Abstract
Miniature inverted-repeat transposable elements (MITEs) are short DNA transposons with terminal inverted repeat (TIR) signals and have been extensively studied in plants and other eukaryotes. But little is known about them in eubacteria. We identified a novel and recently active MITE, Chunjie, when studying the recent duplication of an operon consisting of ABC transporters and a phosphate uptake regulator in the chromosome of Geobacter uraniireducens Rf4. Chunjie resembles the other known MITEs in many aspects, e.g., having TIR signals and direct repeats, small in size, noncoding, able to fold into a stable secondary structure, and typically inserted into A + T-rich regions. At least one case of recent transposition was observed, i.e., the insertion of Chunjie into one copy of the aforementioned operon. As far as we know, this is the first report that the insertion of a MITE does not disrupt the operon structure.
TRANSPOSABLE elements are genetic elements that can relocate themselves within or across genomes, a process called transposition. They are grouped into two major classes, retrotransposons (class I) and DNA transposons (class II), on the basis of whether their transposition intermediates are RNA or DNA molecules (Chandler and Mahillon 2002; Kidwell 2002; Zhou et al. 2008a). Miniature inverted-repeat transposable elements (MITEs) are DNA transposons with no protein-encoding potential and were first observed in plant genomes in the early 1990s (Bureau and Wessler 1992, 1994a,b). Many MITEs have been identified and characterized in the genomes of other eukaryotes (Smit and Riggs 1996; Tu 1997; Feschotte et al. 2002) since then. But our knowledge of MITEs in prokaryotic genomes is very limited (Mazzone et al. 2001; Mennecier et al. 2006; Xu et al. 2006; Filee et al. 2007; Zhou et al. 2008b), considering that a large number of prokaryotic genomes have been sequenced and analyzed.
A MITE is a short noncoding DNA element (100–500 bp) that carries a pair of terminal inverted repeats (TIRs) on the two termini (Feschotte et al. 2002). The TIR signals are essential to the maintenance and proliferation of a MITE, since the transposition of a MITE relies on the transposase encoded by a DNA transposon with the same TIR signals (Jiang et al. 2003; Yang et al. 2007). A MITE will duplicate a short target DNA sequence during its transposition, and the conservation of the two direct repeats (DR) is a representative feature of a recent transposition. Most MITEs are A + T rich and tend to insert into A + T-rich regions (Tu 1997). Their strong tendency to be in the intergenic regions and to fold into stable secondary structures suggests that they might play a role in the transcription regulation of the neighboring genes (Lepetit et al. 2000; Mazzone et al. 2001; Zhou et al. 2008b). And a MITE could proliferate to have as many as 100,000 copies in the host genome, e.g., the human MITE Mer1 (Smit and Riggs 1996).
We report in this work the first recently active MITE, Chunjie, in Geobacter uraniireducens Rf4, which can remove uranium from the polluted groundwater (Anderson et al. 2003), and postulate that it might have been proliferated through the transposase encoded by ISGur4 with very similar TIR signals, since both of them were identified in G. uraniireducens Rf4 and have almost identical copies of the DRs. The recent transposition of Chunjie was further confirmed computationally by identification of one insertion of Chunjie into an operon that was duplicated after the divergence of G. uraniireducens Rf4 and its two close relatives with complete genomes, i.e., G. metallireducens GS-15 and G. sulfurreducens PCA. It is interesting to note that the structure of the operon does not seem to be disrupted by the insertion of Chunjie, compared with the other copy of the operon that was duplicated before the insertion.
RESULTS
Comparison of the two copies of an operon in G.uraniireducens Rf4:
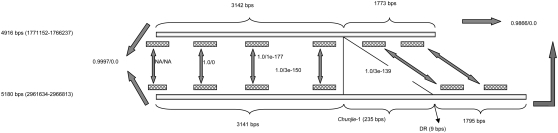
The insertion of an element of 235 bp was observed in a genomic fragment consisting of five genes in the chromosome of G. uraniireducens Rf4, through sequence comparison with its almost identical copy in the same chromosome. The insertion occurred between the third gene (a phosphate ABC transporter, the inner membrane subunit PstA) and the fourth gene (a phosphate ABC transporter, the ATPase subunit). There is no observable sequence similarity between the flanking regions of the two fragments. Both fragments cover four ABC transporter genes and one regulator gene for phosphate uptake, as shown in Figure 1. The five genes are organized in the same order between the two fragments and the sequences of their encoded proteins are identical to each other, respectively. Furthermore, the five genes are predicted to constitute an operon in the two fragments, respectively, using the program OFS with a probability cutoff of 0.85 (Westover et al. 2005). The 5′ end of both fragments partially overlaps with one more gene, but no sequence similarity can be detected at the amino acid level and the nucleotide level of the regions of the two genes that are outside the two fragments. Both genes were annotated as “hypothetical proteins,” and they were predicted not to be in the same operon with the neighboring genes. They might have been misannotated as genes, and the two fragments are actually two operons.
