is derived from an ancient hybridization event between M. castaneus and M. musculus. Likewise, C57BL/6J and other classic laboratory strains of house mice include substantial genetic contributions from both M. musculus and M. castaneus (∼70–90% M. domesticus; Wade et al. 2002; Frazer et al. 2007; Yang et al. 2007). Both Storchová et al. (2004) and Oka et al. (2004) mapped multiple X-linked quantitative trait loci (QTL) contributing to hybrid male sterility in later backcross generations. QTL confidence intervals overlapped between the studies in some instances, including the identification of a major sterility QTL in the same general central region of the X chromosome that shows low introgression across the southern Germany transect (Payseur et al. 2004). However, since the genetic composition of classic inbred strains appears to have been strongly influenced by selection against hybrid incompatibilities (Payseur and Hoekstra 2005), the relevance of incompatibilities mapped using C57BL/6J to reproductive isolation in the wild is uncertain.
Here, we report the results of a large experiment focused on the introgression of the X chromosome between M. musculus and M. domesticus. Our ultimate goal is to elucidate the genetic basis and architecture of X-linked hybrid male sterility between these species. Our results extend previous studies on X-linked reproductive isolation in mice in three important ways. First, our introgression design is reciprocal. These data provide the first direct test for hybrid incompatibilities on the M. domesticus X chromosome and allow us to fully evaluate basic predictions of the D–M model of epistatic incompatibilities (Muller 1942; Coyne and Orr 2004). Second, this is the first study to examine the genetic basis of X-linked hybrid sterility using inbred strains exclusively derived from wild mice. Third, whereas previous studies introgressed the entire X chromosome, we primarily focused on smaller chromosomal segments. By selecting recombinants, we evaluated the independent contributions of specific introgressed regions spanning the X chromosome.
MATERIALS AND METHODS
Animals:
Two breeding colonies were established using wild-derived inbred strains purchased from the Jackson Laboratory (http://www.jax.org). We used the LEWES/EiJ strain to represent M. domesticus (hereafter domesticusLEWES). This strain was originally isolated from natural populations in Lewes, Delaware, and is thought to descend from recent human-mediated dispersal of M. domesticus from western Europe. M. domesticus from eastern North America are genetically indistinguishable from European M. domesticus in allozyme studies (Selander et al. 1969a,b). For M. musculus, we used the PWK/PhJ strain isolated from the central region of the Czech Republic (hereafter musculusPWK, Gregorová and Forejt 2000). In genetic analyses both strains cluster most closely with other wild-derived strains of M. musculus and M. domesticus, respectively (Petkov et al. 2004) and have a standard house mouse karyotype (2n = 40). Recently, some putative cases of introgression between M. domesticus and M. musculus have been reported for a few standard wild-derived strains of mice, including musculusPWK (Yang et al. 2007). The contribution of M. domesticus to the genome of musculusPWK has not been quantified, but levels of introgression are likely to be fairly low on the basis of available data (Gregorová and Forejt 2000). All mice were maintained in accordance with the Institutional Animal Care and Use Committee regulations at the University of Arizona Central Animal Facility.
Experimental design:
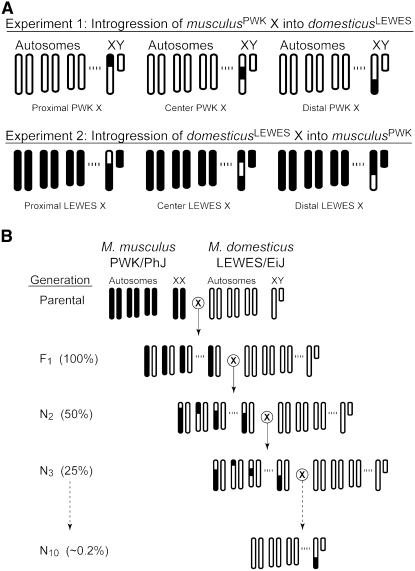
We performed two parallel introgression experiments using a standard backcross design (Figure 1). In experiment one, three genomic regions spanning the M. musculusPWK X chromosome were introgressed into the M. domesticusLEWES autosomal background. Following an initial intercross between ♀ musculusPWK × ♂ domesticusLEWES, F1 female offspring were backcrossed to ♂ domesticusLEWES for a total of nine generations (i.e., to generation N10). Each generation female offspring were screened at 18 diagnostic X-linked microsatellite markers (see below) and females heterozygous for at least one of the three targeted regions were used for the next backcross generation. In experiment two, we repeated this general design to introgress the same three genomic regions of the M. domesticusLEWES X chromosome into the M. musculusPWK genomic background. A total of seven backcross generations (i.e., to generation N8) were performed in experiment two.
Figure 1.—
Experimental design for the reciprocal introgression of the X chromosome. (A) Three targeted X chromosome congenic genotypes (proximal, center, and distal) for experiment 1 and experiment 2. For each male genotype, the genetic composition of three autosomes, the X chromosome, and the Y chromosome is given. White chromosomes represent domesticusLEWES and black chromosomes represent musculusPWK. For both experiments the mitochondrial DNA derives from musculusPWK. (B) Example of congenic strain construction for introgression of the distal musculusPWK X chromosome over 10 generations of breeding. Expected levels of heterozygosity for each generation are given in parentheses; at the N10 generation the genomic background will be ∼99.8% homozygous for domesticusLEWES alleles.
Data from previous studies were used to guide our decision on how to partition the X chromosome for our introgression experiments. Both genetic mapping (Oka et al. 2004; Storchová et al. 2004) and hybrid zone data (Payseur et al. 2004) suggest that the middle of the X chromosome is involved in reproductive isolation, including at least one QTL of major effect on hybrid male sterility, Hstx1, that maps to near marker DXMit119 (68.7 Mb, 29.5 cM; Storchová et al. 2004). We targeted three overlapping regions corresponding to the proximal (marker interval 6.8–62.3 Mb), central (62.3–100.3 Mb), and distal (100.3–162.6 Mb) regions of the X chromosome.
Treatment of experimental males:
Males were weaned into same-sex sibling groups of up to four and split into individual cages at ∼60 days old. All males were sacrificed as adults (all males ≥70 days old and ≥ 11.0 g body weight).
Quantification of male reproductive phenotypes:
While the ability to sire offspring may be taken as the most direct assessment of male fertility, this criterion provides little insight into the underlying phenotypic basis of reproductive failure. To provide a broad assessment of male reproductive condition we considered several general male reproductive phenotypes, including testis weight, sperm count, sperm motility, sperm head morphology, and seminal vesicle weight. In addition, we examined the reproductive success of some males using two different mating assays (see below).
Our methods for quantifying sperm count and motility have been described in detail previously (Good et al. 2008). Briefly, we evaluated sperm motility and count using a Makler counting chamber (Sefi-Medical Instruments, Haifa, Israel) and a light microscope at 200× magnification. Sperm from the caudal epididymides were incubated at 37° in modified phosphate-buffered saline for 10 min. Following incubation, 5 μl of mixed sperm suspension were transferred to the Makler chamber and the number of motile and immotile sperm was determined over a fixed area and observation time. Sperm counts were estimated with a Makler chamber using heat-shocked aliquots of the same sperm suspension.
To examine sperm head morphology, 25 μl of sperm suspension were spread onto a microscope slide and allowed to air dry. The slides were then fixed using a 1% acetic acid solution in 95% ethanol for two min and allowed to dry. Fixed preparations were then stained in 1% Eosin Yellow (Sigma) for five min, rinsed three times in 70% ethanol, dried, and mounted with 30 μl of Permount (Fisher). Mounted preparations were evaluated at 400× magnification using a phase contrast microscope. Sperm head morphology was classified into four different categories on the basis of the degree of abnormalities observed: (1) normal head morphology with a long apical hook (Russell et al. 1990), (2) heads with a shortened hook and/or flattened shape, (3) shortened heads with a very short or absent apical hook, and (4) a general class of severely abnormal shape. We considered a minimum of 100 individual sperm heads per male.
