Abstract
The evolutionary process leading to the fixation of newly duplicated genes is not well understood. It was recently proposed that the fixation of duplicate genes is frequently driven by positive selection for increased gene dosage (i.e., the gene dosage hypothesis), because haploinsufficient genes were reported to have more paralogs than haplosufficient genes in the human genome. However, the previous analysis incorrectly assumed that the presence of dominant abnormal alleles of a human gene means that the gene is haploinsufficient, ignoring the fact that many dominant abnormal alleles arise from gain-of-function mutations. Here we show in both humans and yeast that haploinsufficient genes generally do not duplicate more frequently than haplosufficient genes. Yeast haploinsufficient genes do exhibit enhanced retention after whole-genome duplication compared to haplosufficient genes if they encode members of stable protein complexes, but the same phenomenon is absent if the genes do not encode protein complex members, suggesting that the dosage balance effect rather than the dosage effect is the underlying cause of the phenomenon. On the basis of these and other results, we conclude that selection for higher gene dosage does not play a major role in driving the fixation of duplication genes.
GENE duplication is the primary source of new genes (Ohno 1970) and duplicate genes are prevalent in virtually every sequenced genome in every domain of life (Zhang 2003). The likelihood of gene duplication during evolution is measured by gene duplicability, which is the product of the rate of mutation producing duplicate genes and the probability that the duplicates are fixed and retained in the genome (He and Zhang 2005a). Gene duplicability, especially the fixation and retention probability, is known to be correlated with many biological factors, such as gene importance (He and Zhang 2006), gene complexity (He and Zhang 2005a), gene functional category (Conant and Wagner 2002; Marland et al. 2004; Davis and Petrov 2005; Prachumwat and Li 2006), protein evolutionary rate (Davis and Petrov 2004), number of alternatively spliced forms (Kopelman et al. 2005), connectivity in protein interaction networks (Li et al. 2006; Prachumwat and Li 2006), membership in protein complexes (Papp et al. 2003), protein underwrapping (Liang et al. 2008), and organismal complexity (Yang et al. 2003).
Generally speaking, a duplicate gene may be fixed in a population by genetic drift or positive selection. Recently, it was proposed that the fixation process is frequently driven by positive selection for enhanced gene dosage brought about by gene duplication (Kondrashov and Koonin 2004; Kondrashov and Kondrashov 2006). This gene dosage hypothesis is supported by several case studies. For example, having additional copies of the salivary amylase gene is known to be advantageous to humans with high starch diets, due simply to the increased amount of gene product (Perry et al. 2007). In cases like this, gene duplication may enhance the organismal fitness immediately, driving the adaptive fixation of duplicate genes. Kondrashov and Koonin (2004) conducted a genomic test of the gene dosage hypothesis. They assumed that if halving the amount of gene product is deleterious to an organism (i.e., haploinsufficiency), doubling the amount would be beneficial. Under this assumption, the probability of fixation of a duplicate of a haploinsufficient gene should be higher than that for a haplosufficient gene. Consequently, haploinsufficient genes should have higher duplicabilities than haplosufficient genes, which was reported to be true in humans (Kondrashov and Koonin 2004). However, in their analysis, Kondrashov and Koonin (2004) incorrectly assumed that the presence of dominant abnormal alleles at a human gene means that the gene is haploinsufficient, ignoring the fact that many dominant abnormal alleles arise from gain-of-function mutations rather than loss-of-function mutations (Jimenez-Sanchez et al. 2001; Veitia 2002). For example, pituitary dwarfism due to isolated growth hormone deficiency [Online Mendelian Inheritance in Man (OMIM) 173100] has an autosomal dominant mode of inheritance, but it is caused by splice site or missense mutations in the growth hormone gene that have dominant-negative effects, because the mutated hormone competitively binds to the hormone receptor, hampering the wild-type hormone's ability to bind to the receptor (Binder et al. 1996; Takahashi et al. 1996). In this work, we analyze the relationship between gene haploinsufficiency and gene duplicability in humans and yeast and discuss why the gene dosage hypothesis is unlikely to explain the fixations of most duplicate genes.