Figure 1.—
The insertion of a genetic element of 235 bp into an operon of ABC transporters and an uptake regulator on the chromosome of Geobacter uraniireducens Rf4 (NC_009483). The similarity between each pair of sequences is measured by “identity percentage/E-value,” using the NCBI Blast.
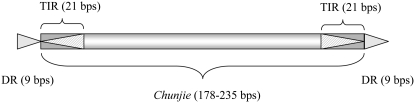
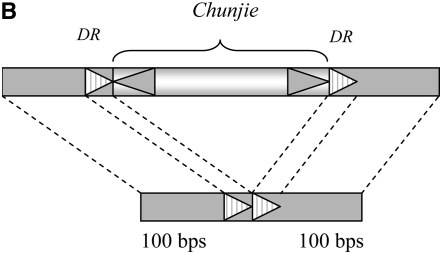
The inserted element has a pair of perfectly reverse complementary TIR signals 21 bp long on the two termini, as shown in Figure 2A. A pair of identical DRs of 9 bp long was also observed to flank the region inserted into the above fragment. Both the DRs and the flanking region are A + T-rich, as shown in Table 1. This inserted element is considered as the first copy, Chunjie-1, of a putative MITE, Chunjie. The structure of MITE Chunjie is illustrated in Figure 3.
Figure 2.—
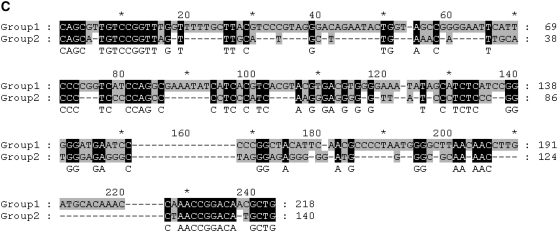
(A) The alignment of the flanking regions and the inserted region of Chunjie-1 in the two operons; (B) the alignment of the 10 full copies of ISGur4 with 10-bp flanking sequences; and (C) the alignment of the consensus sequences of the two groups of full copies of Chunjie, i.e., group 1, {Chunjie-1, …, Chunjie-32}, and group 2, {Chunjie-33, …, Chunjie-38}. TIR signals are in boldface type, and the DRs are shaded.
TABLE 1.
Some of the putative 38 full copies of Chunjie (detailed information of all 38 full copies is in supplemental Table S1)
| Name | Length | TIR | DR | AT(DR) | AT(F) | AT | I(TIR) | I(DR) | MFE | AvgMFE | Overlap |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chunjie-1 | 235 | 21 | 9 | 0.78 | 0.65 | 0.47 | 1.00 | 1.00 | −129.9 | −0.55 | — |
| Chunjie-9 | 219 | 21 | 9 | 0.67 | 0.47 | 0.47 | 1.00 | 1.00 | −126.96 | −0.58 | — |
| Chunjie-12 | 219 | 21 | 9 | 0.78 | 0.64 | 0.47 | 1.00 | 1.00 | −126.96 | −0.58 | 148264141 |
| Chunjie-14 | 219 | 21 | 9 | 0.33 | 0.53 | 0.47 | 1.00 | 1.00 | −126.96 | −0.58 | — |
| Chunjie-15 | 219 | 21 | 9 | 0.56 | 0.68 | 0.48 | 1.00 | 1.00 | −125.86 | −0.57 | 148265944 |
| Chunjie-16 | 219 | 21 | 9 | 0.33 | 0.48 | 0.47 | 1.00 | 1.00 | −126.96 | −0.58 | — |
| Chunjie-31 | 187 | 21 | 9 | 0.39 | 0.50 | 0.47 | 0.95 | 0.89 | −97.5 | −0.52 | — |
| Chunjie-32 | 216 | 21 | 9 | 0.44 | 0.66 | 0.48 | 0.81 | 1.00 | −110.81 | −0.51 | — |
| Chunjie-35 | 179 | 21 | 9 | 0.78 | 0.66 | 0.45 | 0.86 | 1.00 | −110.6 | −0.62 | 148264620 |
AT(F) is the A + T content of the two 50-bp regions flanking the DRs and one DR. AT(DR) and AT are the A + T content of the DRs and the MITE itself. Ident(TIR) and Ident(DR) are the percentages of reversely complementary and identical nucleotides, respectively. MFE and AvgMFE are the minimum folding energy and the MFE divided by the length of the element. Overlap lists the genes' overlap with this element. Underlined numbers are discussed in the text.