For a subset of males we examined fertility on the basis of two different mating assays. First, we considered fecundity on the basis of reproductive output following pairing with musculusPWK females. Males (>90 days old) were caged with a single female for 17 days and, when possible, each male was paired successively with at least two different females. Females were sacrificed and the number of embryos was determined. Second, we performed an in vivo assay of male fertilization potential by pairing males with females that were treated with hormones to induce ovulation (following Nagy et al. 2003). Because musculusPWK females do not respond to the hormone regiment, we used F1 females from a cross between musculusPWK females and M. musculus males from the CZECHII/EiJ wild-derived inbred strain (Jackson Laboratory, http://www.jax.org). Female progeny (5 weeks old) were intraperitoneally injected with 5 units of pregnant mare serum gonadotropin (PMSG, Calbiochem), followed 48 hr later with 5 units of human chorionic gonadotropin (hCG, Calbiochem). Ovulation occurs ∼12 hr after administration of hCG. Immediately following injection with hCG, we paired females individually with males. Females were sacrificed after 20 hr and cumulus masses were dissected from the oviducts and treated with hyaluronidase to isolate oocytes. We counted the number of oocytes with two pronuclei, two polar bodies, or neither. The two pronuclei stage occurs ∼6 hr after fusion of sperm and egg, while the two polar body stage occurs ∼8 hr after fusion (Nagy et al. 2003). Either stage was taken as evidence of fertilization. If we did not observe a vaginal plug, semen contents in the uterus, sperm around the oocytes, or any fertilized embryos, we assumed that mating did not occur.
Genotyping:
Each generation we screened females at 18 simple sequence-length polymorphism (SSLP) markers across the X chromosome (Dietrich et al. 1994, 1996). The screened loci, physical locations in megabases (Mb) based on NCBI mouse build 36 (Ensembl release 46, August, 2007; www.ensembl.org), and the consensus genetic location in centimorgans (cM) based on the Mouse Genome Database (MGI 3.54; Eppig et al. 2005) were as follows: DXMit26 (6.8 Mb, 1.5 cM); DXMit137 (14.9 Mb, 5.7 cM); DXMit81 (32.6 Mb, 9.25 cM); DXMit50 (37.1 Mb, 14.1 cM); DXMit166 (47.9 Mb, 15.5 cM); DXMit141 (55.8 Mb, 19.0 cM); DXMit46 (62.3 Mb, 24.5 cM); DXMit119 (68.7 Mb, 29.5 cM); DXMit195 (81.8 Mb, 32.0 cM); DXMit128 (89.5 Mb, 34.7 cM); DXMit158 (100.3 Mb, 43.6 cM); DXMit65 (104.9 Mb, 48.5 cM); DXMit97 (115.5 Mb, 49.0 cM); DXMit173 (125.7 Mb, 50.5 cM); DXMit67 (136.7 Mb, 60.0 cM); DXMit153 (146.4 Mb, 62.2 cM); DXMit121 (156.8 Mb, 67.0 cM); and DXMit160 (162.6 Mb, 73.3 cM). Experimental males from the N2, N4, N6, N7, N8, N9, and N10 generations were also screened with the same 18 markers.
DNA was extracted from fresh tissue (tail tip, ear notch, or liver) using a PureGene DNA isolation kit (Gentra Systems, Minneapolis). We used 10-μl reactions and, when possible, loci were multiplexed into groups of two or three. The forward primer of each locus was labeled with 6-FAM or HEX. We followed standard polymerase chain reaction (PCR) protocols for these loci (Dietrich et al. 1994, 1996) and we optimized annealing temperature, primer concentration, and the number of amplification cycles as necessary. Primer sequences were retrieved directly from Ensembl (http://www.ensembl.org), and specific PCR conditions and primer combinations are provided in the supplemental material. Genotypes were resolved on an ABI 3100 (Applied Biosystems) with a 500 ROX ladder and scored using Genotyper v2.0-3.7 (Applied Biosystems).
QTL analysis:
A total of 346 males from late backcross generations were used to map quantitative trait loci for several male reproductive phenotypes. For experiment one, we considered 204 males from the N6–N8 generations. For experiment two, 142 males from the N4, N6, and N7 generations were used. Most males included in QTL analyses were resolved for their entire 18-locus genotype and all males were successfully genotyped at a minimum of 14 markers.
Two complementary approaches on the basis of interval mapping were implemented using the QTL Cartographer software package. First, we used the program Zmapqtl to identify X-linked QTL using composite interval mapping (CIM; Zeng 1994). This general method combines background markers as cofactors with standard interval mapping (Lander and Botstein 1989) to increase the power and resolution of QTL identification. For each phenotype, we calculated the likelihood L1 that a given position contributes an additive effect (a ≠ 0) and compared this model to the likelihood L0 of the null hypothesis of no additive effect (a = 0) at the test site. Significance of additive effects was determined using a standard likelihood ratio test statistic [LRS = −2ln(L0/L1)] and critical values for the experiment were calculated with Zmapqtl using 1000 permutations (Churchill and Doerge 1994). The most informative markers for controlling genetic background were selected on the basis of the results of a forward stepwise regression conducted with SRmapqtl. All analyses were performed using a window size of 10 cM and a walking distance of 1 cM. Dominance effects could not be evaluated because males were hemizygous for all X-linked markers.
Following CIM analyses, we estimated a multiple QTL model using multiple interval mapping (MIM; Kao et al. 1999). Our MIM model was constructed using a stepwise approach (Kao et al. 1999; Zeng et al. 2000): (1) an initial model of multiple putative QTL was defined on the basis of the results of CIM; (2) the significance of individual QTL in the model was evaluated, and QTL with LRS scores below the CIM experimentwise critical values of α = 0.05 were removed; (3) positions of significant QTL were refined; (4) the X chromosome was scanned for additional additive QTL; and (5) pairs of all identified QTL were tested for significant epistatic interactions using a forward stepwise search. In addition to CIM and MIM analyses, we also applied simple linear regression and standard interval mapping (IM; Lander and Botstein 1989) to our data. Because our inferences were generally corroborated by these simpler models, albeit with less resolution, we focus on the results of CIM and MIM. A graphical comparison of results for CIM and IM are provided in supplemental Figure 1.
Analysis of candidate genes on the X chromosome:
We evaluated potential candidate genes for hybrid male sterility using a variety of sources. X-linked protein-coding genes were identified from Ensembl (release 46, August 2007; www.ensembl.org) on the basis of NCBI mouse build 36. Gene expression during spermatogenesis was based on data from a recent Affymetrix microarray analysis (Affymetrix 430 2.0) on four distinct classes of germ cells: premeiotic type A and B spermatogonia, pachytene spermatocytes, and postmeiotic round spermatids (Namekawa et al. 2006). Affymetrix probes were matched to protein-coding genes using the Ensembl Biomart tool. Testis specificity was calculated as the amount of testis expression relative to the median expression in a panel of 61 mouse tissues (GNF Symatlas, symatlas.gnf.org; Su et al. 2004).
RESULTS
Male reproductive phenotypes in the parental inbred strains:
Data from musculusPWK and domesticusLEWES for a suite of standard male reproductive phenotypes are presented in Table 1. In general, musculusPWK and domesticusLEWES were similar in overall body weight, sperm count, sperm motility, and paired seminal vesicle weight. A significant reduction in testis weight was observed in domesticusLEWES (Wilcoxon P < 0.001). This difference was strain specific and is not related to divergence between these species (Good et al. 2008). In general, testis weight and sperm count were both significantly correlated with body weight in both strains (Good et al. 2008). We used two different approaches to account for this correlation. First, for all comparisons we report testis weight relative to body weight (relative testis weight, RTW, milligrams of testis per gram of body weight). Second, in all quantitative genetic analyses we used residual trait scores on the basis of a standard least squares regression on body weight.
TABLE 1.
Male reproductive parameters of M. musculusPWK and M. domesticusLEWES
| Strain | n | Body weighta (SD) | Testis weightb (SD) | RTWc (SD) | Sperm count (×106) | Proportion motile sperm | Seminal vesicle |
|---|---|---|---|---|---|---|---|
| musculusPWK | 12 | 14.7 (1.2) | 78.2 (5.6) | 5.4 (0.4) | 15.3 (3.2) | 91.2% (3.2) | 69.8 (10.4) |
| domesticusLEWES | 10 | 14.2 (1.1) | 63.2 (3.0)* | 4.5 (0.3)* | 12.0 (3.5) | 91.8% (3.6) | 75.3 (23.0) |
*Wilcoxon P < 0.001.
All males between 74 and 103 days old. Body weight is in grams.
Single average testis weight in milligrams.
Relative testis weight in milligrams of single average testis per gram of body weight.
Asymmetric sterility in F1 hybrid males:
A previous study showed that crosses between a female musculusPWK and a male domesticusLEWES yield hybrid male offspring with abnormal spermatogenesis while the reciprocal cross produces reproductively normal F1 males (Good et al. 2008). These abnormal hybrid males were characterized by significant reductions in testis weight (Table 2) and sperm count (see also Table 2 in Good et al. 2008). Histological analyses revealed primarily postmeiotic abnormalities in spermatogenesis with a general reduction in the number of maturing spermatids (Good et al. 2008). Because the reciprocal F1 hybrids shared the same autosomal genotype, these data suggested that one or more genes on the M. musculus X chromosome, the M. musculus mitochondrial DNA, and/or the M. domesticus Y chromosome influenced spermatogenesis in F1 hybrid males. A reduction in testis weight was observed in both young males (60-day old) and older males (∼120-day old), demonstrating that this difference does not represent a developmental delay (Table 2).
