Abstract
Aminoacylation of transfer RNAs establishes the rules of the genetic code. The reactions are catalyzed by an ancient group of 20 enzymes (one for each amino acid) known as aminoacyl tRNA synthetases (AARSs). Surprisingly, the etiology of specific diseases—including cancer, neuronal pathologies, autoimmune disorders, and disrupted metabolic conditions—is connected to specific aminoacyl tRNA synthetases. These connections include heritable mutations in the genes for tRNA synthetases that are causally linked to disease, with both dominant and recessive disease-causing mutations being annotated. Because some disease-causing mutations do not affect aminoacylation activity or apparent enzyme stability, the mutations are believed to affect functions that are distinct from aminoacylation. Examples include enzymes that are secreted as procytokines that, after activation, operate in pathways connected to the immune system or angiogenesis. In addition, within cells, synthetases form multiprotein complexes with each other or with other regulatory factors and in that way control diverse signaling pathways. Although much has been uncovered in recent years, many novel functions, disease connections, and interpathway connections of tRNA synthetases have yet to be worked out.
Keywords: AIMP, multifunctional protein
Aminoacyl tRNA synthetases catalyze the ligation of amino acids to their cognate tRNAs. Their catalytic activities determine the genetic code and for that reason, they are essential for protein synthesis and cell viability. The basic reaction is:
Because the reactions require the capacity to recognize tRNAs as well as small chemicals such as amino acids and ATP, the structures of AARSs are well equipped for interacting with diverse molecules that may be associated with their functional versatility. Thus, the catalytic activities for glycyl-, lysyl-, and tryptophanyl-tRNA synthetase have been adapted to synthesize diadenosine polyphosphates (ApnA), which are believed to regulate glucose metabolism (1, 2), cell proliferation, and death (3). Similarly, the tRNA recognition capacity of bacterial threonyl- and human glutamyl-prolyl-tRNA synthetases has been adapted for regulating translation by interacting with the 5′ (4) and 3′ UTR regions (5) of gene-specific transcripts.
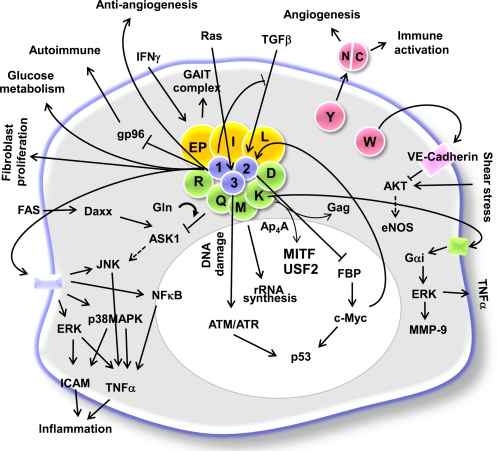
In higher eukaryotes, AARSs form macromolecular complexes via multivalent protein–protein interactions (6, 7). Fig. 1 depicts a single cell with many of the connections made by tRNA synthetases with each other and with other cell signaling molecules in the cytoplasm (light blue) and nucleus (white). Single letter abbreviations are used for each synthetase, such as R for arginyl-tRNA synthetase, and L for leucyl-tRNA synthetase. As shown, some of the synthetases are tightly bound together in a large multisynthetase complex, with three tRNA-synthetase associated proteins at the core. These are known as aminoacyl tRNA synthetase-interacting multifunctional protein (AIMP) 1, 2, and 3, and are simply designated as 1, 2, and 3. Two of the enzymes in the complex are covalently fused together (glutamyl-prolyl-tRNA synthetase) and are designated as EP in Fig. 1. Thus, at least nine synthetases and three synthetase-associated proteins are bound together. It is not known whether other synthetases are more loosely bound with the complex, and therefore not detected as bound (with existing methods), or whether they are not associated whatsoever.
Fig. 1.
Signaling network mediated by mammalian AARSs. Nine different AARSs (EP: glutamyl-prolyl-, I: isoleucyl-, L: leucyl-, M: methionyl-, Q: glutaminyl-, R: arginyl-, K: lysyl-, and D: aspartyl-tRNA synthetase) form a macromolecular complex with three nonenzymatic factors, AIMP1/p43, AIMP2/p38 and AIMP3/p18 (10). Among the components of the complex, EPRS is activated by IFN-γ and dissociates from the complex to form a new complex named GAIT (IFN-gamma-activated inhibitor of translation) that binds to the 3′UTR of the ceruloplasmin transcript for translational silencing (5). KRS is secreted to induce the inflammatory process (18), and QRS binds to apoptosis signal-regulating kinase 1 (ASK1) to regulate apoptosis in a glutamine-dependent manner (89). MRS is translocated to nucleoli to stimulate rRNA synthesis on growth stimuli (90). Among the nonenzyme factors, AIMP2 is translocated to the nucleus to suppress c-Myc via ubiquitin-dependent degradation of far upstream element binding protein (FBP) (64), whereas AIMP3 is mobilized by DNA damage (66) or an oncogenic stimulus (67) to activate p53 via ATM/ATR. AIMP1 is secreted as an extracellular multifunctional ligand active in inflammation (91–93), angiogenesis (61, 62), wound healing (94, 95), and glucose metabolism (83), and it influences the autoimmune response via its association with gp96 (78). AIMP1 also down-regulates the signal mediated by TGF-β through the stabilization of SMAD specific E3 ubiquitin protein ligase 2 (Smurf2) (96). KRS-generated Ap4A releases microphthalmia-associated transcription factor (MITF) and upstream stimulatory factor (USF2) from their bound complexes for transcriptional control of target genes (97). KRS is packaged into the HIV virion via its interaction with the gag protein (98). Among noncomplex forming AARSs, WRS (tryptophanyl-tRNA synthetase) is secreted and its N-terminal truncation results in the formation of an active angiostatic cytokine that works via VE-cadherin (13, 14, 25, 99–102). The active fragment of WRS also inhibits the response of endothelial cells to shear stress (100). YRS (tyrosyl-tRNA synthetase) is also secreted and cleaved into two distinct cytokines that work in angiogenesis as well as in the immune response (14, 15, 25, 103).