Figure 3.—
Schematic of Chunjie. TIR is the terminal inverted repeat and DR is the direct repeat.
A recently active MITE in G.uraniireducens Rf4:
MITEs were proposed to transpose through the transposases encoded by DNA transposons with the same TIR signals and DRs of the same length. We define a region on the genome as a full copy of Chunjie, if this region has a pair of the highly conserved TIR signals to that of Chunjie-1 on the two termini and it is flanked by a pair of DRs of the same length as that of Chunjie-1. We scanned the genome of G. uraniireducens Rf4 for regions with similar sequence and structure to that of Chunjie-1. Thirty-seven additional full copies were found to share highly similar TIR signals with those of Chunjie-1 and were flanked by DRs of the same lengths as Chunjie-1. Their lengths range from 178 to 235 bp, as shown in Table 1 and supplemental Table S1 (supplemental data are available at the Genetics web site and at http://csbl.bmb.uga.edu/publications/materials/ffzhou/chunjie). All 38 copies reside in the intergenic regions, except for Chunjie-12, Chunjie-15, and Chunjie-35, which overlap predicted protein-encoding genes, as shown in Table 1. All three overlapping genes (148264141, 148265944, and 148264620) encode hypothetical proteins and have no homologous proteins in the NCBI NR protein database, as of February 2008. All 38 copies of Chunjie were predicted to have no coding potential, using Glimmer with default parameters (Delcher et al. 2007). The DRs of 34 of the 38 copies (∼89.47%) are A + T rich, and the flanking regions of all but three copies (∼92.11%) are A + T rich, as shown in Table 1, although the A + T content of the copies of Chunjie is only 47% on average. No full copies of Chunjie are identified in any other sequenced prokaryotic genomes.
A MITE could be transposed through the enzymes encoded by DNA transposons (Tu 2001). And the most prevalent DNA transposons in the prokaryotic genomes are insertion sequence (IS) elements (Chandler and Mahillon 2002; Filee et al. 2007). So we have scanned the 38 copies of Chunjie in ISfinder, the most comprehensive database of IS elements (Siguier et al. 2006). No significant sequence similarity was detected between Chunjie and any IS elements in ISfinder, except for ISGur4. ISGur4 shares very similar TIR signals with Chunjie, as shown in Figure 2B. If Chunjie was transposed through the transposase encoded by ISGur4, they should have DRs of the same length. But ISGur4 was proposed to have DRs of 10 bp long in the ISfinder database, whereas the DRs of Chunjie are 9 bp long. ISGur4 was identified in G. uraniireducens Rf4, and it has 10 identical copies and 1 partial copy in the chromosome of G. uraniireducens Rf4, using the NCBI Blast with E-value cutoff e-5 (McGinnis and Madden 2004). The DRs of all the full copies are 9 bp long, which are the same as those of Chunjie, as shown in Figure 2B and supplemental Table S2. Moreover, the A + T contents of both the DRs and the flanking regions of ISGur4 are very similar to those of Chunjie, as shown in supplemental Tables S1 and S2. So Chunjie could have been proliferated through the transposase encoded by ISGur4.
Chunjie has no sequence similarity to any known MITEs:
We searched RepBase (Jurka et al. 2005) (version 12.05; released of July 13, 2007) for homologous repetitive elements of the 38 full copies of Chunjie. No homologous elements were found using the NCBI Blast with E-value cutoff e-5. We then compared Chunjie with all known MITEs in eukaryotes (Feschotte et al. 2002) and found that only one known MITE in rice, Mutator-like Os-mMu, has similar lengths of TIR and DR to those of Chunjie. We then searched the genome of rice, Oryza sativa L. ssp. Indica, downloaded from the NCBI Ftp Server (Yu et al. 2002), for the 38 copies of Chunjie using the NCBI Blast. The best match has an E-value as high as 0.19, which is too high to be considered as a match as a homologous region of Chunjie. We then further compared Chunjie with the known MITEs in bacterial (Mazzone et al. 2001; Mennecier et al. 2006; Xu et al. 2006; Zhou et al. 2008b) and archaeal (Filee et al. 2007) genomes. No known MITEs there have TIRs and DRs of similar lengths to those of Chunjie of G. uraniireducens Rf4.