TABLE 2.
Relative testis weights for reciprocal F1 hybrid males at different ages
| Cross | n | Agea (SD) | Body weightb (SD) | RTWc (SD) |
|---|---|---|---|---|
| ♀ musculusPWK × ♂ domesticusLEWES | 12d | 60 | 15.1 (1.4) | 3.3 (0.2)* |
| ♀ musculusPWK × ♂ domesticusLEWES | 29 | 122.5 (8.8) | 18.4 (2.2) | 3.5 (0.4)* |
| ♀ domesticusLEWES × ♂ musculusPWK | 11d | 60 | 17.1 (0.9) | 4.9 (0.3) |
| ♀ domesticusLEWES × ♂ musculusPWK | 18 | 123.5 (21.1) | 19.8 (1.3) | 4.7 (0.3) |
*Significantly lower vs. the same age class in the reciprocal cross, Wilcoxon P < 0.0001.
Age in days after birth.
Body weight is in grams.
Relative testis weight in milligrams of testis per gram of body weight.
Data for 60-day-old males from Good et al. (2008).
Experiment 1: introgression of the musculusPWK X chromosome into domesticusLEWES
Overview:
Introgression of the M. musculusPWK X chromosome into the M. domesticusLEWES autosomal background caused severe reductions in male fertility. We considered several male reproductive phenotypes in a total of 306 males sampled across nine backcross generations (N2–N10). We begin by reporting the general qualitative patterns observed during the early backcross generations and then provide a detailed account of the genetic basis of sterility in later backcross generations.
X-linked male sterility during the early backcross generations:
We examined X chromosome genotypes and several male reproductive phenotypes for 35 males from the first backcross generation (N2). These N2 males demonstrated some recovery of testis weight (RTW) relative to F1 males from the ♀ musculusPWK × ♂ domesticusLEWES (RTW = 4.2 mg/g, SD = 0.7; Wilcoxon P < 0.0001). Only 3 N2 males showed dramatic reductions in testis size (average RTW = 2.5 mg/g, SD = 0.6) and all 3 were hemizygous for the entire or most of the musculusPWK X chromosome. However, we did observe some variation in X-linked sterility in this first backcross generation, including multiple males with large introgressions of the musculusPWK X chromosome that had relatively normal fertility parameters. By the N4 generation (n = 55 total males, RTW = 4.2 mg/g, SD = 0.9), males with large introgressions of the musculusPWK X chromosome (>100 Mb of musculusPWK X chromosome, n = 8) were characterized by reduced testis weight compared to control backcross males (n = 12) with a domesticusLEWES X chromosome (Wilcoxon P = 0.012). There were two striking exceptions to this trend: the two most severely abnormal males (testis weight <25 mg and no sperm in their caudal epididymides) in this generation both had a domesticusLEWES X chromosome. The cause of this is unclear, but may be due to particular autosomal interactions that were uncovered in this but not subsequent generations. Dramatic reductions in male fertility were not observed in males with the domesticusLEWES X chromosome genotype in any of the later backcross generations.
Multiple factors on the musculusPWK X chromosome contributed to reduced testis weight, sperm count, and abnormal sperm head morphology:
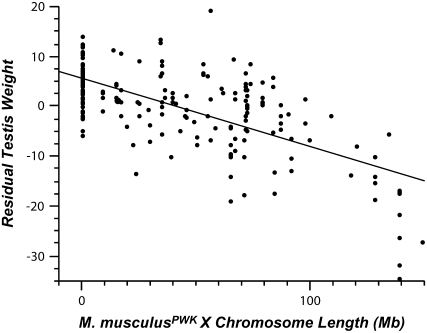
By the N6 generation none of the introgressed males in our experiment possessed a complete musculusPWK X chromosome. Nevertheless, it became apparent that multiple loci on the musculusPWK X chromosome were involved in male sterility and that the severity of reproductive disruption depended on the number of incompatibilities present in a given male. One simple pattern consistent with a multigenic basis to sterility was a significant negative linear correlation between the residual testis weight and the approximate length of the introgressed musculusPWK X chromosome, regardless of the location of the introgressed region (Figure 2; n = 202 N6–8 males, r2 = 0.45, P < 0.0001). Residual sperm count also showed a significant, albeit weaker, negative correlation with musculusPWK X chromosome introgression length (n = 198 males, r2 = 0.12, P < 0.0001). Body weight, seminal vesicle weight, and sperm motility were all uncorrelated with introgression length (all P > 0.40).
Figure 2.—
Negative correlation between length of musculusPWK X chromosome introgression and relative testis weight (n = 202 N6–8 males, r2 = 0.45, P < 0.0001). All males are from the N6, N7, or N8 generations of experiment 1. The abscissa depicts the length of introgressed regions regardless of their location on the X chromosome.
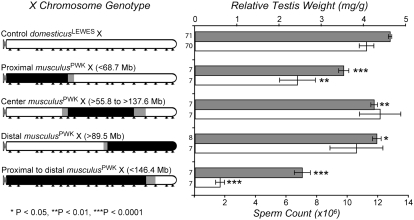
Of the three targeted chromosomal regions (Figure 1), introgression of the proximal portion of the musculusPWK X chromosome had the largest individual impact on male reproduction and resulted in a significant reduction in both testis weight and sperm count relative to control backcross males (Wilcoxon P < 0.01; Figure 3). Only slight reductions in testes weight and no reductions in sperm count were observed for males with either the central or distal musculusPWK X chromosome introgressions. Nevertheless, larger introgressions spanning the proximal, central, and distal portions of the musculusPWK X chromosome resulted in an even greater reduction in both testis weight and sperm count than introgression of the proximal segment alone (Figures 3).
Figure 3.—
Reduction in relative testis weight and sperm count for four different musculusPWK X chromosome introgressions. All males are from the N6, N7, or N8 generations of experiment 1. Bars indicate average values for relative testis weight (shaded) and sperm count (open), error bars indicate ±1 SE. Different genotypic classes are represented on the left margin with open chromosomal regions deriving from domesticusLEWES and solid chromosomes deriving from musculusPWK. The control genotype (all open) represents backcross males with a domesticusLEWES X chromosome. Sample sizes are given on the left margin. The locations of screened markers are indicated with arrowheads along the bottom. Shading for each genotype indicates uncertainty in recombination breakpoints between surveyed markers. Significance was based on Wilcoxon pairwise comparisons vs. the control backcross males.
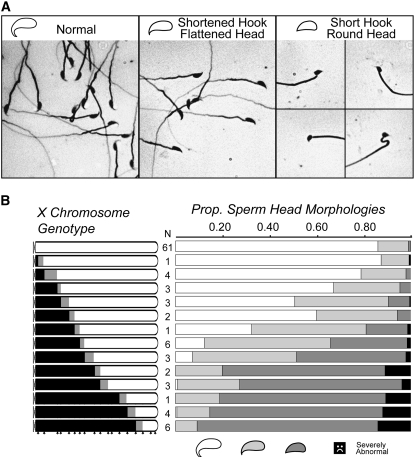
Sperm head morphology was even more strongly influenced by the genotype of the X chromosome in the later backcross generations. In mice, a rounded head shape with a long curved apical hook defines normal sperm morphology (Russell et al. 1990). We observed normal head morphology for the majority of sperm from either parental strain (domesticusLEWES, musculusPWK) and for control backcross males with a domesticusLEWES X chromosome (Figure 4). In contrast, most males with large portions of the musculusPWK X chromosome had sperm with severely abnormal head morphology; smaller introgressions produced less severe abnormalities, including a noticeable shortening of the apical hook and/or a flattened, less rounded head shape (Figure 4). Overall, we observed a very significant correlation between the proportion of sperm with abnormal head morphology and musculusPWK X chromosome introgression length (arcsine square-root transformed, n = 174, r2 = 0.75, P < 0.0001). As with testis weight, the proximal half of the musculusPWK X chromosome had the strongest influence on sperm head morphology. Figure 4B shows the negative influence of progressively larger introgressions of the musculusPWK X chromosome (proximal to distal) on sperm head morphology. Males with ≥81.9 Mb of the proximal musculusPWK X chromosome produced essentially no sperm with normal head morphology (Figure 4B).
