Abstract
Hybrid sterility is a major form of postzygotic reproductive isolation. Although reproductive isolation has been a key issue in evolutionary biology for many decades in a wide range of organisms, only very recently a few genes for reproductive isolation were identified. The Asian cultivated rice (Oryza sativa L.) is divided into two subspecies, indica and japonica. Hybrids between indica and japonica varieties are usually highly sterile. A special group of rice germplasm, referred to as wide-compatibility varieties, is able to produce highly fertile hybrids when crossed to both indica and japonica. In this study, we cloned S5, a major locus for indica–japonica hybrid sterility and wide compatibility, using a map-based cloning approach. We show that S5 encodes an aspartic protease conditioning embryo-sac fertility. The indica (S5-i) and japonica (S5-j) alleles differ by two nucleotides. The wide compatibility gene (S5-n) has a large deletion in the N terminus of the predicted S5 protein, causing subcellular mislocalization of the protein, and thus is presumably nonfunctional. This triallelic system has a profound implication in the evolution and artificial breeding of cultivated rice. Genetic differentiation between indica and japonica would have been enforced because of the reproductive barrier caused by S5-i and S5-j, and species coherence would have been maintained by gene flow enabled by the wide compatibility gene.
Keywords: subspecies of rice, hybrid sterility, wide compatibility, aspartic protease
Reproductive isolation plays important roles in speciation and maintaining species identity and thus has remained a key issue in evolutionary biology for many decades in a wide range of organisms (1, 2). According to the stage of the barriers, reproductive isolation can be divided into two general categories: prezygotic reproductive isolation and postzygotic reproductive isolation. Hybrid sterility is a major form of postzygotic reproductive isolation. For decades, the Dobzhansky–Muller model of deleterious allelic interactions for hybrid sterility has been the subject of intensive investigations, especially in hybrids between Drosophila species (2). However, only very recently, a few genes for interspecific hybrid sterility in Drosophila have been identified (2–7). In plants, a receptor-like kinase was identified in Arabidopsis as mediating male–female interactions, which may play a role in reproductive isolation in interspecific crosses (8). An epistatic interaction was also identified in an intraspecific cross of Arabidopsis as inducing hybrid necrosis, another type of postzygotic isolation (9).
The Asian cultivated rice (Oryza sativa L.) grown worldwide is divided into two major subspecies, indica (O. sativa L. ssp. indica) and japonica (O. sativa L. ssp. japonica). It has been observed for nearly a century that hybrids between indica and japonica usually show low fertility (10–12), which is probably one of the best known examples of reproductive barriers in plants. In contrast, a special group of rice germplasm, referred to as wide-compatibility varieties (WCVs), is able to produce highly fertile hybrids when crossed to both indica and japonica varieties (11).
Genetic analyses have identified a large number of loci affecting hybrid fertility, which are further resolved into ones causing female gamete abortion (13–16) and ones inducing pollen sterility (16–19). It was established that S5 located on chromosome 6 is a major locus for indica–japonica hybrid sterility. Ikehashi and Araki (13) proposed that there are three alleles at the S5 locus: an indica allele (S5-i), a japonica allele (S5-j), and a neutral allele (S5-n) also referred to as a wide-compatibility gene (WCG). Plants from zygotes formed of S5-n with either S5-i or S5-j would be fully fertile, whereas plants genotypically S5-i/S5-j would be highly sterile. Results of several studies support this proposal (20–25). Furthermore, it was determined that the sterility of the S5-i/S5-j hybrid was primarily caused by female gamete abortion (16, 22). However, nothing is known at present about the genes controlling reproductive barriers in rice.
Aspartic proteases (APs) constitute one of the four superfamilies of proteolytic enzymes and are distributed in a wide variety of organisms including animals, plants, fungi, bacteria, and viruses (24, 25). Much of our knowledge on the roles of APs comes from microbial and animal studies. A striking example found in humans is that a membrane-anchored AP has Alzheimer's disease β-secretase activity (26). In plants, APs have been identified from a number of tissues in diverse species including rice (27, 28). Results from Arabidopsis studies indicated that they are involved in disease resistance signaling (29) and programmed cell death in reproductive tissues (30). However, definite biological functions of these enzymes in vivo for any process in any plant have yet to be established.