Although these eukaryotic AARSs appear to get together to improve aminoacylation efficiency by “channeling” substrates to the ribosome (8), their propensity to form complexes appears to be related in part to the eukaryotic cell's need for a reservoir of regulatory factors that are based on the alternative functions of these enzymes (9). The complex-forming AARSs are involved in the regulation of transcription, translation, and various signaling pathways as summarized in Fig. 1 (7, 10). Eukaryotic AARSs contain unique extensions and domains, which endow them with functional diversity through the interactions with various cellular partners (6, 11). The expanded functions of tRNA synthetases are not simply correlated with their being tightly associated with the complex. For example, tryptophanyl (WRS)- and tyrosyl (YRS)-tRNA synthetases are well characterized procytokines (12–17), but are not generally associated with the complex. On the other hand, the unusual EP fusion protein is associated with the multisynthetase complex and is released when cells are stimulated by γ-IFN (the EP fusion enzyme then acts to regulate translation of mRNAs associated with the inflammatory response) (5). In general, any specific synthetase may have more than one subcellular location—including being bound to the multisynthetase complex, free in the cytosol, in the nucleus, and mobilized for exocytosis (12, 18, 19). Considering the functional versatility of these enzymes, their expanded functions and expression may be pathologically associated with various human diseases. Here we describe specific cases in which AARSs are implicated in human disease.
AARSs in Neuronal Diseases
Charcot–Marie–Tooth (CMT) Disease Caused by Heritable Mutations in GRS and YRS.
CMT disease provides a clear example of heritable mutations in genes for tRNA synthetases being causally associated with a specific pathological condition. CMT disease has a frequency of ≈1 in 2,500, which makes it the most common heritable disorder of the peripheral nervous system. CMT disease is induced by axonal demyelination (type I) or decreased amplitudes of evoked motor and sensory nerve responses (type II), with no demyelination (20). Patients with CMT disease display muscular weakness and atrophy in the distal extremities, stoppage gait, high arched foot (pes cavus), absent or diminished deep-tendon reflexes, and impaired sensation.
The genes coding for glycyl-tRNA synthetase (GRS) and YRS are among the different genetic loci causally linked to the disease (21, 22). The disease-causing mutations are dominant and the mutant proteins have been shown in specific instances to be fully active for aminoacylation (23). At least 11 distinct mutant alleles in the human population have been reported for GRS. These mutations cause CMT2D, a subtype of the disease characterized by a slowly progressing neuropathy affecting mostly the distal extremities.
A mouse model developed by Seburn et al. encodes a mutant GRS that is fully active for aminoacylation (24). The mutant mice have the CMT2D phenotype of reduced nerve conduction velocities, a loss of axons with large diameters, and have no defects in myelination. Consistent with a deficit of aminoacylation function per se not being the cause of CMT, a mouse harboring a loss-of-function (aminoacylation) allele created by a gene trap insertion was normal.
The x-ray crystallographic structures of the homodimeric human YRS (25) and GRS (26) proteins and a disease-causing G526 mutant allele of GRS have been solved (27). Placing 11 disease-causing mutations onto the structure of human GRS showed them within a band encompassing both sides of the dimer interface (23). Strikingly, two CMT-causing mutations are complementary partners of a “kissing” contact across the dimer interface. Thus, mutation of either of these two residues causes CMT. Further analyses showed that most mutations affect dimer formation (i.e. either enhance or weaken it). A subset of seven mutant proteins and the wild-type protein were expressed in transfected neuroblastoma cells that sprout primitive neurites. Whereas the wild-type protein distributed into the nascent neurites and was associated with normal sprouting, all mutant proteins were distribution-defective. Thus, the CMT-causing mutations of GRS have a common defect that may be connected in some way to a change in the surfaces at the dimer interface (23).
Significantly, YRS is located prominently at axonal termini of differentiating primary motor neurons (22). CMT-associated missense or deletion mutations of YRS were transported to termini of differentiating neuronal cells and induced axonal degeneration. These results are consistent with both GRS and YRS having a specific role in the development or homeostasis (or both) of the peripheral nervous system. This role appears to be in addition to its function in protein synthesis.
Editing-Defective tRNA Synthetase Causally Associated with Ataxia in the Mouse.
A subset of synthetases sometimes catalyze the linkage of noncognate amino acids to tRNAs by mistake. For enzymes that make occasional errors, they have a second active site that clears the mischarged tRNA (28–31). An example is alanyl-tRNA synthetase ARS, which deacylates Ser-tRNAAla and Gly-tRNAAla. If these mischarged tRNAs are not cleared, then the wrong amino acid is inserted at the codons for alanine. Even a small amount of mischarging that is not corrected can, in principle, lead to the synthesis of proteins with errors that cause local or global misfolding. Over time, mistranslation leads to the gradual accumulation of these misfolded proteins (32–34). The sti/sti mouse harbors a mutation in the editing domain of ARS, which results in a approximately two-fold decrease in the activity to clear Ser-tRNAAla (35). The aminoacylation activity of the mutant enzyme is normal. Despite the small reduction in editing activity, the sti/sti mouse shows a marked loss of Purkinje cells in the cerebellum, and develops severe ataxia. Significantly, intracellular unfolded proteins accumulate in neurons, accompanied by up-regulation of cytoplasmic protein chaperones and induction of the unfolded protein response.
Unlike the dominant CMT-causing mutations in GRS and YRS, the sti mutation in the gene for ARS is recessive. Most likely, stronger editing-defective mutations would be lethal and possibly dominant. On this point, it is worth noting that a dominant phenotype (apoptosis-like response) was observed when a stronger editing-defective mutation (in the case of VRS) was introduced into mammalian cells. In this instance, the unfolded protein response was also triggered (36).
Possible Connection of KRS to Amyotrophic Lateral Sclerosis.