RESULTS
Genomic test of the gene dosage hypothesis in humans:
Kondrashov and Koonin (2004) identified 685 haploinsufficient and 422 haplosufficient human genes by searching for diseases with dominant and recessive inheritances, respectively, in the database OMIM (http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim). Because dominance is not necessarily caused by haploinsufficiency and can arise from dominant-negative mutations, we decided to use a better search strategy. We searched OMIM with the terms “haploinsufficiency” and “haploinsufficient” and identified 222 haploinsufficient genes at the time of this study (October 2007). However, we could not search for haplosufficient genes using the terms “haplosufficiency” and “haplosufficient” because the vast majority of genes are haplosufficient and OMIM flags only haploinsufficient genes. Following Kondrashov and Koonin (2004), we identified 780 genes from OMIM by searching for diseases of recessive inheritance. Among them, 51 are known to be haploinsufficient, and the remaining 729 are regarded as haplosufficient. A haploinsufficient gene could cause a recessive disease if the disease-causing mutation does not completely abolish the gene function but only reduces it. Thus, it is possible that the above 729 genes still include some unknown haploinsufficient genes. Nonetheless, the separation of haploinsufficient and haplosufficient genes should be much better using our approach than using that of the earlier study.
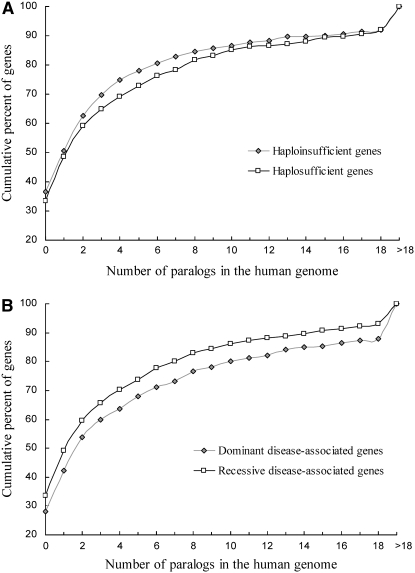
We searched for the paralogs of a given gene in the human genome by using its protein sequence as BLASTP query against all human genes (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome). The longest peptide was used if multiple splicing variants exist for a human gene. To be rigorous, we used an E-value cutoff of 10−10. For a hit to be considered valid, we further required that the length of the alignable region be at least 50% of the longer of the query and the hit. Contrary to the prediction of the gene dosage hypothesis, we found that haploinsufficient genes have fewer paralogs than haplosufficient genes in the human genome, although the difference is not statistically significant (P = 0.29, two-tailed Mann–Whitney U-test; P = 0.66, two-tailed t-test; Figure 1A). Many changes occurred in OMIM between 2004 and our study, so it is possible that our results differ due to changes in the database. However, we were able to confirm that dominant disease-associated genes have significantly more paralogs than recessive disease-associated genes (905 dominant and 493 recessive genes after exclusion of those with both types of inheritance; P = 0.003, two-tailed U-test; P = 0.02, two-tailed t-test; Figure 1B). Thus, the difference between our result and that of Kondrashov and Koonin (2004) is due to improved retrieval of haploinsufficient genes in our study, but not to changes in OMIM that occurred between the two studies. Because the primary reason for a haplosufficient gene causing dominant disease is a dominant-negative mutation, our result implies that genes with dominant-negative mutations tend to have more paralogs in the genome. The exact cause of this phenomenon is unclear, but it may be due to the specific functional categories to which genes with dominant-negative mutations belong. For example, we found that 29.7% of haploinsufficient genes encode enzymes, whereas the fraction is 38.9% among dominant but not haploinsufficient genes (P = 0.01, χ2 test). Because enzyme genes have higher duplicabilities than nonenzyme genes (Marland et al. 2004), dominant but not haploinsufficient genes are expected to have greater duplicabilities than haploinsufficient genes.
Figure 1.—
Human haploinsufficient genes do not have more paralogs than haplosufficient genes. (A) Cumulative distributions of the number of paralogs of haploinsufficient and haplosufficient genes in the human genome. No significant difference between haploinsufficient and haplosufficient genes is found (P = 0.29, two-tailed Mann–Whitney U-test; P = 0.66, two-tailed Student's t-test). (B) Cumulative distributions of the number of paralogs of dominant disease-associated genes and recessive disease-associated genes in the human genome. Dominant disease genes have significantly more paralogs than recessive disease genes do (P = 0.003, two-tailed U-test; P = 0.02, two-tailed t-test).