Figure 4.—
Influence of the musculusPWK X chromosome on abnormal sperm head morphology in experiment 1. (A) Examples of three types of sperm head morphologies observed in N6–8 males from experiment 1. (B) Sperm head morphologies observed for males with progressively large introgressions of musculusPWK X chromosome (solid chromosomes). The first genotypic class represents control backcross males with a domesticusLEWES X chromosome (all open). Sample sizes are given on the left margin and the locations of screened markers are indicated with arrowheads along the bottom. Shading for each genotype indicates uncertainty in recombination breakpoints between surveyed markers. Sperm head morphology is partitioned into four phenotypic classes proceeding from normal to abnormal: (1) normal, (2) flattened head, shortened hook, (3) round head, short or absent hook, and (4) severely abnormal. Severely abnormal head morphologies were grouped together and took on a variety of dramatically altered shapes.
We localized four nonoverlapping intervals in recombinant males (0–37.1 Mb; 47.9–81.8 Mb; 89.5–125.7 Mb; and 125.7 Mb–telomere) that resulted in abnormal head morphology. Each interval represents the minimal genotype in each region that resulted in at least one male with proportionally fewer sperm with normal head morphology relative to the entire range of values observed in all control N6–8 males (range = 0.67–0.96 normal sperm, n = 61). Thus, at least four independent factors on the musculusPWK X chromosome contributed to abnormal sperm head morphology.
QTL mapping of sterility factors:
We used a combination of CIM and MIM to further resolve the number, location, and phenotypic effects of X-linked QTL contributing to reduced testis weight (n = 204), sperm count (n = 200), and abnormal sperm head morphology (n = 174) in males from the N6, N7, and N8 generations. Residual sperm count followed a normal distribution (Shapiro-Wilk W-test, P = 0.880). However, both testis weight and sperm head morphology were not normally distributed due to strong left skews. To reduce the influence of outliers, we performed a Box-Cox transformation on testis weight and an arcsine square-root transformation on sperm head morphology. In both cases the distributions were dramatically improved but still deviated from normality (both characters Shapiro-Wilk W-test, P < 0.03).
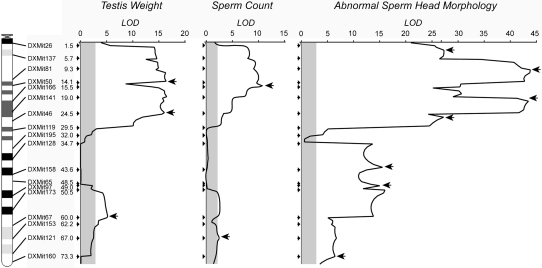
Residual testis weight, residual sperm count, and the proportion of abnormal sperm head morphology all showed evidence of multiple significant QTL on the basis of CIM. Figure 5 shows the likelihood of odds scores (LOD ≈ 0.217 likelihood ratio static) for all three traits as a function of genetic position on the X chromosome. For testis weight, LOD scores exceeded critical values (LOD = 2.82, α = 0.05) for much of the first third of the X chromosome (DXMit26–DXMit119) and between DXMit173 and DXMit67 on the distal end (Figure 5). The QTL with the largest effect on testis weight was estimated to occur near (position 18.5 cM) marker DXMit141 and explained 21.7% of the total phenotypic variance (LOD = 16.52). A similar pattern was found for sperm count. The QTL with the largest LOD score for sperm count (LOD = 10.69) was estimated to occur at position 15.1 cM and explained 21.9% of the total phenotypic variance.
Figure 5.—
Results for QTL mapping of hybrid male sterility phenotypes in experiment 1. LOD scores are based on CIM analysis of testis weight, sperm count, and the proportion of abnormal sperm in males from the N6, N7, and N8 generations. The physical location, identity, and genetic position (cM) of screened markers are given along the left margin. The significance threshold (α = 0.05) for each trait is shaded and is based on 1000 permutations. Arrows indicate the position of individual QTL that remained significant in MIM models.
The likelihood surface was more peaked in our analysis of sperm head morphology (proportion of abnormal sperm heads, arcsine square root). As with our analysis of testis weight and sperm count, the strongest influence was found for loci from the first third of the chromosome. Within this region, there were two distinct peaks and multiple peaked shoulders. The QTL with the largest influence on sperm head morphology accounted for 55.5% of the total phenotypic variation and was located at position 20.0 cM (LOD = 43.4). The central and distal end of the X chromosome also showed evidence of QTL for abnormal sperm head morphology, including one or more QTL near the center of the X chromosome where QTL were not observed for testis weight and sperm count. We also considered abnormal sperm head morphology using an index for each male on the basis of the proportion of different classes of abnormalities within a given male (index modified from Oka et al. 2004) and this alternative quantification produced very similar results in CIM analysis (analysis not shown).
Inspection of Figure 5 suggested that several of the QTL influencing testis weight, sperm count, and sperm morphology were nonindependent. We detected significant pairwise correlations between all three characters (all P < 0.0001; testis weight × sperm count, r2 = 0.26; testis weight × sperm head morphology, r2 = 0.50; sperm count × sperm head morphology, r2 = 0.18). The first principle component (PC1) of the covariance of these three traits accounted for 85.1% of the total variation and CIM analysis of PC1 produced qualitatively similar results (analysis not shown).
Resolution of the location and phenotypic effects of individual QTL identified by CIM was clearly influenced by the occurrence of multiple closely linked QTL. To help address this issue we constructed a multiple QTL model using MIM (Table 3 and Figure 5; Kao et al. 1999). In the MIM framework, R2 is the sum of the contributions of all singly significant QTL to the total genotypic variance divided by the total phenotypic variance and provides an estimate of broad sense heritability. Three QTL remained significant for testis weight, and heritability was moderately high (R2 = 0.631) while only two QTL remained significant for sperm count and heritability was fairly low (R2 = 0.241). For both traits, all QTL identified represented musculusPWK alleles that resulted in hybrid abnormalities and all loci of large effect (R2 > 0.10) were localized proximal to DXMit46 (24.5 cM, 62.3 Mb). By contrast, seven positions remained significant for abnormal sperm head morphology and these loci explained most of the phenotypic variation observed in the trait (R2 = 0.944). Three QTL of major effect were located proximal to DXMit119 (29.5 cM, 68.7 Mb) and one was found distal to DXMit97 (49.0 cM, 115.5 Mb). In addition, one locus contributed an additive effect in the opposite direction (Table 3), consistent with a positive influence of a musculusPWK allele within this region. We found no evidence for significant epistatic interactions between the identified QTL for any of the three characters.
TABLE 3.
Hybrid sterility QTL on the musculusPWK X chromosome identified using a MIM framework
| Trait and QTLa | Position (CI)b | Phenotypic contribution (%)c |
|---|---|---|
| Reduced testis weight | ||
| 1 | 13.3 (5.7–14.1) | 31.5 |
| 2 | 24.0 (15.1–25.5) | 25.2 |
| 3 | 59.5 (50.5–63.2) | 6.4 |
| Total | 63.1 | |
| Low sperm count | ||
| 1 | 15.1 (5.7–15.5) | 20.9 |
| 2 | 67.0 (50.5–UD) | 3.2 |
| Total | 24.1 | |
| Abnormal sperm head | ||
| 1 | 2.5 (0.1–UD) | 9.7 |
| 2 | 10.3 (7.7–14.1) | 23.6 |
| 3 | 19.0 (18.5–24.5) | 26.9 |
| 4 | 25.5 (24.5–27.5) | 22.6 |
| 5 | 42.7 (40.7–43.6) | −9.7 |
| 6 | 49.0 (47.6–50.0) | 21.1 |
| 7 | 73.3 (UD–75.3) | 0.2 |
| Total | 94.4 |
Significant QTL in MIM analyses.
Position in centimorgans estimated by MIM. CI, 2-LOD confidence interval based on CIM analysis; UD, undefined interval based on complete overlap with neighboring QTL or extending to the telomere/centromere.
Estimate of R2 in the MIM model.
Male fecundity and fertility of select genotypes:
We used two different mating assays to evaluate the relative reproductive success of males from three genotypic classes from the N9 or N10 generations: control (domesticusLEWES), proximal (<DXMit119, 29.5 cM, 68.7 Mb), and all major QTL (<DXMit153, 62.2 cM, 146.4 Mb; see Table 3). The average fecundity (measured as the average number of embryos produced per male) of proximal males was only marginally reduced relative to control backcross males (Wilcoxon P < 0.04; Table 4). However, we observed a dramatic and consistent reduction between these two genotypic classes for in vivo fertilization rate (Fisher's exact test, P < 0.0001; Table 4). Both males containing all major QTL from the musculusPWK X chromosome were completely sterile in our fecundity and fertilization assays and failed to sire offspring or fertilize eggs in repeated trials.