In some patients, a mutation in Cu/Zn superoxide dismutase 1 (SOD1) is causally associated with amyotrophic lateral sclerosis 1 (ALS1) (37–41). Interestingly, lysyl-tRNA synthetase (KRS) associates with mutant but not wild-type SOD1 (42). The SOD1 mutation enhances oligomerization of SOD1 to form aggregates with other proteins, which induces apoptosis of motor neurons, thus leading to the onset of neurodegeneration that is the hallmark of ALS. Whether the oligomerization or aggregation of SOD1 with KRS contributes to the inhibition of the normal activity of KRS is not known. If so, then the neuronal degeneration seen in patients with a SOD1 mutation might be caused or enhanced by a defect in protein synthesis.
Mutations in Mitochondrial DRS Associated with Leukoencephalopathy.
Specific mutations in the gene for mitochondrial aspartyl-tRNA synthetase (DRS) cause leukoencephalopathy, which has brainstem and spinal cord involvement and lactate elevation (LBSL) (43). LBSL is diagnosed by a characteristic constellation of abnormalities that can be observed by magnetic resonance imaging and spectroscopy. The disease is caused by autosomal recessive mutations that induce progressive cerebella ataxia. Most of the disease-causing mitochondrial DRS mutations are located within exons three and five. These mutations can alter normal splicing and lead to frame shifts, premature termination of translation, or exon skipping. Although mutant mitochondrial DRSs have reduced aminoacylation activity, the mitochondrial complex, critical for electron transport, is not affected. Thus, development of LBSL appears to be caused by a defect in mitochondrial DRS that is related to subtle effects on metabolic events within mitochondria.
Possible Connection of AIMP2 to Parkinson's Disease.
The aminoacyl-tRNA synthetase-interacting factor, AIMP2 (also known as p38), was shown to be the substrate of Parkin, an E3 ubiquitin–protein ligase (44, 45). Parkin promotes ubiquitination and proteasome degradation of specific protein substrates. The control of expression of Parkin substrates through ubiquitination and degradation is critical for dopaminergic cell survival (45). Loss of Parkin function by mutation results in the accumulation of abnormal or toxic proteins, leading to Parkinson's disease (PD). In Parkin null mice, AIMP2 is overexpressed in the ventral midbrain/hindbrain and over expression of AIMP2 induces apoptosis in neuronal cells and the formation of aggresome-like inclusions (45). These observations raise the possibility of a pathological connection of AIMP2 to neurodegenerative disease.
AARSs in Cancer
Possible Role for MRS.
Several components of the translation apparatus show abnormal up- or down-regulation in hepatomas, colon cancer, Burkett's lymphoma, prostate adenocarcinoma, breast cancer, sarcoma, colorectal adenocarcinoma and pituitary adenoma (46). This abnormal regulation could be partly because of differences in the quantity and quality of cellular protein synthesis in tumors. The aminoacylation activity of methionyl-tRNA synthetase (MRS), which is required for translation initiation, is increased in human colon cancer (47). Coincidently, the 3′ untranslated region (UTR) of MRS contains a 56 base pair complementary sequence to that of the 3′ UTR of C/EBP homologous protein (CHOP) that is critically connected to the onset of specific tumors (48–51). Thus, the transcripts for MRS and CHOP have at least the potential to associate via 56 base pairs. Whether this complementary relationship has any connection to the etiology of cancer is still to be determined.
Potential Involvement of Other tRNA Synthetases in the Etiology of Cancer.
In another vein, genes encoding different AARSs have been highly expressed or modified in association with a variety of cancers. For example, cysteinyl-tRNA synthetase (CRS) is expressed as fusion proteins to anaplastic lymphoma kinase (ALK) (52, 53). As another example, the promoter region of mitochondrial isoleucyl-tRNA synthetase (IRS), encoded by nuclear DNA, was mutated to modify its expression in hereditary nonpolyposis colorectal cancer (HNPCC) and Turcot syndrome (54). Because expression of glutamyl-prolyl-tRNA synthetase (EPRS) and IRS is controlled by the c-myc proto-oncogene (55), abnormal expression of EPRS and IRS under oncogenic conditions is not surprising. Preferential expression of the α-subunit of phenylalanyl-tRNA synthetase (FRS) was observed in lung solid tumors and acute phase chronic myeloid leukemia (56, 57). GRS up-regulation was also reported in papillary thyroid carcinoma (PTC) (58, 59), and KRS is overexpressed in breast cancer (18). It would be interesting to see whether cancer-specific overexpression of these enzymes is also observed in other cancers. Whereas YRS and tryptophanyl-tRNA synthetase (WRS) directly regulate angiogenesis as pro- and antiangiogenic cytokines (Fig. 1), EPRS negatively regulates this process via translational suppression of vascular endothelial growth factor-A (VEGF-A) (60). Because angiogenesis is associated with cancer development, the unique roles of these enzymes in the control of angiogenesis may have some implications in tumorigenesis.
Involvement of Aminoacyl-tRNA Synthetase-Interacting Factors.
Three AARS-interacting factors associated with the multisynthetase complex (AIMPs) have each been linked to signaling pathways relevant to cancers. For example, AIMP1/p43 enhances phosphorylation of Jun N-terminal kinase (JNK) and sequentially induces activation of caspase 3 to stimulate apoptosis of endothelial cells that are linked to tumor angiogenesis (61). As expected, systemic administration of AIMP1 suppresses cancer progression (62).
AIMP2/p38 is essential for the stability of the multisynthetase complex (63). Interestingly, transforming growth factor-β (TGF-β) induces nuclear localization of AIMP2, where it binds to FUSE-binding protein (FBP), which is the transcriptional activator of the c-myc proto-oncogene (64). AIMP2 enhances ubiquitin-dependent degradation of FBP, which results in down-regulation of c-Myc. Consistent with this observation, genetic depletion of mouse AIMP2 resulted in overexpression of c-Myc and caused neonatal lethality for the hyperplasia of lung epithelial cells. Because members of the Myc family of proteins are overexpressed in the majority of cancers, and TGF-β is a critical tumor suppressive signal, the role of AIMP2 in linking c-Myc to TGF-β implies its potential role as a tumor suppressor.