Here we show that S5 encodes an AP. The indica (S5-i) and japonica (S5-j) alleles differed by two nucleotides. The WCG (S5-n) had a large deletion in the N terminus of the predicted S5 protein, which would cause subcellular mislocalization of the protein and thus is presumably nonfunctional. This triallelic system has a profound implication in the evolution and artificial breeding of cultivated rice.
Results
Map-Based Cloning of S5.
Qiu et al. (22) delimited S5 to a 40-kb DNA fragment containing five ORFs (ORFs1–5), whereas Ji et al. (23) mapped this locus to a 50-kb region. The targeted S5-containing regions of the two mapping results overlapped by a fragment of 24.8 kb [supporting information (SI) Fig. S1], where two complete ORFs (ORF4 and ORF5) were predicted.
Because the sterility occurs only when the plant is genotypically S5-i/S5-j, or having S5-i and S5-j alleles simultaneously, the strategy that we adopted for verifying the candidate gene was transformation of a japonica variety (homozygous for S5-j) with S5-i, such that sterility would be expected in the transformants (genotypically S5-j/S5-j + S5-i). To do this, transformation constructs were prepared for ORF3, ORF4, and ORF5, with the constructs for ORF3 and ORF4 obtained by cutting the corresponding genomic DNA fragments of a clone from a bacterial artificial chromosome library of Zhenshan 97 and the construct for ORF5 by PCR amplification of the genomic DNA from Nanjing 11 (Fig. S2). Both Zhenshan 97 and Nanjing 11 are indica, and their hybrids with japonica varieties show greatly reduced fertility (12). The constructs were introduced into the japonica variety Balilla, resulting in 43, 74, and 33 independent T0 plants for ORF3, ORF4, and ORF5, respectively.
Examination of the spikelet fertility of the T0 plants under natural field conditions showed that there was not a statistically significant difference between the transgene-positive and -negative plants of ORF3 or ORF4 (Table S1), indicating that neither of them is a likely candidate for S5. By contrast, transgene-positive and -negative plants of the ORF5 construct (hereafter referred to as ORF5-NJ11) showed a highly significant difference in spikelet fertility; the average spikelet fertility (53.3%) of the negative plants was much higher than that (2.8%) of the positive plants (Fig. 1 and Table S1).
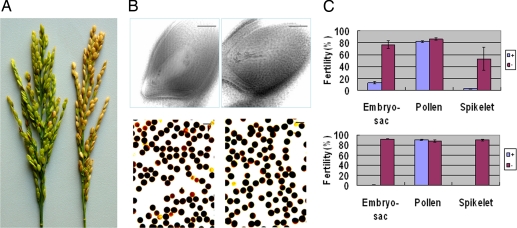
Fig. 1.
Fertility of ORF5-NJ11 transgenic-positive and -negative plants with Balilla as the recipient. (A) Spikelet fertility of positive (left, sterile) and negative (right, fertile) T1 segregants. (B) Top, embryo sacs of negative (left, normal) and positive (right, abortive) plants; Bottom, pollen of negative (left) and positive (right) plants. Scale bars: 50 μm. (C) Fertility scores (error bar = 1 SEM) of transgene positive and negative plants in T0 (Top) and a T1 family (Bottom). There are 29 positive and 4 negative plants in T0 and 15 positive and 14 negative segregants in the T1 family (B10).
As hybrid sterility imparted by S5 is due to embryo-sac abortion (16), we examined the embryo sac as well as pollen fertility of the ORF5-NJ11-transformed T0 plants. Whereas pollen fertility was not significantly different between the transgene-positive and -negative plants, a highly significant difference in embryo-sac fertility was observed between them (Fig. 1 and Table S1), strongly suggesting ORF5 to be the candidate for S5.