Distribution of Chunjie in microbial genomes:
We studied the distribution of Chunjie in the genome of G. uraniireducens Rf4. Fifty-seven additional copies were identified by scanning the 38 copies of Chunjie in the genome of G. uraniireducens Rf4 using theNCBI Blast with E-value cutoff e-5. These are either partial copies or possibly inactive copies without the conserved TIR signals or DRs.
We further searched the Chunjie copies against all the 988 prokaryotic genomes that are being or have been sequenced at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi (February 2008), using the NCBI Blast with E-value cutoff e-5. Only Chunjie-35, Chunjie-36, Chunjie-37, and Chunjie-38 have a homologous region in Geobacter sp. FRC-32, whose genome is currently being assembled by the U.S. Department of Energy Joint Genome Institute, respectively. The complementary strand of the region from 24,240 to 24,324 of contig 110549156 has >90% sequence identity to the internal regions of these four copies of Chunjie, respectively.
Recent transpositions of Chunjie in G.uraniireducens Rf4:
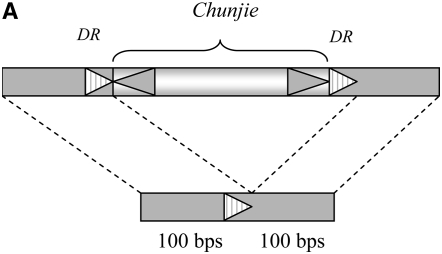
We reconstructed the flanking regions of each copy of Chunjie before its transposition into the current location using the two 100-bp regions flanking the DRs and a DR, as shown in Figure 4A. We then searched these sequences of the 38 copies of Chunjie in the 988 prokaryotic genomes at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi (February 2008), using the NCBI Blast with E-value e-5. Only one copy of Chunjie, Chunjie-1, was found to be inserted inside the operon containing ABC transporters and a phosphate-uptake regulator, as shown in Figures 1 and 2A. Since one copy of the operon has the sequence signals before the transposition of Chunjie and Chunjie is in the other copy of the operon, we infer that Chunjie-1 was inserted into one copy of the operon after it was duplicated. Note that a MITE will leave two copies of the DRs at the original site when it is transposed from the site. So we reconstructed the flanking regions of the target site of a copy of Chunjie after its transposition from the current location, using the two 100-bp regions flanking the DRs and two DRs, as shown in Figure 4B. But no homologous regions were found in the 988 prokaryotic genomes, using the NCBI Blast with E-value e-5.
Figure 4.—
Flanking regions of the target site of a copy of Chunjie (A) before its transposition into the site and (B) after its transposition from the site.
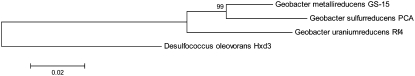
We then studied the distribution of the aforementioned operon consisting of four ABC transporters and a phosphate-uptake regulator in three completely sequenced Geobacter, i.e., G. metallireducens GS-15, G. sulfurreducens PCA, and G. uraniireducens Rf4 (Figure 5). The operon was observed to have one copy in G. metallireducens GS-15 and have no homologous region in G. sulfurreducens PCA, using the NCBI Blast with E-value cutoff e-5. The operon should have been in the common ancestor of the three Geobacteria. Alternatively, Chunjie might have invaded G. metallireducens GS-15 and G. uraniireducens Rf4, independently, which is probably much less likely, provided that the operon was also observed in two other Geobacteria whose genomes are being sequenced, i.e., Geobacter sp. FRC-32 and G. bemidjiensis Bem. The duplication of the operon in G. uraniireducens Rf4 might have occurred after the divergence of G. uraniireducens Rf4 and the common ancestor of G. metallireducens GS-15 and G. sulfurreducens PCA. The alternative hypothesis should be much less likely that the duplication occurred before their divergence, based on the evidence that (a) the whole regions of the two copies of the operon in G. uraniireducens Rf4 are almost identical, including their intergenic regions, as shown in Figure 1, and (b) it should be much less likely to independently delete one copy of the operon from G. metallireducens GS-15 and two copies from G. sulfurreducens PCA than to duplicate the operon in G. uraniireducens Rf4.