AIMP3/p18 harbors the glutathione S-transferase (GST) homology domain and associates with MRS within the multisynthetase complex (65). AIMP3 is activated by DNA damage (66) and oncogenic stresses (67), and translocates to the nucleus to interact with ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia and Rad-3 related (ATR) kinases, thereby leading to the activation of p53. Based on these activities, AIMP3 was thought to be a novel haploinsufficient tumor suppressor. Homozygous depletion of AIMP3 caused embryonic lethality in the mouse, whereas mice with a single allelic loss of AIMP3 developed breast adenocarcinoma, a sarcoma of unknown origin, adenocarcinoma in seminal vesicles, hepatocarcinoma, and lymphoma (66). A few mutations of AIMP3 that affect its interaction with ATM and ability to activate p53 have been found in human chronic myeloid leukemia patients (68), further supporting its relationship to the initiation or progression of human leukemia.
AARSs in Autoimmune Diseases
Autoantibodies Directed Against a Subset of Synthetases in Autoimmune Disorders.
Autoantibodies against different AARSs including histidyl-tRNA synthetase (HRS), threonyl-tRNA synthetase (TRS), ARS, IRS, FRS, GRS and asparaginyl-tRNA synthetase (NRS) have been found in ≈30% of all autoimmune patients (69, 70). The “antisynthetase syndrome” includes, among others, idiopathic inflammatory myopathies (IIM), interstitial lung diseases (ILD), rheumatoid and erosive arthritis, and Reynaud's phenomenon. IIM is a group of systemic diseases characterized by chronic muscle inflammation and can be classified into three distinct clinicopathologic subgroups – polymyositis (PM), dermatomyositis (DM) and inclusion body myositis (IBM). Both PM and DM patients slowly develop proximal and often symmetrical muscle weakness over weeks to months. The frequent disease types correlated with anti-synthetase antibodies are PM and DM. HRS (Jo-1) is the most frequently targeted autoantigen in PM. Anti-HRS autoantibodies are produced in 15 to 25% of all patients with PM. The patients identified with anti-NRS antibodies have ILD (71).
Rationale for Autoantibodies Directed Against AARSs.
The reason why AARSs are targeted as autoantigens is not understood. Because there is a general similarity in conformation between certain tRNA synthetases, common structural motifs are speculated as important for autoantibody generation. In that connection, AARSs can be divided into two classes—class I and class II–based on their characteristic motifs and conformational architecture. Except for IRS, most autoantigenic AARSs belong to class II. However, the epitope of HRS for autoantibody generation in most patients is found at the N-terminus of the protein (72), spanning ≈60 amino acids that make up a coiled-coil structure. An N-terminal deletion mutant is not recognized by the HRS autoantibodies that are found in patients (72). It is worth noting that the peptides that are homologous to this antigenic N-terminal peptide of HRS are also present in the N-terminal extension of EPRS and GRS, which are other autoantigenic AARSs. The N-terminal extension is not part of the structure that is conserved and shared by all class II enzymes. This coiled-coil structure is commonly found in ≈40% of autoantigens associated with a variety of autoimmune diseases, including non-tRNA synthetase autoantigens. For example, autoantibodies directed against NRS recognize a similar coiled-coil structure.
Many autoantigens redistribute to and cluster on the surface of cells undergoing apoptosis (73). Because autoantigens are commonly generated by apoptotic proteases (during apoptosis) to generate antigenic fragments (74, 75), the native synthetases are not likely to be the autoantigen responsible for the autoimmune response. Recent studies suggest that most autoantigens targeted in systemic autoimmune diseases are substrates of granzyme B, a serine protease that is involved in the induction of apoptosis during lymphocyte cytotoxicity (76). IRS, HRS and ARS are efficiently cleaved by granzyme B, releasing the fragments that contain the epitopes for the autoantibodies. Interestingly, HRS and NRS induce migration of CD4+ and CD8+ T cells, monocytes, and immature dendritic cells (iDC) via the CCR5 and CCR3 receptors, respectively (77). iDCs express these receptors on their surface, and infiltrate muscle tissue in patients burdened with myositis. Thus, AARSs might be cleaved by granzyme B to generate autoantigenic fragments with chemokine activity. Uptake and presentation of antigenic fragments by antigen presenting cells (APCs) initiates the primary immune response against the self-antigen. Self-reactive CD8+ T cells might further generate additional fragments, thereby amplifying both chemo attraction and immune responses.
AIMP1/p43 Involvement in the Autoimmune Response.
AIMP1 is also implicated in the control of the autoimmune response. A portion of the protein is located in the endoplasmic reticulum (ER) where it can hold the ER-resident chaperone gp96, which belongs to the hsp90 family (78). Surface translocation or extracellular release of gp96 induces activation or maturation of dendritic cells (DCs) via its interaction with CD91 and Toll-like receptor 2/4 of DCs. Thus, chronic surface presentation of gp96 could result in abnormal activation of DCs, leading to a lupus-like autoimmune condition (79). Aberrant surface presentation of gp96 hyperactivates DCs that then secrete TNF-α, IL-1β, and IL-12, resulting in systemic immune activation (78). Because AIMP1 prevents surface presentation of gp96, genetic depletion of AIMP1 in antigen-presenting cells such as splenocytes, fibroblasts, and macrophages enhances surface localization of gp96, thereby leading to the phenotypes that are similar to the chronic surface presentation of gp96 (78). In summary, AIMP1 may act as a regulator for the ER retention of gp96 and provide a regulatory mechanism for immune responses.
Potential Connections of AARSs with Diabetes
Mitochondrial leucyl-tRNA synthetase (Mito-LRS), like all mitochondrial tRNA synthetases in humans, is encoded by the nuclear genome. In diabetes, both hyperglycemia and hyperinsulinemia stimulate accumulation of mitochondrial tRNA mutations by inducing oxidative stress. In addition, a single nucleotide polymorphism of mito-LRS leading to an amino acid substitution (H324Q) was found in patients afflicted with type 2 diabetes mellitus (80). Because H324Q mito-LRS has normal aminoacylation and editing activities, the causal relationship between the mito-LRS H324Q mutation and type 2 diabetes is not yet understood. Likewise, mutations in mitochondrial tRNAs, the substrates of AARSs, appear to have a causal relationship to multiple diseases including diabetes (81, 82).