Genomic test of the gene dosage hypothesis in yeast:
One disadvantage of the above analysis is that haploinsufficient genes identified from human diseases may not accurately represent all haploinsufficient genes in the human genome (Jimenez-Sanchez et al. 2001). In this regard, yeast (Saccharomyces cerevisiae) is a better organism for testing the gene dosage hypothesis because 184 haploinsufficient yeast genes in rich media (YPD) have been identified through a genomewide experiment measuring the fitness values of heterozygous gene-deletion strains relative to the homozygous wild type (Deutschbauer et al. 2005). Because gene importance affects gene duplicability (He and Zhang 2006), to compare fairly with the YPD haploinsufficient genes, we define YPD haplosufficient genes as those with a statistically significant fitness deduction when both alleles are deleted but without a significant fitness deduction when only one allele is deleted in YPD (Deutschbauer et al. 2005). A total of 1826 haplosufficient genes were identified using this definition. Although a haploinsufficient gene may become haplosufficient and vice versa when the environment changes, a recent study showed that haploinsufficiency is quite stable, at least among the nutritional environments tested (Delneri et al. 2008). In other words, genes that are haploinsufficient in YPD are likely to be haploinsufficient in the yeast's natural environment as well. The S. cerevisiae lineage experienced a whole-genome duplication (WGD) event ∼100 million years ago (Wolfe and Shields 1997). On the basis of conserved synteny, it can be deduced that at least 450 pairs of duplicate genes generated by the WGD are still present in the S. cerevisiae genome (Kellis et al. 2004). Because WGD and individual gene duplication have different consequences for among-gene dosage balance (Papp et al. 2003; Liang et al. 2008), we separate duplicate genes generated by WGD from those generated by individual duplication events. The operational definition of paralogs used here is the same as used for human genes.
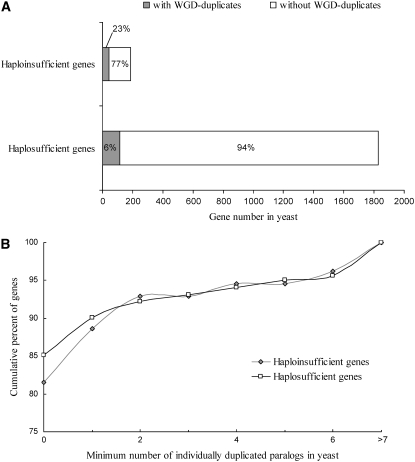
We first separated the WGD-generated duplicate gene pairs that are still retained in S. cerevisiae (WGD duplicates) from those genes whose duplicate copies from the WGD have been lost (WGD singletons). The WGD duplicates used here were previously identified through conserved synteny (Kellis et al. 2004) and thus do not include those retained duplicates that have moved to different chromosomal locations since the WGD. Rather, these relocated WGD duplicates are likely to have been erroneously included in the group of WGD singletons. Despite these potential errors that would reduce the difference between the two groups, we found that the proportion of genes with WGD duplicates is significantly greater among haploinsufficient genes than among haplosufficient genes (P < 10−15, χ2 test; Figure 2A). This observation is consistent with a previous study that used a different method to identify haploinsufficient and haplosufficient genes (Sugino and Innan 2006), although our interpretation of the observation differs (see below).
Figure 2.—
Duplicability of yeast haploinsufficient and haplosufficient genes in the WGD and individual gene duplications. (A) The proportion of haploinsufficient genes with WGD duplicates is significantly greater than the proportion of haplosufficient genes with WGD duplicates (P < 10−15, χ2 test). (B) Cumulative distributions of the minimum number of individually duplicated paralogs of haploinsufficient and haplosufficient genes. Haploinsufficient genes have significantly fewer paralogs than haplosufficient genes do in a two-tailed t-test (P = 0.022), but not in a two-tailed U-test (P = 0.26).
We then considered paralogs generated by individual gene duplication events. Because the above defined group of WGD singletons potentially still contains WGD duplicates, we decided to consider only n − 1 paralogs for a gene with n ≥ 1 paralogs, which would guarantee that paralogs generated from the WGD are not counted. This treatment would potentially favor the gene dosage hypothesis, because we undercount individually duplicated paralogs more for haplosufficient genes than for haploinsufficient genes, as there are fewer WGD-generated paralogs for haplosufficient genes than for haploinsufficient genes (Figure 2A). However, we found that haplosufficient genes have more individually duplicated paralogs than haploinsufficient genes do (Figure 2B). This difference is statistically significant in the two-tailed t-test (P = 0.022) but not significant in the U-test (P = 0.26). Thus, there is no evidence for higher duplicabilities of haploinsufficient genes compared to haplosufficient genes during individual gene duplications in yeast.