TABLE 4.
Fecundity and fertilization rates of males with the proximal portions of the musculusPWK X chromosome
| Fecundity
|
Fertilization rate
|
|||||||
|---|---|---|---|---|---|---|---|---|
| X genotype | Malesa | Crosses | Average no. embryosb (SD) | Attempted matings | Successful matingsc | Fertilized eggs | Unfertilized eggsd | % fertilized |
| Control domesticusLEWES X | 5 | 10 | 7.5 (0.7) | 15 | 11 | 84 | 271 | 23.7 |
| Proximal musculusPWK X (<68.7 Mb) | 5 | 10 | 5.4 (1.3)* | 20 | 9 | 13 | 433 | 2.9** |
| Proximal to distal musculusPWK X (<146.4 Mb) | 2 | 4 | 0 | 6 | 0 | — | — | — |
*Significantly lower fecundity, Wilcoxon P < 0.04; **Significantly lower fertilization rate, Fisher's exact test, P < 0.0001.
All males from the N9 or N10 generation.
Average number of embryos per male.
Matings where a vaginal plug, semen contents in the uterus, sperm around the oocytes, or fertilized embryos were observed.
Number of unfertilized eggs observed in successful matings.
Experiment 2: introgression of the M. domesticusLEWES X chromosome into M. musculusPWK
Overview:
Asymmetric sterility in the reciprocal F1 crosses between M. domesticusLEWES and M. musculusPWK suggests that the domesticusLEWES X chromosome was not involved in F1 hybrid male sterility (Good et al. 2008). Nevertheless, this asymmetric pattern does not preclude the existence of factors on the M. domesticusLEWES X chromosome contributing to hybrid male sterility because epistatic interactions between hemizygous X-linked loci and recessive autosomal incompatibilities are masked in the F1 generation. Therefore, we introgressed three genomic regions spanning the domesticusLEWES X chromosome onto the musculusPWK genomic background. We considered a total of 227 males in four backcross generations (N2, N4, N6, and N7). In contrast to experiment 1, we observed no introgressed hybrid males with strongly reduced reproductive parameters relative to the parental strains. However, M. musculusPWK had significantly larger testis than M. domesticusLEWES (Table 1) and our experiment identified two X-linked QTL contributing to differences in testis size between these strains.
X-linked loci contributed to differences in testis size between M.musculusPWK and M. domesticusLEWES:
The average RTW of male progeny from the first backcross generation (N2) was intermediate between M. domesticusLEWES and M. musculusPWK, increased across later backcross generations, and approached the values characteristic of M. musculusPWK (Table 5). To determine if X-linked loci contributed to differences in testis weight between M. domesticusLEWES and M. musculusPWK we genotyped 142 males from the N4, N6, and N7 generations. In these males, relative testis weight showed a significant negative correlation with the approximate length of the introgressed domesticusLEWES X chromosome (n = 142, r2 = 0.21, P < 0.0001), consistent with multiple X-linked loci contributing to differences in testis weight between the strains.
TABLE 5.
Relative testis weights for backcross males from experiment 2
| Generation | No. males | RTWa (SD) |
|---|---|---|
| musculusPWK | 12 | 5.4 (0.4) |
| domesticusLEWES | 10 | 4.5 (0.3) |
| F1b | 18 | 4.7 (0.3) |
| N2 | 47 | 4.8 (0.6) |
| N4 | 61 | 5.0 (0.6) |
| N6 | 72 | 5.4 (0.4) |
| N7 | 47 | 5.2 (0.4) |
Relative testis weight in milligrams of testis per gram of body weight.
F1 males from a ♀ domesticusLEWES × ♂ musculusPWK cross.
QTL mapping of X-linked factors influencing testis weight:
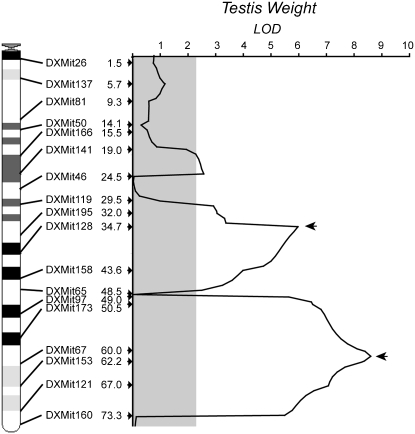
CIM and MIM analysis of testis weight (residuals of Box-Cox transformed values) of 142 males identified two major QTL in the central (34.7 cM, 34.0–44.6 2-LOD confidence interval) and distal (61.0 cM, 50.5–68.0) regions of the X chromosome, respectively (Figure 6). A third putative QTL was borderline significant (LOD = 2.55) on the basis of a critical value of LOD = 2.26 (α = 0.05, 1000 permutations) in CIM but was not significant in our MIM model. Overall, the additive effects of these two major QTL explained 46.8% of the variance in testis weight in our MIM model (R2 = 0.468), with the largest contribution coming from distal most QTL (61.0 cM, R2 = 0.278). We did not detect any significant epistatic interactions between these two QTL.
Figure 6.—
Results for QTL mapping of differences in testis weight between domesticusLEWES and musculusPWK in experiment 2. LOD scores are based on CIM analysis of testis weight in males from the N4, N6, and N8 generations. The physical location, identity, and genetic position (cM) of screened markers are given along the left margin. The significance threshold (α = 0.05; LOD = 2.25) is indicated with shading and is based on 1000 permutations. Arrows indicate the position of individual QTL that remained significant in MIM models.
DISCUSSION
Introgression of the M. musculus X chromosome into a wild-derived M. domesticus genetic background produced male-limited sterility, consistent with previous experiments using classic inbred strains to represent M. domesticus. QTL analysis of late backcross generation males implicated multiple hybrid incompatibilities and thus a fairly complex genetic basis to X-linked sterility. In some instances our patterns were in agreement with previous mapping experiments, while in other cases we found evidence for apparently novel hybrid incompatibilities. No sterility factors were uncovered during the reciprocal introgression of a wild-derived M. domesticus X chromosome onto a M. musculus genetic background. However, we mapped two loci involved in differences in testis weight between the two wild-derived strains we considered, including one QTL coincident with a sterility factor on the M. musculus X chromosome. Below we discuss the broader significance of our findings in relation to the evolution of D–M incompatibilities during the early stages of speciation. We then contrast our results with previous studies looking at X-linked reproductive isolation between these species, including patterns of gene flow in the hybrid zone. Finally, we discuss specific X-linked genes that are potential candidates for hybrid male sterility in mice.
The evolution of X-linked hybrid male sterility:
The simplest form of the D–M model involves epistatic interactions between two loci with alternative alleles fixed between species (e.g., AAbb and aaBB). Under this scenario, the genetic basis of hybrid dysfunction will be asymmetric and arise from interactions between the A and B alleles (i.e., incompatibilities should not involve interactions between a and b; Muller 1942; Orr 1995). This prediction was uniformly supported in our reciprocal introgression experiments with asymmetric genetic interactions evident for all of the X-linked loci underlying hybrid male sterility between M. domesticus and M. musculus. These data provide strong support for the D–M model and add to data from other reciprocal introgression experiments in animals where individual sets of epistatic interactions have been shown to be asymmetric (e.g., Wu and Beckenbach 1983; Vigneault and Zouros 1986; Orr 1989; Wittbrodt et al. 1989; Johnson et al. 1992).
All of the D–M incompatibilities we identified in later backcross generations involved epistatic interactions between incompatibilities on the M. musculus X chromosome and the M. domesticus genomic background. F1 hybrid male sterility was also asymmetric in crosses between M. domesticusLEWES and M. musculusPWK (Table 2; Good et al. 2008). While asymmetric sterility at the F1 and later backcross generations could reflect the same incompatibilities, the genetic composition of hybrid males was fundamentally different at these two stages of the experiment. In particular, the effects of recessive autosomal hybrid incompatibilities will be masked in F1 males but exposed in later backcross generations. Thus, both the genetic basis of X-linked sterility and the overall sterility phenotype could change over the course of our experiment. We observed no evidence for a shift in the developmental timing of sterility. Hybrid male sterility in F1 males (♀ musculusPWK × ♂ domesticusLEWES) involves primarily postmeiotic disruptions in spermatogenesis, resulting in morphologically abnormal sperm, reduced testis size, and lower sperm count (Good et al. 2008) and a similar phenotype was manifest in N4 and later-generation males with large introgressions of the musculusPWK X chromosome (Figure 4B). However, we did observe some variation in patterns of sterility in the N2 generation that may reflect important differences in the genetic basis of sterility in F1 and later backcross generation males.