Because the ORF5-NJ11 transgene-positive plants showed a very high rate of embryo-sac abortion, T0 plants harboring a single-copy transgene were used as the male parents to cross with the wide-type parent Balilla, and fertility of the resulting F1 (T1) families was again examined. A total of 29 plants were obtained from one of the F1 (T1) families (B10), of which 15 were positive and the remaining 14 negative, and examined in the natural rice field (Fig. 1). Whereas the negative segregants showed normal spikelet fertility (89.4%), no seed was produced by the positive segregants. The embryo-sac fertility of the negative segregants ranged from 83.3 to 97.8% with an average of 92.3%, whereas that of the positive segregants ranged from 0 to 5.0% with an average of 1.3%. There was no significant difference in pollen fertility between the positive and negative segregants. Similar results were also observed in an analysis of 33 F1 (T1) plants obtained from three other families grown in a greenhouse in the winter (Table S1). Such perfect cosegregation between the transgene and embryo-sac fertility (hence spikelet fertility) further confirmed that the product encoded by ORF5 is the cause of the embryo-sac fertility in the indica–japonica hybrid, and we thus concluded that ORF5 is the S5 gene.
The S5 Gene Encodes an AP.
The predicted coding sequence of S5 is 1419 bp based on the annotated genomic sequence of the cultivar Nipponbare (31). The genomic sequences of Balilla and Nipponbare are identical in this region. The S5 transcripts of Nanjing 11 and Balilla determined by rapid amplification of the cDNA ends (RACE) and PCR amplification were 1911 and 1912 bp in length, respectively, with three noticeable differences between them (Fig. 2). Using the Balilla cDNA sequence as the reference, an adenosine (A) was deleted in Nanjing 11 at 172 bp downstream of the predicted translation termination site, decreasing the transcript length by 1 bp. There were two nucleotide substitutions in the predicted coding sequence, both of which caused amino acid substitutions (Fig. 2 and Fig. S3). Alignment of the transcript with the Nipponbare genomic sequence showed that the S5 transcript consisted of three exons, and the coding sequence was located only in two exons (Fig. 2).
Fig. 2.
The sequence features of the S5 transcripts from 02428 (WCV), Balilla (japonica), and Nanjing 11 (indica).
The predicted S5 protein, 472 aa in length, has significant similarity (E = 2e−06) with members of eukaryotic APs (pfam00026) (Fig. S4). Two active sites containing Asp (D) residues characteristic of the APs (25) were identified with a PROSITE analysis (http://www.expasy.ch/cgi-bin/prosite/). A signal peptide was identified using SignalP 3.0 (http://www.cbs.dtu.dk/cgi-bin/) (Fig. S3). However, it does not have the plant-specific sequence found in many plant APs (27).
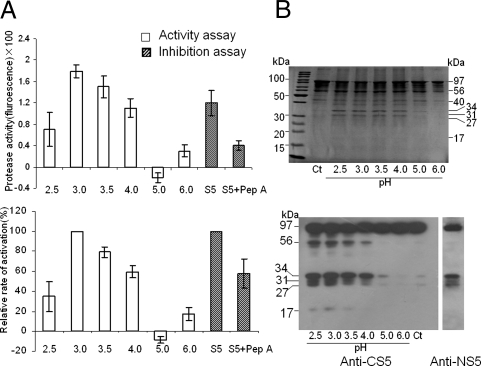
APs have optimal activity at acidic pH and are inhibited by pepstatin A (25). To assess whether S5 is an AP, an in vitro protease assay of the S5 protein obtained by expressing the cDNA from Balilla in Escherichia coli was conducted, which showed that the activity of S5 was low but detectable in acidic pH, the highest at pH 3.0, and greatly reduced with the addition of pepstatin A (Fig. 3A). We also tested the autoproteolytic activity of S5, which showed that the protein could cleave itself especially at pH 2.5–4.0 (Fig. 3B). Similar results were also obtained in a protease assay using the cDNA from Nanjing 11 and an equal mixture of protein from Balilla and Nanjing 11 (data not shown). These results were consistent with a previous in vitro activity assay of eukaryotic AP (28), demonstrating that the S5-encoded protein is indeed an AP.