In another vein, AIMP1 works as regulator of glucose homeostasis (83). Genetic depletion of AIMP1 in the mouse leads to hypoglycemic phenotypes. Yet to be determined is whether this pathological phenotype can also be applied to humans.
Implications and Perspectives
The tRNA synthetases arose early in evolution, being essential for establishing the genetic code that relates nucleotide triplets to specific amino acids. In an RNA world, the aminoacylation reaction is thought to have been catalyzed by ribozymes. Because the aminoacyl linkage is higher in energy than the peptide bond, the production of aminoacyl RNAs provided a straightforward path to peptides (for example, the side-by-side docking of two aminoacyl tRNAs leads to spontaneous peptide bond formation) (84, 85). The present-day genetic code proved so robust that it overwhelmed all other alternatives, and over the eons, gave birth to the tree of life with its three great kingdoms—archae, bacteria, and eukarya. The two classes of tRNA synthetases are present at the root, or base, of this tree and are intimately tied to the historical development of the universal code.
Perhaps because of their presence from the beginning, the synthetases have always been available for adaptation and recruitment to emerging cell signaling pathways. Insertions of new motifs, the carving out of novel fragments by alternative splicing or proteolysis, and posttranslational modifications were all possible and provided a way to link translation to cell signaling pathways and biological networks. Viewed from this perspective, the synthetases may have been among the earliest cytokines and cell signaling molecules. Later developments, such as the formation of the multisynthetase complex in eukaryotes, gathered together many of the tRNA synthetase cytokines in an organized way that, among other features, enabled them to be mobilized by specific cues (such as IFN-γ stimulation) for their expanded functions.
In the future, the major advances in understanding how synthetases are linked to cell signaling pathways will come from elucidation of the many interacting protein partners of tRNA synthetases. Annotation of these interacting partners, and the structural elucidation of the motifs needed for these interactions, will provide some of the most fundamental understanding of how synthetases became so tightly connected with a vast array of cell signaling pathways. These investigations will undoubtedly lead back to the catalytic site itself, and the pockets and determinants for binding the amino acid ligand, ATP, and tRNA. Adaptations of these pockets and determinants for protein–protein and protein–nucleic acid interactions conceptually provide at least one route to expanding the world of the synthetases. These enzymes also have attachments and insertions of peptides that are capable of interacting with diverse partners. These additional domains might have paved another way for functional diversification. Initially, as proteins present in every cell type, they constantly were exposed to many other cellular components with which they had serendipitous and “nonphysiological” contacts. These contacts, over time, could have engendered new activities that eventually became highly specific and even essential for cell growth and development.
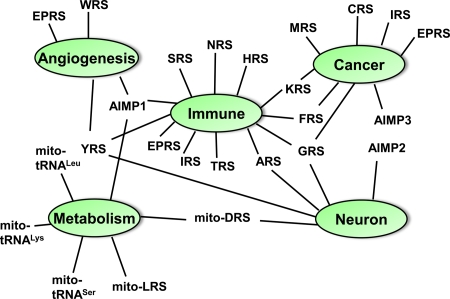
The diverse connections of AARSs with various human diseases make them attractive as targets for the development of therapeutics (Fig. 2). Two of the synthetases—WRS and EPRS—have been studied in detail for their alternative functions in cell signaling pathways but have not yet been linked to a specific disease. Regardless of such disease connections, however, therapeutics based on the potent cytokine fragments of the synthetases like WRS are of great interest. In addition, because the enzymes are essential, their active sites in pathogenic microorganisms are obvious targets for antiinfectives (86, 87). Here, specificity for the pathogen synthetase is possible because of the variations in the evolution of the active site residues, so that a drug targeted to the pathogen does not cross-bind to the human counterpart. Many of the synthetases have a second active site designed for clearing errors of aminoacylation. This site offers yet another target for antiinfectives. Interestingly, a novel antifungal traps tRNALeu in the editing site of yeast cytoplasmic LRS (88). As a consequence, tRNALeu cannot be aminoacylated. Thus, as therapeutic agents in and of themselves, and as targets for drugs, the tRNA synthetases will yield many opportunities for disease intervention.
Fig. 2.
Linkage map of AARSs with various human diseases. Mutants of G/Y/ARS, mitochondrial DRS and AIMP2 are implicated in CMT, leukoencephalopathy and Parkinson's disease, respectively. The M, C, I, EP, F and KRSs are overexpressed in various human cancers, although the potential cause for the overexpression is probably idiosyncratic. AIMP3/p18 is a haploinsufficient tumor suppressor acting on ATM/ATR, thereby leading to activation of p53. The H, T, A, I, F, G, S and NRSs are implicated in the autoimmune diseases collectively designated “antisynthetase syndrome.” AIMP1 controls a lupus-like autoimmune disease via its interaction with gp96. The C-terminal domains of YRS and KRS work as inflammatory cytokines. The secretion of the N-terminal domain of YRS, N-terminal truncated WRS, and full-length AIMP1/p43 regulate angiogenesis. EPRS represses angiogenesis via translational silencing of VEGF-A (60). A mutation of mitochondrial LRS is associated with type 2 diabetes, even though the mutation does not affect its catalytic activity. Reduced activity of mitochondrial DRS is associated with leukoencephalopathy and lactate elevation. Mutations in mitochondrial tRNALys (104, 105), tRNALeu (106), and tRNASer (107) were shown to be associated with diabetes. AIMP1 works as hormone for glucose homeostasis.
Acknowledgments.