The occurrence of multiple linked sterility factors makes it difficult to resolve the total number of incompatibilities on the musculusPWK X chromosome. There were at least four nonoverlapping intervals on the X chromosome that contributed to abnormal sperm head morphology, and MIM analysis implicated a total of seven QTL for this trait (Figure 5, Table 3). Overall, QTL within these four regions made largely additive contributions to hybrid male sterility (Table 3), resulting in complete sterility when all major QTL were combined on the same genotype (see also Storchová et al. 2004). By contrast, males with the proximal portion of the musculusPWK X chromosome were still capable of siring relatively large litters (Table 4). While the majority of sperm produced by these proximal males presented some morphological aberrations, some normal sperm were also observed and abnormalities were generally much less severe than sterile males with larger introgressions (Figure 4B). Interestingly, we did detect a dramatic reduction in the fertilization potential of proximal males relative to control backcross males (Table 4). This discrepancy between a striking reduction in fertilization potential when paired with “super-ovulated” females and a weak reduction in male fecundity (i.e., the number of late-stage embryos produced) when paired with noninduced females suggests that overall litter size was not sperm limited in these benign laboratory conditions and that these subfertile males nevertheless ejaculate enough viable sperm to assure a normal litter size. Nevertheless, it seems likely that these subfertile mutations could strongly reduce male fitness in nature where females often mate with multiple males (Dean et al. 2006), generating intense postcopulatory sperm competition among males.
We did not detect significant epistatic interactions between QTL identified in our MIM models; however, we cannot rule out interactions among tightly linked loci within these regions. Indeed, we observed abnormal sperm head morphology in multiple males containing a relatively small interval in the center of the musculusPWK X chromosome, but we were unable to fine-scale map the phenotype with smaller introgressions (data not shown). Similar patterns have been observed when mapping X-linked sterility in Drosophila (Perez and Wu 1995; Davis and Wu 1996), and complex epistasis within linked regions may be a common pattern of hybrid male sterility (Sawamura et al. 2004). While our data do not speak to the number of interacting partners in specific D–M incompatibilities (but see Oka et al. 2007), X-linked hybrid male sterility in mice does appear to have a relatively complex, multigenic basis overall.
At least two other studies have mapped X-linked QTL underlying differences in testis weight between inbred strains of mice (Le Roy et al. 2001; Bolor et al. 2006), suggesting the X chromosome plays an important role in regulation of mouse spermatogenesis. The QTL that we have identified on the X chromosome further support this view. This is noteworthy for two reasons. First, the X chromosome in mammals comprises a fairly small portion of the overall genome (i.e., ∼4% of known protein-coding genes are X linked in mice) compared to some other species (e.g., 20–40% of the Drosophila genome is X linked). Second, like Drosophila, genes involved in spermatogenesis are proportionally less frequent on the mouse X chromosome than the autosomes due to transcriptional inactivation of most X-linked genes during and after meiosis (Khil et al. 2004). Therefore, X-linked spermatogenesis genes comprise a very small portion of the overall testis transcriptome (∼2.4%; Khil et al. 2004), especially when considering genes involved in postmeiotic spermiogenesis. Thus it seems likely that there are many additional sterility factors that are on autosomes. In Drosophila, X-linked male sterility factors are approximately two and a half times more frequent than autosomal incompatibilities (Tao et al. 2003), a pattern referred to as the “large X effect.” If a similar ratio occurs in mice, assuming a minimum of 4 X-linked sterility factors, we might expect on the order of 65 autosomal recessive hybrid male sterility factors. It is important to bear in mind, however, that a large X effect has not yet been demonstrated in mice, and this calculation is thus very rough. Two recent studies constructed large sets of consomic lines substituting individual chromosomes from M. m. molossinus (MSM/Ms; Takada et al. 2008) or M. musculus (PWK/Ph; Gregorová et al. 2008) on the background of the classic laboratory strain C57BL/6J. Both experiments uncovered putative incompatibilities involving most autosomes, with deleterious effects on strain reproduction and viability. The extent to which these results involve male sterility phenotypes has not been determined, and some evidence exists for postzygotic isolation between M. musculus and M. domesticus due to hybrid inviability (Sage et al. 1986) and female sterility (Britton-Davidian et al. 2005).
Comparison with previous studies on reproductive isolation in mice:
Two previous studies have focused on the genetic basis of X-linked sterility between M. musculus and M. domesticus (Oka et al. 2004; Storchová et al. 2004). Both studies reported male-limited sterility related to the introgression of the M. m. musculus (Storchová et al. 2004) or M. m. molossinus (a hybrid between M. musculus and M. castaneus; Oka et al. 2004) X chromosome onto the genetic background of the classic laboratory strain C57BL/6J. The degree of concordance between our data and these studies depends on which sterility-related phenotypes were considered. Neither of the previous studies detected QTL contributing to testis weight on the proximal portion of the X chromosome (all QTL > 60.2 Mb, Oka et al. 2004; Storchová et al. 2004), the same region where we detected the largest phenotypic effects (Figure 5, Table 3). In general, the predominant influence of the proximal portion of the X chromosome in our study stands as the strongest discrepancy with previous mapping efforts. This difference may reflect genetic differences between the different introgressed M. musculus X chromosomes and/or the genomic background of C57BL/6J vs. domesticusLEWES.
All three studies consistently identified multiple QTL influencing abnormal sperm head morphology spanning the X chromosome. Our minimum estimate of four sterility factors related to abnormal sperm head morphology exceeds the estimates of both previous studies, where the independent influences of specific intervals were not considered in detail (Oka et al. 2004; Storchová et al. 2004). Oka et al. (2004) described three major QTL (sperm head abnormality 1, Sha 2, Sha 3) that overlap with our proximal, central, and distal QTL intervals (Figure 5). Sha 2 also overlapped with the primary hybrid sterility-related QTL (hybrid sterility-X1 or Hstx1) identified by Storchová et al. (2004) and maps to the same region where we identified two QTL for abnormal sperm head morphology and one QTL for reduced testis weight (Table 3). When considered jointly, these data strongly suggest that one or more genes involved in reproductive isolation between M. musculus and M. domesticus reside within this region (see below).
Several parallels between our data on the genetics of hybrid male sterility and patterns of gene flow across the European hybrid zone merit further consideration. First, the two X-linked markers with lowest levels of gene flow across the hybrid zone are located in the central (Pola1, 89.9Mb) and distal (Btk2, 132.1Mb) regions of the chromosome (Payseur et al. 2004). Pola1 is distal to our major central QTL, as well as the Sha 2/Hstx1 interval. However, mapping resolution on the X chromosome has proven to be fairly limited in the hybrid zone and several adjacent markers also show reduced gene flow (Payseur et al. 2004), resulting in a large region of restricted gene flow (64.9–99.8 Mb) that partially overlaps with QTL identified in our study as well as Sha 2/Hstx1. Btk2 is clearly contained within the confidence interval of our distalmost QTL for reduced testis weight (Table 3).
Second, asymmetric introgression has been noted across several hybrid zone transects with a tendency for more introgression of M. domesticus alleles into M. musculus (Vanlerberghe et al. 1986; Tucker et al. 1992; Dod et al. 1993, 2005; Teeter et al. 2008). In light of our data on asymmetric X-linked sterility, it is tempting to speculate that this pattern reflects a general accumulation of more incompatibilities on the M. musculus background. However, the fact that asymmetric introgression in the hybrid zone is seen for loci throughout the genome suggests that it may be driven primarily by demographic processes rather than selection on individual loci.
Finally, levels of gene flow across the entire X chromosome are lower than levels generally found on the autosomes (Tucker et al. 1992; Dod et al. 1993; Payseur et al. 2004; Macholán et al. 2007; Teeter et al. 2008). This pattern would not be expected if the X chromosome harbored only one or a few incompatibilities of major effect. Our mapping study was designed to detect fairly specific abnormalities in male reproduction (of moderate to large effect) while hybrid zone data should be sensitive to a broad range of isolation phenotypes (Payseur et al. 2004). Thus, concordance between our data and hybrid zone studies is consistent with the notion that hybrid male sterility plays an important role in reproductive isolation in nature, but does not address the contribution of other X-linked phenotypes to speciation in house mice.