Figure 5.—
The bootstrapped neighbor-joining phylogenetic tree of three Geobacter genomes, i.e., Geobacter metallireducens GS-15, G. sulfurreducens PCA, and G. uraniireducens Rf4, based on their 16S RNA genes, which is rooted by another delta-proteobacteria, Desulfococcus oleovorans Hxd3. The phylogenetic tree is constructed by MEGA version 4.0 (Tamura et al. 2007).
Recent burst of Chunjie in G.uraniireducens Rf4:
We have built bootstrapped neighbor-joining and minimum evolution phylogenetic trees for the 38 copies of Chunjie, rooted at ISGur4, using MEGA version 4.0 (Tamura et al. 2007). The 38 copies of Chunjie were consistently divided into two groups, (Chunjie-1, …, Chunjie-32) and (Chunjie-33, …, Chunjie-38), as shown in supplemental Figure S1. The consensus sequences of the two groups of full copies of Chunjie were inferred from the multiple sequence alignments of the sequences, using programs ClustalW (Thompson et al. 1994) and GeneDoc (Nicholas et al. 1997), as shown in Figure 2C. The multiple sequence alignment of Chunjie-1, …, and Chunjie-32 shows that these 32 copies are highly similar to each other. Less than 0.46% of nucleotides in the alignment are different from the consensus sequence among Chunjie-1, …, and Chunjie-28, as shown in supplemental Figure S2a. The divergence percentages of the other four copies in the first group to the consensus sequence are 7.83% for Chunjie-29, 10.00% for Chunjie-30, 2.69% for Chunjie-31, and 11.21% for Chunjie-32, respectively. Whereas the copies in the other group are a little less similar to each other with divergence percentages no more than 26.52% to the consensus sequence, as shown in supplemental Figure S2b. This seems to suggest that there was a burst of proliferations of Chunjie in the genome of G. uraniireducens Rf4 very recently, during which all these 32 copies were transposed into their current positions. This speculation is further supported by another piece of evidence that almost all of Chunjie-1, …, and Chunjie-32 have perfect reverse complementary TIRs and identical DRs, as shown in supplemental Table S1.
Chunjie could fold into a very stable RNA molecule:
The vast majority of MITEs were known to fold into stable RNA secondary structures and played a role in regulating the expression of neighboring genes (Washietl et al. 2005; Gruber et al. 2007). The thermodynamic stability of a functional noncoding RNA (ncRNA) molecule is measured by the minimum folding energy (MFE) of its secondary structure. Chunjie could form stable secondary structures with free energy ΔG < −97.3 kcal/mol (Table 1) predicted using RNAfold in the ViennaRNA program package (Mathews et al. 1999).
DISCUSSION
Chunjie is a short DNA element with a TIR and a DR and has no coding potential. It preferably inserts itself into A + T-rich regions and can fold into a very stable secondary structure to carry out its functions. Our study indicates that there is no observable homolog of Chunjie among the eukaryotic repetitive elements, including MITEs in RepBase and other known MITEs in prokaryotes. There is only one MITE in rice, Mutator-like Os-mMu, that has similar lengths of TIR and DR to those of Chunjie, but neither homologous regions of TIR signals nor those of the internal regions of Chunjie could be observed in the rice genome. So we believe that Chunjie is a novel MITE in G. uraniireducens Rf4, and it is one of the MITEs with the longest DRs among the known MITEs (Lepetit et al. 2000; Feschotte et al. 2002) since it is consistent with all the key characteristics of known MITEs.
Chunjie might have undergone a burst of proliferation very recently through the transposase of ISGur4 into G. uraniireducens Rf4. Both Chunjie and ISGur4 were identified in the genome of G. uraniireducens Rf4, and ISGur4 has at least 10 identical copies with a pair of identical DRs, suggesting that ISGur4 was active recently. Their DRs have the same length and share the same TIR signals, which can act as the recognition and cleavage sites of the transposase (Chandler and Mahillon 2002). So the transposase of ISGur4 might have been involved in the proliferation of both Chunjie and ISGur4. The 32 almost identical copies of Chunjie with perfect DRs strongly suggest that they were very recently relocated into the current positions. And the recent activity of Chunjie in the genome of G. uraniireducens Rf4 was clearly demonstrated by our identification of the insertion of Chunjie-1 into an operon after the divergence of G. uraniireducens Rf4 and the common ancestor of G. metallireducens GS-15 and G. sulfurreducens PCA.