Fig. 3.
In vitro assay of protease activity of S5. (A) Absolute activity measured as the spectrophotometer reading with that of water control set to zero (Top), and relative activity by using the highest activity (pH 3.0) as the reference (Bottom). The open bars are the readings of protease activity assays and the shaded bars are the readings of the pepstatin inhibition assays. Error bars = 1 SEM. (B) SDS-PAGE showing the protein bands of the autolytic products in a series of pH buffers (Top), and immunoblot confirmation of the S5 autolytic products by using Anti-CS5 and Anti-NS5 as the probes (Bottom). The lanes marked Ct are the controls. The molecular weight on the right side of the top section and the left side of the bottom section was estimated on the basis of size markers (as shown on the left side of the top section).
Expression and Subcellular Localization.
To investigate the expression profile of S5, we searched a dataset of whole-genome expression arrays of two indica cultivars Zhenshan 97 and Minghui 63 with RNA samples from 25 tissues spanning the entire life cycle of the rice plants (Fig. S5). It can be seen that expression of S5 is extremely low throughout the life cycle, only at the background level in almost all of the stages.
We analyzed the expression of S5 in the developing panicles during stages 3–8 (see legend of Fig. S6 for description), in comparison with the leaves harvested at the corresponding stages. S5 expression was not detectable in leaves, whereas variable levels of expression were observed in the developing panicles in all three genotypes, Balilla, Nanjing 11, and 02428 (Fig. S6A). Real-time PCR was also performed using developing panicles at stage 6 to compare the S5 expression levels in the three parents and their hybrids. The analysis revealed significant differences in S5 expression among the three parents as well as among the hybrids (Fig. S6B). Obviously these differences were not correlated with the fertility levels.
RNA in situ hybridization showed that S5 has a localized expression in the ovule tissues including nucellus, integument, megasporocytes, and megaspores during megaspore development (Fig. 4 A–C), consistent with the observed effect on embryo-sac abortion.
Fig. 4.
Expression of the S5 gene and subcellular localization of S5. (Top) In situ hybridization using an antisense probe showing expression in nucellar and archesporial cells (02428) (A), integuments and megasporocytes (Balilla) (B), and integuments and megaspores (Nanjing 11) (C), with the sense probe as the negative control (D). A is a transection, and B–D are longitudinal sections. Scale bars: 25 μm. (Bottom) Subcellular localization of the S5 protein in nucellar cells by immunogold and TEM techniques showing a large quantity of gold particles in the cell wall of Nanjing 11 (E) and Balilla (F) and few particles in the cell wall of 02428 (G). The one without the primary antibody is the negative control (H). Scale bars: 0.5 μm. Arrows indicate the gold particles. C, cytosol; CW, cell wall; M, mitochondrion; V, vacuole.
Posttranslational transport and activation of APs involves complex processing (32), which makes it difficult to determine the subcellular localization of S5 by using a transgenic approach by fusing the protein with a reporter. We used immunogold labeling and transmission electron microscopy (TEM) techniques to assay the subcellular localization of S5, using sections of nucellar tissues where the gene showed relatively high expression. It was revealed that the gold particles were distributed in large quantity in the cell walls, whereas in very small quantity in other subcellular compartments (Fig. 4 E–H and Fig. S7 A–F). Thus, the final destination of this protein is the cell wall, consistent with the nature of a secreted protein as predicted by the signal peptide.
The WCG.
To gain insight into the WCG S5-n, the genomic sequence of the WCV 02428 was compared with Nanjing 11 and Balilla, with the features illustrated in Fig. 2. In the region of transcribed sequences, there was a major difference among the three genotypes resulting from two deletions in 02428 relative to the other two genotypes, a total of 136 bp that were separated by a TAAT motif in the first exon. Of the 136-bp sequence, 67 bp occurred downstream of the predicted the translation start site (TSS) ATG and the other 69 bp upstream the TSS (Fig. 2). The rest of the sequence in 02428 was the same as Nanjing 11 except an AT insertion in the 3′UTR of 02428 relative to the other two genotypes.