Candidate genes for hybrid male sterility:
Our QTL analysis indicated that one or more loci involved in reduced testis weight and abnormal sperm head morphology occur within the 12.88 Mb interval between DXMit141 and DXMit119. This general QTL interval represents the clearest region of overlap between the current work and previous studies (Oka et al. 2004; Storchová et al. 2004). In the work of Storchová et al. (2004), the proximal boundary of major QTL for reduced testis weight and sperm count is clearly demarcated at DXMit144 (60.24 Mb). If we further restrict our interval to this proximal boundary, then the region of interest is reduced to 8.44 Mb (DXMit144 at 60.24 Mb to DXMit119 at 68.66). Of the 1026 protein-coding genes currently annotated on the mouse X chromosome, 41 are located within this region. However, only 9 of these loci (Table 6) were expressed during spermatogenesis in a recent survey of X-linked expression in male germ cells (Namekawa et al. 2006). Of these 9 genes, 3 (Ctag2, 4933436I01Rik, and 1700020N15Rik) show high expression in testis relative to other tissues (>3× median across 61 tissues; Su et al. 2004). All 3 genes are expressed primarily in postmeiotic round spermatids (Namekawa et al. 2006) and have no known function. 1700020N15Rik is a paralog to ENSMUSG00000056815 and both genes match the same probe on the Affymetrix 430 2.0 chip used by Namekawa et al. (2006). Thus, putative spermatogenesis expression of these 2 genes was ambiguous.
TABLE 6.
Nine candidate genes for X-linked hybrid male sterility in mice
| Gene symbol | Position (Mb)a | Spermatogenic expressionb | Relative testis expressionc | dN:dSd |
|---|---|---|---|---|
| 3830417A13Rik | 60.43 | Postmeiotic | 2.1 | 0.43 |
| Ctag2 | 61.31 | Postmeiotic | 8.3 | 0.43 |
| 4933436I01Rik | 64.18 | Postmeiotic | 4.2 | 0.85 |
| Fmr1nb | 65.02 | Pre and post | 1.9 | 0.64 |
| ENSMUSG00000056815 | 65.15 | Postmeiotice | NA | NA |
| 1700020N15Rik | 66.21 | Postmeiotice | 14.8 | NA |
| BC023829 | 66.72 | Pre and post | 1.0 | 0.00 |
| Cd99l2 | 67.68 | Premeiotic | 0.1 | 0.40 |
| Hmgb3 | 67.82 | Premeiotic | 1.2 | NA |
Physical position based on NCBI mouse build 36 (Ensembl 46, August 2007).
Expression in spermatogenic germ cells (Namekawa et al. 2006).
Relative expression in adult testis vs. median value expression across 61 tissues (Su et al. 2004).
Rate of protein evolution based on pairwise comparison between one-to-one orthologs in the rat (Dean et al. 2008).
ENSMUSG00000056815 and 1700020N15Rik are paralogs that were interrogated with the same Affymetrix probes.
Multiple genes involved in reproductive isolation in Drosophila have been shown to be rapidly evolving (Ting et al. 1998; Barbash et al. 2003, 2004; Presgraves et al. 2003; Brideau et al. 2006) and positive selection is thought to be an important process in the evolution of D–M incompatibilities (Coyne and Orr 2004). The two testis-specific genes with unambiguous orthologs in rat (4933436I01Rik and Ctag2) show elevated rates of protein evolution, as is common for testis- and other tissue-specific genes in mice (Winter et al. 2004; Good and Nachman 2005; Dean et al. 2008). In particular, 4933436I01Rik is among the most rapidly evolving genes on the X chromosome (highest 3% dN:dS values based on pairwise comparison with one-to-one rat orthologs) and was originally identified in a genomewide cDNA screen of adult testis (Bono et al. 2003). Inspection of known SNPs (dbSNP version 26) in wild-derived inbred strains of Mus revealed three nonsynonymous substitutions between M. musculus (sequenced strain PWD/PhJ) and M. domesticus (sequenced strain WSB/EiJ). Further population genetic analyses will be needed to evaluate whether 4933436I01Rik and other testis-expressed genes within this interval have evolved due to positive directional selection.
Finally, hybrid male sterility in mice may be influenced by locus-specific or X chromosomewide transcription misregulation in hybrid testis. Disrupted genomic imprinting has been put forth as a general mechanism for asymmetric isolation in mammals (Vrana 2007) and stands as a formal possibility for X-linked sterility in house mice (Storchová et al. 2004; Homolka et al. 2007). Likewise, expression divergence between M. musculus and M. domesticus of specific X-linked loci could also be involved in the disruption of spermatogenesis in hybrids. Abnormal patterns of male-biased gene expression are common in Drosophila F1 hybrids and may contribute to male-limited sterility (Michalak and Noor 2003; Ranz et al. 2004). Interestingly, a recent study found that the testis shows high levels of expression divergence between M. musculus and M. domesticus relative to liver and brain (Rottscheidt and Harr 2007). Rottscheidt and Harr (2007) did not find strong evidence for misexpression in testis of F1 hybrid males from reciprocal crosses between M. musculus and M. domesticus. However, they did not evaluate the fertility of F1 males in their experiment and F1 hybrid male sterility is polymorphic between these species (Forejt and Iványi 1975; Britton-Davidian et al. 2005; Vyskocilová et al. 2005; Good et al. 2008). Thus, the potential relevance of these data to reproductive isolation remains speculative. We are in the process of examining patterns of testis expression in M. musculus, M. domesticus, reciprocal F1 hybrids, and select congenic models for which detailed male reproductive data are also available.
Acknowledgments
We thank T. Shiroishi for sharing published data on the genetics of hybrid male sterility in mice. M. A. Handel, C. W. Birky, B. Payseur, A. Doyle, and T. Quill provided useful advice on experimental design and methodology. C. W. Birky and M. Worobey provided equipment used during the course of this experiment. We are grateful to T. Shiroishi, A. Oka, R. Storchová, B. Payseur, M. Macholán, two anonymous reviewers, and members of the Nachman lab for critical comments on this manuscript. We thank B. Gibson, M. Longhi, K. Smith, M. Sans-Fuentes, M. Rand, and the staff of the University of Arizona Central Animal Facility for assistance with mouse husbandry. J.M.G. received support from the National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship (IGERT) Program in Genomics at the University of Arizona, an NSF Doctoral Dissertation Improvement grant (DEB0608452), and small research grants from the Department of Ecology and Evolutionary Biology, University of Arizona. This work was also supported by NSF grants (DEB0213013, DEB0642778) and a National Institutes of Health grant (RO1 GM-074245) to M.W.N.
References
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash, D. A., P. Awadalla and A. M. Tarone, 2004. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson, W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by A. C. Seward. Cambridge University Press, Cambridge, UK.