This work was supported by the Korea Science and Engineering Foundation through Acceleration Research Grant R17-2007-020-01000-0, Seoul Research and Development Program 11125, by National Institutes of Health Grants GM15539, 23562, and CA92577, and by a fellowship from the National Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Edgecombe M, Craddock HS, Smith DC, McLennan AG, Fisher MJ. Diadenosine polyphosphate-stimulated gluconeogenesis in isolated rat proximal tubules. Biochem J. 1997;323:451–456. doi: 10.1042/bj3230451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verspohl EJ, Hohmeier N, Lempka M. Diadenosine tetraphosphate (Ap4A) induces a diabetogenic situation: Its impact on blood glucose, plasma insulin, gluconeogenesis, glucose uptake and GLUT-4 transporters. Pharmazie. 2003;58:910–915. [PubMed] [Google Scholar]
- 3.Nishimura A, et al. Diadenosine 5′, 5‴-P1, P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells. 1997;2:401–413. doi: 10.1046/j.1365-2443.1997.1300328.x. [DOI] [PubMed] [Google Scholar]
- 4.Romby P, Springer M. Bacterial translational control at atomic resolution. Trends Genet. 2003;19:155–161. doi: 10.1016/S0168-9525(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 5.Sampath P, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: Gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Rho SB, et al. Genetic dissection of protein–protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci USA. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: Beyond translation. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 8.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29:419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: New perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 13.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem Sci. 2004;29:250–256. doi: 10.1016/j.tibs.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Wakasugi K, et al. Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J Biol Chem. 2002;277:20124–20126. doi: 10.1074/jbc.C200126200. [DOI] [PubMed] [Google Scholar]
- 16.Otani A, et al. Bone marrow-derived stem cells target retinal astrocytes and can promote or inhibit retinal angiogenesis. Nat Med. 2002;8:1004–1010. doi: 10.1038/nm744. [DOI] [PubMed] [Google Scholar]
- 17.Kise Y, et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 18.Park SG, et al. Human lysyl-tRNA synthetase is secreted to trigger pro-inflammatory response. Proc Natl Acad Sci USA. 2005;102:6356–6361. doi: 10.1073/pnas.0500226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor M, et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J Biol Chem. 2008;283:2070–2077. doi: 10.1074/jbc.M706028200. [DOI] [PubMed] [Google Scholar]
- 20.Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. II. Neurologic, genetic, and electrophysiologic findings in various neuronal degenerations. Arch Neurol. 1968;18:619–625. doi: 10.1001/archneur.1968.00470360041003. [DOI] [PubMed] [Google Scholar]
- 21.Antonellis A, et al. Glycyl-tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordanova A, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 23.Nangle LA, Zhang W, Xie W, Yang XL, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci USA. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Yang XL, Skene RJ, McRee DE, Schimmel P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc Natl Acad Sci USA. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W, Schimmel P, Yang XL. Crystallization and preliminary X-ray analysis of a native human tRNA synthetase whose allelic variants are associated with Charcot-Marie-Tooth disease. Acta Crystallogr F. 2006;62:1243–1246. doi: 10.1107/S1744309106046434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cader MZ, et al. Crystal structure of human wild type and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy. FEBS Lett. 2007;581:2959–2964. doi: 10.1016/j.febslet.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 28.Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J Biol Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- 29.Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- 30.Yarus M. Phenylalanyl-tRNA synthetase and isoleucyl-tRNAPhe: A possible verification mechanism for aminoacyl-tRNA. Proc Natl Acad Sci USA. 1972;69:1915–1919. doi: 10.1073/pnas.69.7.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. [Google Scholar]
- 32.Doring V, et al. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 33.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Williams AM, Martinis SA. Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity. Proc Natl Acad Sci USA. 2006;103:3586–3591. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 36.Nangle LA, De Crecy Lagard V, Doring V, Schimmel P. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J Biol Chem. 2002;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- 37.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim YJ, Nakatomi R, Akagi T, Hashikawa T, Takahashi R. Unsaturated fatty acids induce cytotoxic aggregate formation of amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutants. J Biol Chem. 2005;280:21515–21521. doi: 10.1074/jbc.M502230200. [DOI] [PubMed] [Google Scholar]
- 39.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrekopoulos C, Zhang H, Joseph J, Kalivendi S, Kalyanaraman B. Bicarbonate enhances alpha-synuclein oligomerization and nitration: Intermediacy of carbonate radical anion and nitrogen dioxide radical. Biochem J. 2004;378:435–447. doi: 10.1042/BJ20031466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valentine JS, Hart PJ. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunst CB, Mezey E, Brownstein MJ, Patterson D. Mutations in SOD1 associated with amyotrophic lateral sclerosis cause novel protein interactions. Nat Genet. 1997;15:91–94. doi: 10.1038/ng0197-91. [DOI] [PubMed] [Google Scholar]
- 43.Scheper GC, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 44.Corti O, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: Linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 45.Ko HS, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SW, Kang YS, Kim S. Multi-functional proteins in tumorigenesis: Aminoacyl-tRNA synthetases and translational components. Curr Proteomics. 2006;3:233–247. [Google Scholar]