Because the 136-bp deletion in 02428 involved the predicted TSS, the first 115 amino acids in the N terminus would be lost, assuming the next in-frame ATG would serve as the TSS leading to read-through. This would result in the loss of the entire signal peptide plus a fragment of 87 amino acids. Consequently, the encoded protein in 02428 would not enter the secretory pathway and thus end in a different destination compared with the indica and japonica varieties. To assess whether this is indeed the case, subcellular localization of S5 in 02428, Balilla, and Nanjing 11 was assayed by immunogold and TEM techniques using sections of the nucellar tissues (Fig. 4 E–H and Fig. S7 A–I). It was revealed that the gold particles occurred in large quantity in the cell walls, but were hardly detectable in the cytoplasm in Nanjing 11 and Balilla. In contrast, very few gold particles were observed in cell walls of 02428, but occurred in relatively large quantity in the cytoplasm, especially in mitochondria. It is highly likely that such mislocalization of the protein in the cellular compartments impairs the normal function of the protein.
On the basis of the hypothesis that S5-n is a nonfunctional allele, it was predicted that transgenic plants produced by transformation of 02428 with S5-i would behave like an indica genotype with respect to its indica–japonica compatibility. To test this hypothesis, the ORF5-NJ11 construct was introduced to 02428. Two plants with a single-copy transgene, C02 and C04, were obtained, both of which showed normal fertility (spikelet fertility 70 and 86%, respectively). T1 progenies derived from these two plants were test-crossed with Balilla and Nanjing 11, and the resulting progenies were examined for embryo-sac fertility and spikelet fertility. As expected, no significant fertility difference was detected between the transgene-positive and -negative segregants in progenies from the cross with Nanjing 11, whereas, in progenies from the cross with Balilla, fertility of the positive plants was much lower than that of the negative plants (Table S2). These results strongly support the hypothesis that S5-n is a nonfunctional allele.
Generality of the Triallele System.
To inspect whether the above-described features of sequence differences exist in a broader range of rice germplasms, we obtained S5 genomic sequences for a total of 16 varieties representative of indica, japonica, and WCVs from a wide geographical area and with no related pedigrees according to the available information (Table S3). The predicted protein sequences of these varieties fell naturally into three groups: indica, japonica, and WCV (Fig. S3). All of the japonica varieties have identical sequences featured by the variant amino acid combination of Leu-273 (L) and Val-471 (V), and all indica varieties shared the Phe (F) and Ala (A) combination at the corresponding sites, although another variant amino acid was also detected at amino acid 423 in an indica cultivar 9311 that has a Ser (S) instead of a Pro (P) as in all other varieties. All of the WCVs had the 115-aa deletion at the N terminus. Such sequence differences are consistent with the functional classification of the varieties, suggesting that this triallelic system is a widespread phenomenon in the species O. sativa.
Discussion
The coexistence of indica, japonica, and WCVs is a unique and spectacular outcome of the evolution in rice, in which the S5 locus would have played a significant role. Genetic differentiation between indica and japonica varieties has been frequently reported, which appears to be a major source of genetic diversity in the cultivated rice gene pool (33–36). The present results clearly imply that the S5-i/S5-j hybrid sterility has been an important promoting factor for the indica–japonica differentiation during the evolution of rice O. sativa. In contrast, the WCG provides an opposing force allowing for hybridization and gene flow, at least in artificial crosses. The counteractive effects of the triallelic system suggest a dynamic relationship between adaptive differentiation and genetic coherence of the cultivated rice species during evolution and artificial selection. Thus, although S5 may not be essential for growth, development, or reproduction as indicated by the complete normality of the plants homozygous for S5-n, it nonetheless provides a coherent force for holding different parts together at the species level.