- Besansky, N. J., J. Krzywinski, T. Lehmann, F. Simard, M. Kern et al., 2003. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolor, H., N. Wakasugi, W. D. Zhao and A. Ishikawa, 2006. Detection of quantitative trait loci causing abnormal spermatogenesis and reduced testis weight in small testis (Smt) mutant mouse. Exp. Anim. 55 97–108. [DOI] [PubMed] [Google Scholar]
- Bono, H., K. Yagi, T. Kasukawa, I. Nikaido, N. Tominaga et al., 2003. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res. 13 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot, P., J.-C. Auffray, J. Britton-Davidian and F. Bonhomme, 1993. The evolution of house mice. Ann. Rev. Ecol. Syst. 24 119–152. [Google Scholar]
- Brideau, N. J., H. A. Flores, J. Wang, S. Maheshwari, X. Wang et al., 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 1292–1295. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian, J., F. Fel-Clair, J. Lopez, P. Alibert and P. Boursot, 2005. Postzygotic isolation between two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. 84 379–393. [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. H. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130 113–146. [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. Two rules of speciation, pp. 180–207 in Speciation and Its Consequences, edited by D. Otte and J. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., and H. A. Orr, 1997. Patterns of speciation in Drosophila revisited. Evolution 51 295–303. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Davis, A. W., and C. I. Wu, 1996. The broom of the sorcerer's apprentice: the fine structure of a chromosomal region causing reproductive isolation between two sibling species of Drosophila. Genetics 143 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, M. D., K. G. Ardlie and M. W. Nachman, 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol. Ecol. 15 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, M. D., J. M. Good and M. W. Nachman, 2008. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Mol. Biol. Evol. 25 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, W. F., J. C. Miller, R. G. Steen, M. Merchant, D. Damron et al., 1994. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat. Genet. 7 220–245. [DOI] [PubMed] [Google Scholar]
- Dietrich, W. F., J. Miller, R. Steen, M. A. Merchant, D. Damron-Boles et al., 1996. A comprehensive genetic map of the mouse genome. Nature 380 149–152. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Dod, B., L. S. Jermiin, P. Boursot, V. H. Chapman, J. T. Nielsen et al., 1993. Counterselection on sex chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 6 529–546. [Google Scholar]
- Dod, B., C. Smadja, R. C. Karn and P. Boursot, 2005. Testing for selection on the androgen-binding protein in the Danish mouse hybrid zone. Biol. J. Linn. Soc. 84 447–459. [Google Scholar]
- Eppig, J. T., C. J. Bult, J. A. Kadin, J. E. Richardson and J. A. Blake, 2005. The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 33 D471–D475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forejt, J., and P. Iványi, 1975. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24 189–206. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., E. Eskin, H. M. Kang, M. A. Bogue, D. A. Hinds et al., 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448 1050–1053. [DOI] [PubMed] [Google Scholar]
- Good, J. M., and M. W. Nachman, 2005. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol. Biol. Evol. 22 1044–1052. [DOI] [PubMed] [Google Scholar]
- Good, J. M., M. A. Handel and M. W. Nachman, 2008. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorová, S., and J. Forejt, 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies: a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. 46 31–41. [DOI] [PubMed] [Google Scholar]
- Gregorová, S., P. Divina, R. Storchová, Z. Trachtulec, V. Fotopulosova et al., 2008. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 18 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12 101–109. [Google Scholar]
- Homolka, D., R. Ivanek, J. Capkova, P. Jansa and J. Forejt, 2007. Chromosomal rearangement interferes with meiotic X chromosome inactivation. Genome Res. 17 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, N. A., D. E. Perez, E. L. Cabot, H. Hollocher and C.-I. Wu, 1992. A test of reciprocal X-Y interactions as a cause of hybrid sterility in Drosophila. Nature 358 751–753. [DOI] [PubMed] [Google Scholar]
- Kao, C.-H., Z.-B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil, P. P., N. A. Smirnova, P. J. Romanienko and R. D. Camerini-Otero, 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36 642–646. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics 147 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy, I., S. Tordjman, D. Migliore-Samour, H. Degrelle and P. Roubertoux, 2001. Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán, M., P. Munclinger, M. Sugerková, P. Dufková, B. Bímová et al., 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61 746–771. [DOI] [PubMed] [Google Scholar]
- Masly, J. P., and D. C. Presgraves, 2007. High-resolution genome-wide dissection of two rules of speciation in Drosophila. PLoS Biol. 5 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak, P., and M. A. F. Noor, 2003. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20 1070–1076. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520–562. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Munclinger, P., E. Boziková, M. Sugerková, J. Piálek and M. Macholán, 2002. Genetic variation in house mice (Mus, Muridae, Rodentia) from the Czech and Slovak Republics. Folia Zool. 51 81–92. [Google Scholar]
- Nagy, A., M. Gertsenstein, K. Vintersten and R. R. Behringer, 2003. Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Namekawa, S. H., P. J. Park, L.-F. Zhang, J. E. Shima, J. R. McCarrey et al., 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16 660–667. [DOI] [PubMed] [Google Scholar]
- Oka, A., A. Mita, N. Sakurai-Yamatani, A. Yamamoto, N. Takagi et al., 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, A., T. Aoto, Y. Totsuka, R. Takahashi, M. Ueda et al., 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1987. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and Drosophila persimilis. Genetics 116 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1989. Genetics of sterility in hybrids between two subspecies of Drosophila. Evolution 43 180–189. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1997. Haldane's rule. Ann. Rev. Ecol. Syst. 28 195–218. [Google Scholar]
- Orth, A., T. Adama, W. Dinn and F. Bonhomme, 1998. Natural hybridization of two subspecies of house mice, Mus musculus domesticus and Mus musculus castaneus, near Lake Casitas (California). Genome 41 104–110. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and H. E. Hoekstra, 2005. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics 171 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur, B. A., J. G. Krenz and M. W. Nachman, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58 2064–2078. [DOI] [PubMed] [Google Scholar]
- Perez, D. E., and C. I. Wu, 1995. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov, P. M., Y. Ding, M. A. Cassell, W. Zhang, G. Wagner et al., 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., and H. A. Orr, 1998. Haldane's rule in taxa lacking a hemizygous X. Science 282 952–954. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 715–719. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., K. Namgyal, G. Gibson and D. L. Hartl, 2004. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottscheidt, R., and B. Harr, 2007. Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics 177 1553–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, L. D., R. A. Ettlin, A. P. Sinha Hikin and E. D. Clegg, 1990. Histological and Histopathological Evaluation of the Testis. Cache River Press, Clearwater, FL.
- Sage, R. D., D. Heyneman, K. C. Lim and A. C. Wilson, 1986. Wormy mice in a hybrid zone. Nature 324 60–63. [DOI] [PubMed] [Google Scholar]
- Sawamura, K., J. Roote, C.-I. Wu and M. T. Yamamoto, 2004. Genetic complexity underlying hybrid male sterility in Drosophila. Genetics 166 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander, R. K., W. G. Hunt and S. Y. Yang, 1969. a Protein polymorphism and genetic heterozygosity in two European subspecies of house mouse. Evolution 23 379–390. [DOI] [PubMed] [Google Scholar]
- Selander, R. K., S. Y. Yang and W. G. Hunt, 1969. b Polymorphism in esterases and hemoglobin in wild populations of the house mouse (Mus musculus). Univ. Texas Publ. 6918 271–338. [Google Scholar]
- She, J. X., F. Bonhomme, P. Boursot, L. Thaler and F. Catzeflis, 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP Data. Biol. J. Linn. Soc. 41 83–103. [Google Scholar]
- Shifman, S., J. T. Bell, R. R. Copley, M. S. Taylor, R. W. Williams et al., 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4 2227–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová, R., S. Gregorová, D. Buckiová, V. Kyselová, P. Divina et al., 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mammal. Genome 15 515–524. [DOI] [PubMed] [Google Scholar]
- Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching et al., 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, T., A. Mita, A. Maeno, T. Sakai, H. Shitara et al., 2008. Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res. 18 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., S. N. Xhen, D. L. Hartl and C. C. Laurie, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter, K. C., B. A. Payseur, L. W. Harris, M. A. Bakewell, L. M. Thibodeau et al., 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur, M. L. Wu and C.-I. Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282 1501–1504. [DOI] [PubMed] [Google Scholar]
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, P. K., R. D. Sage, J. Warner, A. C. Wilson and E. M. Eicher, 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46 1146–1163. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis, and the genetics of postzygotic isolation. Genetics 154 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, F., P. Boursot, M. Bellis, L. Thaler and F. Bonhomme, 1986. Genetic aspect of the semi-specific interaction between Mus musculus domesticus and Mus musculus musculus in eastern Europe. Genet. Res. 47 222–223. [DOI] [PubMed] [Google Scholar]
- Vigneault, G., and E. Zouros, 1986. The genetics of asymmetrical male sterility in Drosophila mojavensis and Drosophila arizonensis hybrids: interaction between the Y chromosome and autosomes. Evolution 40 1160–1170. [DOI] [PubMed] [Google Scholar]
- Vrana, P. B., 2007. Genomic imprinting as a mechanism of reproductive isolation in mammals. J. Mammal. 88 5–23. [Google Scholar]
- Vyskocilová, M., Z. Trachtulec, J. Forejt and J. Piálek, 2005. Does geography matter in hybrid sterility in house mice? Biol. J. Linn. Soc. 84 663–674. [Google Scholar]
- Wade, C. M., E. J. Kulbokas, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420 574–578. [DOI] [PubMed] [Google Scholar]
- Winter, E. E., L. Goodstadt and C. P. Ponting, 2004. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 14 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt, J., D. Adam, B. Malitschek, W. Maueler, F. Raulf et al., 1989. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341 415–421. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I., 1992. A note on Haldane's rule: hybrid inviability versus hybrid sterility. Evolution 46 1584–1587. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I., and A. T. Beckenbach, 1983. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of Drosophila pseudoobscura and Drosophila persimilis and identification of hybrid sterility factors. Genetics 105 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.-I., and A. W. Davis, 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane's rule and its genetic bases. Am. Nat. 142 187–212. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I., N. A. Johnson and M. F. Palopoli, 1996. Haldane's rule and its legacy: Why are there so many sterile males? Trends Ecol. Evol. 11 281–284. [DOI] [PubMed] [Google Scholar]
- Wu, C.-I., and C. T. Ting, 2004. Genes and speciation. Nat. Rev. Genet. 5 114–122. [DOI] [PubMed] [Google Scholar]
- Yang, H., T. A. Bell, G. A. Churchill and F. Pardo-Manuel de Villena, 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 39 1100–1107. [DOI] [PubMed] [Google Scholar]
- Zeng, Z.-B., 1994. Precision mapping of quantitative trait loci. Genetics 136 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z.-B., J. J. Liu, L. F. Stam, C. H. Kao, J. M. Mercer et al., 2000. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]