- 47.Kushner JP, Boll D, Quagliana J, Dickman S. Elevated methionine-tRNA synthetase activity in human colon cancer. Proc Soc Exp Biol Med. 1976;153:273–276. doi: 10.3181/00379727-153-39526. [DOI] [PubMed] [Google Scholar]
- 48.Forus A, Florenes VA, Maelandsmo GM, Fodstad O, Myklebost O. The protooncogene CHOP/GADD153, involved in growth arrest and DNA damage response, is amplified in a subset of human sarcomas. Cancer Genet Cytogenet. 1994;78:165–171. doi: 10.1016/0165-4608(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 49.Nilbert M, Rydholm A, Mitelman F, Meltzer PS, Mandahl N. Characterization of the 12q13–15 amplicon in soft tissue tumors. Cancer Genet Cytogenet. 1995;83:32–36. doi: 10.1016/s0165-4608(95)00016-x. [DOI] [PubMed] [Google Scholar]
- 50.Palmer JL, Masui S, Pritchard S, Kalousek DK, Sorensen PH. Cytogenetic and molecular genetic analysis of a pediatric pleomorphic sarcoma reveals similarities to adult malignant fibrous histiocytoma. Cancer Genet Cytogenet. 1997;95:141–147. doi: 10.1016/s0165-4608(96)00243-9. [DOI] [PubMed] [Google Scholar]
- 51.Reifenberger G, et al. Refined mapping of 12q13–q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer Res. 1996;56:5141–5145. [PubMed] [Google Scholar]
- 52.Cools J, et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 53.Debelenko LV, et al. Identification of CARS-ALK fusion in primary and metastatic lesions of an inflammatory myofibroblastic tumor. Lab Invest. 2003;83:1255–1265. doi: 10.1097/01.lab.0000088856.49388.ea. [DOI] [PubMed] [Google Scholar]
- 54.Miyaki M, et al. Alterations of repeated sequences in 5′ upstream and coding regions in colorectal tumors from patients with hereditary nonpolyposis colorectal cancer and Turcot syndrome. Oncogene. 2001;20:5215–5218. doi: 10.1038/sj.onc.1204578. [DOI] [PubMed] [Google Scholar]
- 55.Coller HA, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 57.Rodova M, Ankilova V, Safro MG. Human phenylalanyl-tRNA synthetase: Cloning, characterization of the deduced amino acid sequences in terms of the structural domains and coordinately regulated expression of the alpha and beta subunits in chronic myeloid leukemia cells. Biochem Biophys Res Commun. 1999;255:765–773. doi: 10.1006/bbrc.1999.0141. [DOI] [PubMed] [Google Scholar]
- 58.Wasenius VM, et al. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: A cDNA and tissue microarray study. Clin Cancer Res. 2003;9:68–75. [PubMed] [Google Scholar]
- 59.Scandurro AB, Weldon CW, Figueroa YG, Alam J, Beckman BS. Gene microarray analysis reveals a novel hypoxia signal transduction pathway in human hepatocellular carcinoma cells. Int J Oncol. 2001;19:129–135. doi: 10.3892/ijo.19.1.129. [DOI] [PubMed] [Google Scholar]
- 60.Ray PS, Fox PL. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 2007;26:3360–3372. doi: 10.1038/sj.emboj.7601774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SG, et al. Dose-dependent biphasic activity of tRNA synthetase-associating factor, p43, in angiogenesis. J Biol Chem. 2002;277:45243–45248. doi: 10.1074/jbc.M207934200. [DOI] [PubMed] [Google Scholar]
- 62.Lee YS, et al. Antitumor activity of the novel human cytokine AIMP1 in an in vivo tumor model. Mol Cells. 2006;21:213–217. [PubMed] [Google Scholar]
- 63.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: Implications for its physiological significance. Proc Natl Acad Sci USA. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim MJ, et al. Downregulation of fuse-binding protein and c-myc by tRNA synthetase cofactor, p38, is required for lung differentiation. Nat Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 65.Quevillon S, Mirande M. The p18 component of the multisynthetase complex shares a protein motif with the beta and gamma subunits of eukaryotic elongation factor 1. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- 66.Park BJ, et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. 2005;120:209–221. doi: 10.1016/j.cell.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 67.Park BJ, et al. AIMP3 haploinsufficiency disrupts oncogene-induced p53 activation and genomic stability. Cancer Res. 2006;66:6913–6918. doi: 10.1158/0008-5472.CAN-05-3740. [DOI] [PubMed] [Google Scholar]
- 68.Kim KJ, et al. Determination of three dimensional structure and residues of novel tumor suppressor, AIMP3/p18, required for the interaction with ATM. J Biol Chem. 2008;283:14032–14040. doi: 10.1074/jbc.M800859200. [DOI] [PubMed] [Google Scholar]
- 69.Mathews MB, Bernstein RM. Myositis autoantibody inhibits histidyl-tRNA synthetase: A model for autoimmunity. Nature. 1983;304:177–179. doi: 10.1038/304177a0. [DOI] [PubMed] [Google Scholar]
- 70.Mathews MB, Reichlin M, Hughes GR, Bernstein RM. Anti-threonyl-tRNA synthetase, a second myositis-related autoantibody. J Exp Med. 1984;160:420–434. doi: 10.1084/jem.160.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirakata M, et al. Anti-KS: Identification of autoantibodies to asparaginyl-transfer RNA synthetase associated with interstitial lung disease. J Immunol. 1999;162:2315–2320. [PubMed] [Google Scholar]
- 72.Raben N, et al. A motif in human histidyl-tRNA synthetase which is shared among several aminoacyl-tRNA synthetases is a coiled-coil that is essential for enzymatic activity and contains the major autoantigenic epitope. J Biol Chem. 1994;269:24277–24283. [PubMed] [Google Scholar]
- 73.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrade F, et al. Granzyme B directly and efficiently cleaves several downstream caspase substrates: Implications for CTL-induced apoptosis. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- 75.Casciola-Rosen L, Rosen A. Ultraviolet light-induced keratinocyte apoptosis: A potential mechanism for the induction of skin lesions and autoantibody production in LE. Lupus. 1997;6:175–180. doi: 10.1177/096120339700600213. [DOI] [PubMed] [Google Scholar]