Evidence from the current study suggests that the function of S5, although not fully understood, is centered on megaspore formation or survival. Analysis of the molecular variations may shed light on the functional differences of the three alleles. Crystal structure analysis established that an AP has three domains, the central domain, the N-terminal lobe, and the C-terminal lobe (27, 37). The central domain consists of residues from three regions, the N-terminal end, the central part, and the C-terminal end. By sequence alignment, it can be deduced that both of the mutant sites in S5, amino acids 273 and 471, are located in the central domain. Of the two variant residues, Phe-273 (F) is conserved across a large range of organisms from plants to animals and humans, whereas amino acid 471 is highly variable (Fig. S4, ref. 37). However, the conserved Phe-273 (hydrophobic and aromatic) is replaced by Leu (hydrophobic but nonaromatic) in S5-j. This change may reduce the stability and activity of the enzyme. Moreover, if S5-n started at the second Met (Fig. S3), the N-terminal segment of the central domain would be completely lost, which would greatly affect the stability and activity of the enzyme, in addition to the mislocalization due to the loss of the signal peptide. However, how such likely reduced activity is related to the embryo-sac fertility remains to be characterized.
The molecular nature of the WCG elucidated in this study has important implications in rice genetic improvement. Hybrids between indica and japonica rice often show strong heterosis in yield-related traits, compared to intra-subspecific hybrids. However, it has been difficult to use such heterosis in breeding programs because of hybrid sterility. With the discovery and molecular characterization of the WCGs, exploitation of the inter-subspecific heterosis by developing inter-subspecific hybrids has become a reality. The present results also provided functional markers for WCG germplasm screening and WCV development in rice breeding programs.
Materials and Methods
Rice Varieties, Transformation, and Assay of Transgenic Plants.
A total of 16 rice varieties (Table S3), including 5 indica, 4 japonica, and 7 WCVs, were used in this study.
Transformation was conducted according to a published protocol (38). For preparing the constructs, genomic fragments were ligated into the vector pCAMBIA1301 (39) and transformed into the Agrobacterium strain EHA105. For the ORF5 construct (Fig. S2), a DNA fragment of 5465 bp was PCR amplified from the genomic DNA of Nanjing 11, using primers ASPHF and ASPHR (Table S4).
Copy number of the transgene was determined using Southern blot hybridization. For segregation analysis, the progeny plants were stained for GUS activity (40) and also PCR amplified for the GUS gene fragment, using primers GUS1.6F and GUS1.6R (Table S4). An internal marker in the S5 sequence was obtained by amplifying the genomic DNA with primers dCAPS-F and dCAPS-R (Table S4), resulting in a fragment of 130 bp in length from both Balilla and Nanjing 11. Digestion of the Balilla fragment with HindIII resulted in two fragments of 110 and 20 bp that can be well separated by agarose gel electrophoresis.
Plant Growth and Fertility Examination.
Transgenic plants were grown in the field in the normal rice growing season in Wuhan, China, and in a greenhouse in the winter time. During flowering time, 30–80 florets per plant from the middle and upper parts of the panicles were sampled for examining embryo-sac fertility, using the whole stain-clearing method (16). Anthers of 6–9 mature flowers from each plant were sampled for examining pollen fertility, using the I2-KI staining method. A total of 300–800 pollen grains were observed per plant. The whole plants were harvested for examining spikelet fertility scored as the ratio of the number of filled grains to the total spikelets.
Determining the Full-Length Transcripts.
Leaves and developing young panicles were harvested from field-grown plants and stored in liquid nitrogen. Total RNA was isolated using a RNA extraction kit (TRIzol reagent, Invitrogen). For synthesizing the first-strand cDNA, 1 μg total RNA was reverse transcribed using the protocol provided by the SMART RACE cDNA amplification kit (Clontech) with a final volume to 100 μl.