Because the dosage hypothesis predicts higher duplicabilities for haploinsufficient genes than for haplosufficient genes regardless of whether the duplication is individual or genomewide, the inconsistent observations between the two types of duplication in yeast (Figure 2) imply that the phenomenon of higher duplicabilities of haploinsufficient genes in WGD is unlikely due to the gene dosage effect as was previously suggested (Sugino and Innan 2006).
The dosage balance effect in whole-genome duplication:
Haploinsufficiency can arise from the dosage effect (Kondrashov and Koonin 2004) or the dosage balance effect (Papp et al. 2003). The dosage effect refers to an insufficient amount of gene product when one allele of a gene is deleted, whereas the dosage balance effect refers to the situation where deleting one allele of a gene causes an imbalance between the dosage of that gene and the (normal) dosage of its interacting partner(s). In the latter case, a further deletion of one allele of the interacting partner should rescue the phenotype caused by the first deletion because the dosages are rebalanced. It has been observed that ∼77% of haploinsufficient genes encode components of stable protein complexes, whereas the genomic average is only ∼20% (Deutschbauer et al. 2005), suggesting that haploinsufficiency is primarily caused by the dosage balance effect.
The above fact prompts us to hypothesize that the observation of higher duplicabilities of haploinsufficient genes compared to haplosufficient genes during WGD (Figure 2A) is due to the dosage balance effect rather than to the dosage effect. When a yeast cell with WGD establishes a population, the loss of a haploinsufficient gene that has a dosage balance requirement would be deleterious and thus prohibited by natural selection, while the loss of a haploinsufficient gene that does not have a dosage balance requirement could still be neutral because the gene loss simply returns the gene dosage to the original status before WGD. Therefore, we predict that after WGD (i) haploinsufficient genes have higher retention rates than haplosufficient genes if they encode components of stable protein complexes and (ii) haploinsufficient genes have retention rates similar to haplosufficient genes if they do not encode components of stable protein complexes.
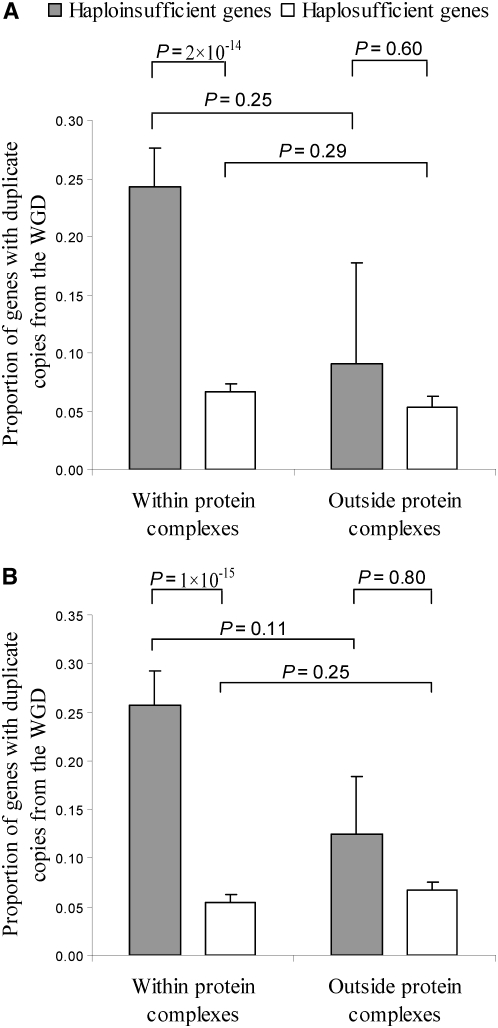
To test these predictions, we compared WGD retention rates of haploinsufficient and haplosufficient genes that are reported to encode components of protein complexes in the Munich Information Center for Protein Sequences (MIPS) database. Indeed, haploinsufficient protein complex genes have a significantly higher retention rate after WGD than haplosufficient protein complex genes (P = 2 × 10−14, χ2 test; Figure 3A). By contrast, when not encoding components of protein complexes, haploinsufficient genes do not have higher post-WGD retentions than haplosufficient genes (P = 0.60, χ2 test; Figure 3A). These results strongly support our hypothesis that the higher duplicabilities of haploinsufficient genes compared to haplosufficient genes during WGD are due to the dosage balance effect rather than to the dosage effect. We did not find a significant difference in retention between haploinsufficient genes within and outside protein complexes (P = 0.25, χ2 test; Figure 3A). Neither did we find a significant difference in retention between haplosufficient genes within and outside protein complexes (P = 0.29, χ2 test; Figure 3A). The lack of significant differences in the above two comparisons is not unexpected, because proteins within and outside complexes tend to belong to different functional categories and thus are not directly comparable. We also repeated our analysis using protein complex annotations in the Gene Ontology (GO) database (ftp://genome-ftp.stanford.edu/pub/yeast/data_download/literature_curation/) and observed virtually identical results (Figure 3B). Overall, of 43 haploinsufficient genes retained after WGD, 42 and 39 encode components of protein complexes annotated in MIPS and GO, respectively. Thus, the vast majority of post-WGD retention of haploinsufficient genes is likely attributable to the dosage balance effect.