- 76.Casciola-Rosen L, Andrade F, Ulanet D, Wong WB, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: Implications for initiation of autoimmunity. J Exp Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howard OM, et al. Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J Exp Med. 2002;196:781–791. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han JM, et al. Aminoacyl-tRNA synthetase-interacting multifunctional protein 1/p43 controls endoplasmic reticulum retention of heat shock protein gp96: Its pathological implications in lupus-like autoimmune diseases. Am J Pathol. 2007;170:2042–2054. doi: 10.2353/ajpath.2007.061266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu B, et al. Cell surface expression of an endoplasmic reticulum resident heat shock protein gp96 triggers MyD88-dependent systemic autoimmune diseases. Proc Natl Acad Sci USA. 2003;100:15824–15829. doi: 10.1073/pnas.2635458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.t Hart LM, et al. Evidence that the mitochondrial leucyl tRNA synthetase (LARS2) gene represents a novel type 2 diabetes susceptibility gene. Diabetes. 2005;54:1892–1895. doi: 10.2337/diabetes.54.6.1892. [DOI] [PubMed] [Google Scholar]
- 81.Florentz C. Molecular investigations on tRNAs involved in human mitochondrial disorders. Biosci Rep. 2002;22:81–98. doi: 10.1023/a:1016065107165. [DOI] [PubMed] [Google Scholar]
- 82.Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park SG, et al. Hormonal activity of AIMP1/p43 for glucose homeostasis. Proc Natl Acad Sci USA. 2006;103:14913–14918. doi: 10.1073/pnas.0602045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tamura K, Schimmel P. Oligonucleotide-directed peptide synthesis in a ribosome- and ribozyme-free system. Proc Natl Acad Sci USA. 2001;98:1393–1397. doi: 10.1073/pnas.98.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tamura K, Schimmel P. Peptide synthesis with a template-like RNA guide and aminoacyl phosphate adaptors. Proc Natl Acad Sci USA. 2003;200:8666–8669. doi: 10.1073/pnas.1432909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S, Lee SW, Choi EC, Choi SY. Aminoacyl-tRNA synthetases and their inhibitors as a novel family of antibiotics. Appl Microbiol Biotechnol. 2003;61:278–288. doi: 10.1007/s00253-003-1243-5. [DOI] [PubMed] [Google Scholar]
- 87.Pohlmann J, Brotz-Oesterhelt H. New aminoacyl-tRNA synthetase inhibitors as antibacterial agents. Curr Drug Targets Infect Disord. 2004;4:261–272. doi: 10.2174/1568005043340515. [DOI] [PubMed] [Google Scholar]
- 88.Rock FL, et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316:1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 89.Ko YG, et al. Glutamine-dependent anti-apoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276:6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 90.Ko YG, Kang YS, Kim EK, Park SG, Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J Cell Biol. 2000;149:567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ko YG, et al. A cofactor of tRNA synthetase, p43, is secreted to up-regulate proinflammatory genes. J Biol Chem. 2001;276:23028–32303. doi: 10.1074/jbc.M101544200. [DOI] [PubMed] [Google Scholar]
- 92.Park H, et al. Monocyte cell adhesion induced by a human aminoacyl-tRNA synthetase associated factor, p43: Identification of the related adhesion molecules and signal pathways. J Leukoc Biol. 2002;71:223–230. [PubMed] [Google Scholar]
- 93.Park H, Park SG, Kim J, Ko YG, Kim S. Signaling pathways for TNF production induced by human aminoacyl-tRNA synthetase-associating factor, p43. Cytokine. 2002;20:148–153. doi: 10.1006/cyto.2002.1992. [DOI] [PubMed] [Google Scholar]
- 94.Park SG, et al. The novel cytokine p43 stimulates dermal fibroblast proliferation and wound repair. Am J Pathol. 2005;166:387–398. doi: 10.1016/S0002-9440(10)62262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han JM, Park SG, Lee Y, Kim S. Structural separation of different extracellular activities in aminoacyl-tRNA synthetase-interacting multi-functional protein, p43/AIMP1. Biochem Biophys Res Commun. 2006;342:113–118. doi: 10.1016/j.bbrc.2006.01.117. [DOI] [PubMed] [Google Scholar]
- 96.Lee YS, et al. AIMP1/p43 downregulates TGF-β signaling via stabilization of smurf2. Biochem Biophys Res Commun. 2008;371:395–400. doi: 10.1016/j.bbrc.2008.04.099. [DOI] [PubMed] [Google Scholar]
- 97.Yannay-Cohen N, Razin E. Translation and transcription: The dual functionality of KRS in mast cells. Mol Cells. 2006;22:127–132. [PubMed] [Google Scholar]
- 98.Halwani R, et al. Cellular distribution of lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J Virol. 2004;78:7553–7564. doi: 10.1128/JVI.78.14.7553-7564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, Shue E, Ewalt KL, Schimmel P. A new gamma-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucl Acids Res. 2004;32:719–727. doi: 10.1093/nar/gkh240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tzima E, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci USA. 2003;100:14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tzima E, et al. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem. 2005;280:2405–2408. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- 102.Banin E, et al. T2-WRS inhibits preretinal neovascularization and enhances physiological vascular regrowth in OIR as assessed by a new method of quantification. Invest Ophthalmol Vis Sci. 2006;47:2125–2134. doi: 10.1167/iovs.05-1096. [DOI] [PubMed] [Google Scholar]
- 103.Ivakhno SS, Kornelyuk AI. Cytokine-like activities of some aminoacyl-tRNA synthetases and auxiliary p43 cofactor of aminoacylation reaction and their role in oncogenesis. Exp Oncol. 2004;26:250–255. [PubMed] [Google Scholar]
- 104.Kameoka K, et al. Novel mitochondrial DNA mutation in tRNA(Lys) (8296A–>G) associated with diabetes. Biochem Biophys Res Commun. 1998;245:523–527. doi: 10.1006/bbrc.1998.8437. [DOI] [PubMed] [Google Scholar]
- 105.Kameoka K, Isotani H, Tanaka K, Kitaoka H, Ohsawa N. Impaired insulin secretion in Japanese diabetic subjects with an A-to-G mutation at nucleotide 8296 of the mitochondrial DNA in tRNA (Lys) Diabetes Care. 1998;21:2034–2035. doi: 10.2337/diacare.21.11.2034. [DOI] [PubMed] [Google Scholar]
- 106.Janssen GM, Maassen JA, van Den Ouweland JM. The diabetes-associated 3243 mutation in the mitochondrial tRNA (Leu(UUR)) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J Biol Chem. 1999;274:29744–29748. doi: 10.1074/jbc.274.42.29744. [DOI] [PubMed] [Google Scholar]
- 107.Choo-Kang AT, et al. Defining the importance of mitochondrial gene defects in maternally inherited diabetes by sequencing the entire mitochondrial genome. Diabetes. 2002;51:2317–2320. doi: 10.2337/diabetes.51.7.2317. [DOI] [PubMed] [Google Scholar]