Chunjie should have invaded into the chromosome of G. uraniireducens Rf4 very recently, since it is observed only in G. uraniireducens Rf4 and its close relative Geobacter sp. FRC-32. Another piece of supporting evidence is that no copies of Chunjie were observed in another two completely sequenced Geobacter, i.e., G. metallireducens GS-15 and G. sulfurreducens PCA. After Chunjie initiated its proliferation in G. uraniireducens Rf4, it relocated one of its copies into an operon of ABC transporters and a phosphate uptake regulator. It is interesting to find that the insertion of Chunjie into this operon does not seem to have disrupted the operon structure, compared with another copy of this operon duplicated before the insertion. As far as we know, this is the first report that a prokaryotic MITE lands safely into an operon.
Acknowledgments
We thank all the Computational Systems Biology Laboratory colleagues for their comments on this work. We extend our special thanks to the anonymous reviewer for the helpful suggestions on our manuscript. This work was supported by the National Science Foundation (NSF/DBI-0354771, NSF/ITR-IIS-0407204, NSF/DBI-0542119, and NSF/CCF0621700), the National Institutes of Health (1R01GM075331 and 1R01GM081682), and a Distinguished Scholar grant from the Georgia Cancer Coalition. This work was also supported in part by grants (60673059, 10631070, and 60373025) from the National Science Foundation of China, the Taishan Scholar Fund from Shandong Province, China, and the State Scholarship Fund of China (20073020).
References
- Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long et al., 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69 5884–5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1992. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1994. a Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, T. E., and S. R. Wessler, 1994. b Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, M., and J. Mahillon, 2002. Insertion sequences revisited, pp. 305–366 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society of Microbiology, Washington, DC.
- Delcher, A. L., K. A. Bratke, E. C. Powers and S. L. Salzberg, 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., X. Zhang and S. R. Wessler, 2002. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons, pp. 1147–1158 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Filee, J., P. Siguier and M. Chandler, 2007. Insertion sequence diversity in archaea. Microbiol. Mol. Biol. Rev. 71 121–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, A. R., R. Neubock, I. L. Hofacker and S. Washietl, 2007. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res. 35 W335–W338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, X. Zhang, H. Hirochika, S. R. Eddy et al., 2003. An active DNA transposon family in rice. Nature 421 163–167. [DOI] [PubMed] [Google Scholar]
- Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany et al., 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110 462–467. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115 49–63. [DOI] [PubMed] [Google Scholar]
- Lepetit, D., S. Pasquet, M. Olive, N. Theze and P. Thiebaud, 2000. Glider and Vision: two new families of miniature inverted-repeat transposable elements in Xenopus laevis genome. Genetica 108 163–169. [DOI] [PubMed] [Google Scholar]
- Mathews, D. H., J. Sabina, M. Zuker and D. H. Turner, 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288 911–940. [DOI] [PubMed] [Google Scholar]
- Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano et al., 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278 211–222. [DOI] [PubMed] [Google Scholar]
- McGinnis, S., and T. L. Madden, 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32 W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennecier, S., P. Servant, G. Coste, A. Bailone and S. Sommer, 2006. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59 317–325. [DOI] [PubMed] [Google Scholar]
- Nicholas, K. B., H. B. Nicholas, Jr. and D. W. I. Deerfield, 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. News 4 14. [Google Scholar]
- Siguier, P., J. Perochon, L. Lestrade, J. Mahillon and M. Chandler, 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F., and A. D. Riggs, 1996. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 93 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., J. Dudley, M. Nei and S. Kumar, 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z., 1997. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 94 7475–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Z., 2001. Eight novel families of miniature inverted repeat transposable elements in the African malaria mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 98 1699–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl, S., I. L. Hofacker and P. F. Stadler, 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 102 2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover, B. P., J. D. Buhler, J. L. Sonnenburg and J. I. Gordon, 2005. Operon prediction without a training set. Bioinformatics 21 880–888. [DOI] [PubMed] [Google Scholar]
- Xu, M., R. Kong and X. Xu, 2006. A miniature insertion element transposable in Microcystis sp. FACHB 854. Prog. Nat. Sci. 16 486–491. [Google Scholar]
- Yang, G., F. Zhang, C. N. Hancock and S. R. Wessler, 2007. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 10962–10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., S. Hu, J. Wang, G. K. Wong, S. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Zhou, F., V. Olman and Y. Xu, 2008. a Insertion sequences show diverse recent activities in Cyanobacteria and Archaea. BMC Genomics 9 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F., T. Tran and Y. Xu, 2008. b Nezha, a novel active miniature inverted-repeat transposable element in cyanobacteria. Biochem. Biophys. Res. Commun. 365 790–794. [DOI] [PubMed] [Google Scholar]