Figure 3.—
Greater post-WGD retention rates of haploinsufficient genes compared to haplosufficient genes in yeast is found only for genes encoding members of stable protein complexes. P-values are from χ2 tests. Error bars show one standard error. Annotations of protein complexes are based on either (A) MIPS or (B) GO.
DISCUSSION
The previous test of the dosage hypothesis of gene duplication assumes that if halving the gene dosage is deleterious (haploinsufficiency), doubling the dosage will be beneficial (Kondrashov and Koonin 2004). This assumption is unwarranted for two reasons. First, haploinsufficiency is often due to the dosage balance effect and duplication of a haploinsufficient gene is consequently often deleterious (Papp et al. 2003). Second, increase of gene dosage has an energy cost associated with production of extra gene product, which may significantly reduce the fitness of the organism if the extra product is not useful (Wagner 2005). The dosage hypothesis assumes that the expression levels of many genes are below the optimal levels, which appears contradictory to experimental results showing that gene expression levels are quickly optimized by natural selection, at least in microbes (Dekel and Alon 2005). The dosage hypothesis further assumes that when the expression level of a gene is below the optimal level, expression-enhancing mutations occur more frequently by gene duplication than by altering the regulatory sequences of the gene. In yeast, the point mutation rate is on the order of 10−10/site/generation (Drake et al. 1998; Lang and Murray 2008), whereas the gene duplication rate is between 10−11 and 10−8/gene/year (Lynch and Conery 2000; Gao and Innan 2004). Even if the effective point mutational target for enhancing the expression of the concerned gene is only one nucleotide, the point mutation rate for enhancing the gene expression is on the order of 10−7/site/year, if we assume that yeast has ∼1000 generations/year in nature. This rate is one to four orders of magnitude greater than the rate of gene duplication. This comparison suggests that when the expression level of a gene is below the optimal level, the beneficial mutations that enhance the expression level are almost always point mutations rather than gene duplications. One may argue that, for already highly expressed genes, further increases in gene expression through point mutations may be difficult if the promoters cannot possibly be stronger. This argument is inconsistent with the observation that the very strong lac promoter in Escherichia coli can become stronger through point mutations (Mayo et al. 2006). The dosage hypothesis of gene duplication predicts low functional divergence between duplicates because functional divergence reduces the effective dosage of the product that is beneficial. This prediction is inconsistent with the observation of generally rapid divergence of gene expression and function after duplication (Wagner 2001; Gu et al. 2002; He and Zhang 2005b). All these considerations, together with our empirical results in humans and yeast that consistently show no evidence for the dosage hypothesis of gene duplication, lead us to conclude that selection for higher gene dosage is not an important force driving the fixations of most duplicate genes. Our conclusion implies that fixations of most duplicate genes are due to neutral genetic drift because dosage-unrelated positive selection would require the improbable emergence of new beneficial functions in duplicate genes during the short fixation period (Lynch and Force 2000). After a duplicate gene is fixed and stably preserved in the genome, there will be ample time for neofunctionalization (He and Zhang 2005b).
Acknowledgments
We thank Meg Bakewell, Ben-Yang Liao, and Zhi Wang for valuable comments. This work was supported by research grants from the National Institutes of Health and the University of Michigan Center for Computational Medicine and Biology to J.Z.
References
- Binder, G., M. Brown and J. S. Parks, 1996. Mechanisms responsible for dominant expression of human growth hormone gene mutations. J. Clin. Endocrinol. Metab. 81 4047–4050. [DOI] [PubMed] [Google Scholar]
- Conant, G. C., and A. Wagner, 2002. GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res. 30 3378–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. C., and D. A. Petrov, 2004. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2 E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. C., and D. A. Petrov, 2005. Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 21 548–551. [DOI] [PubMed] [Google Scholar]
- Dekel, E., and U. Alon, 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436 588–592. [DOI] [PubMed] [Google Scholar]
- Delneri, D., D. C. Hoyle, K. Gkargkas, E. J. Cross, B. Rash et al., 2008. Identification and characterization of high-flux-control genes of yeast through competition analyses in continuous cultures. Nat. Genet. 40 113–117. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer et al., 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L. Z., and H. Innan, 2004. Very low gene duplication rate in the yeast genome. Science 306 1367–1370. [DOI] [PubMed] [Google Scholar]
- Gu, Z., D. Nicolae, H. H. Lu and W. H. Li, 2002. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18 609–613. [DOI] [PubMed] [Google Scholar]
- He, X., and J. Zhang, 2005. a Gene complexity and gene duplicability. Curr. Biol. 15 1016–1021. [DOI] [PubMed] [Google Scholar]
- He, X., and J. Zhang, 2005. b Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., and J. Zhang, 2006. Higher duplicability of less important genes in yeast genomes. Mol. Biol. Evol. 23 144–151. [DOI] [PubMed] [Google Scholar]
- Jimenez-Sanchez, G., B. Childs and D. Valle, 2001. Human disease genes. Nature 409 853–855. [DOI] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428 617–624. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F. A., and A. S. Kondrashov, 2006. Role of selection in fixation of gene duplications. J. Theor. Biol. 239 141–151. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F. A., and E. V. Koonin, 2004. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20 287–290. [DOI] [PubMed] [Google Scholar]
- Kopelman, N. M., D. Lancet and I. Yanai, 2005. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat. Genet. 37 588–589. [DOI] [PubMed] [Google Scholar]
- Lang, G. I., and A. W. Murray, 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Y. Huang, X. Xia and Z. Sun, 2006. Preferential duplication in the sparse part of yeast protein interaction network. Mol. Biol. Evol. 23 2467–2473. [DOI] [PubMed] [Google Scholar]
- Liang, H., K. R. Plazonic, J. Chen, W. H. Li and A. Fernandez, 2008. Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genet. 4 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marland, E., A. Prachumwat, N. Maltsev, Z. Gu and W. H. Li, 2004. Higher gene duplicabilities for metabolic proteins than for nonmetabolic proteins in yeast and E. coli. J. Mol. Evol. 59 806–814. [DOI] [PubMed] [Google Scholar]
- Mayo, A. E., Y. Setty, S. Shavit, A. Zaslaver and U. Alon, 2006. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 4 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin.
- Papp, B., C. Pal and L. D. Hurst, 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424 194–197. [DOI] [PubMed] [Google Scholar]
- Perry, G. H., N. J. Dominy, K. G. Claw, A. S. Lee, H. Fiegler et al., 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachumwat, A., and W. H. Li, 2006. Protein function, connectivity, and duplicability in yeast. Mol. Biol. Evol. 23 30–39. [DOI] [PubMed] [Google Scholar]
- Sugino, R. P., and H. Innan, 2006. Selection for more of the same product as a force to enhance concerted evolution of duplicated genes. Trends Genet. 22 642–644. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., H. Kaji, Y. Okimura, K. Goji, H. Abe et al., 1996. Brief report: short stature caused by a mutant growth hormone. N. Engl. J. Med. 334 432–436. [DOI] [PubMed] [Google Scholar]
- Veitia, R. A., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24 175–184. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2001. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol. Biol. Evol. 18 1283–1292. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2005. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22 1365–1374. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713. [DOI] [PubMed] [Google Scholar]
- Yang, J., R. Lusk and W. H. Li, 2003. Organismal complexity, protein complexity, and gene duplicability. Proc. Natl. Acad. Sci. USA 100 15661–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18 292–298. [Google Scholar]