For 5′-RACE, the first round of amplification was conducted according to the protocol provided with the Advantage 2 PCR kit (Clontech), using primer S5-GSP1 (Table S4) and the UPM adapter primer from the kit. For the second round of amplification, the PCR product from the first round was diluted ×50, and 1 μl of the dilution was used as the template. The amplification was conducted using the nested primer S5-R1 together with the adapter primer NUPM provided in the kit. The 3′-RACE was essentially the same except that primer S5-GSP2 was used with UPM in the first round of reaction, S5-RACE 4 was used with NUPM in the second round of reaction, and the dilution was ×30.
To recover the internal sequence of the transcripts, RNA isolated from leaves at the young panicle development stage was reverse transcribed in a 20-μl reaction by using DNase I and SuperScript II (Invitrogen) according to the manufacturer's instruction. Primers were designed according to the sequences within the boundaries defined by the cDNA ends (Table S4 and Fig. S8) for PCR amplification of the reverse-transcription products. The PCR was in a 20-μl volume containing 1 μl template.
RNA in Situ Hybridization.
Tissue preparation, in situ hybridization, and detection were as described previously (41). For preparing the probe, the predicted ORF5 was amplified from Nipponbare by PCR using a pair of primers with added BglII and XhoI restriction sites and ligated to pGEM-T (Promega). This clone (named cDORF5) was digested with NcoI and self-ligated to recover 532 bp unique sequence of S5. The sense and antisense RNA probes were produced by T7 and SP6 transcriptase labeled with digoxigenin (Roche).
Preparation of the Antibodies.
The antibodies were made by Eurogentec (Belgium). Two peptides, PIFDPGRSYTSRRVRC and IGYHRTSRARQESYIC near the N and C termini of the S5 protein (Fig. S3), were synthesized for immuno-injection, and the corresponding antibodies were named anti-NS5 and anti-CS5, respectively. The specificity of the antibodies was tested by a competition experiment, in which the antibody was mixed with an excessive amount of the antigen polypeptide. The mixture was used as the probe to hybridize with membrane blotted with E. coli-expressed S5 proteins, compared with ones using the antibody as the probe. The results showed that both antibodies are highly specific.
Expression and Purification of S5 in E. coli.
The insert in cDORF5 described above was subcloned into pMAL-c2x expression vector (Biolabs) to generate pMAL-S5, in which S5 is fused at the C terminus of the maltose-binding protein. pMAL-S5 was introduced into E. coli strain BL21(DE3), and expression of S5 was induced by the addition of isopropyl 1-thio-β-D-galactopyranoside (IPTG; final concentration, 0.5 mM) when the A600 of the cells grown at 37°C had reached 0.6. After 3 h, the cells were harvested and resuspended in column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, pH 7.0) and stored overnight at −20°C. The fused protein was purified according to the pMAL Protein Fusion and Purification Instruction Manual (Biolabs).
Assay of Protease Activity.
Proteolytic activity of the S5 protein was assayed using a fluorescein isothiocyanate (FITC)-casein derivative from the Protease Fluorescent Detection kit (Sigma) according to the manufacturer's instructions. Briefly, 20 μl purified pMAL-S5 (10 μg) was incubated with an equal volume of each pH buffer (0.1 M Gly-HCl, pH 2.5; 0.1 M sodium citrate, pH 3.0, pH 3.5, pH 4.0; 0.1 M sodium acetate, pH 5.0; and 0.1 M sodium phosphate, pH 6.0) and 20 μl of FITC-casein solution. For pepstatin A inhibition assay, pepstatin A was added to a final concentration 0.15 mM, and an equal volume 0.1 M sodium citrate (pH 3.0) was added. The reactions were at 37°C for 15 h and stopped by adding 100 μl of 0.6 N tricarboxylic acid. After centrifugation at 12,000 × g for 10 min, 100 μl of supernatant of each sample were transferred to a second tube, to which 2 ml of 0.5 M Tris-HCl (pH 8.5) were added. Enzyme activity was obtained by measuring fluorescence using a fluorescence spectrophotometer (Hitachi 850). A control assay was conducted simultaneously to provide the background reading, using water in place of the S5 protein.
Autolysis of pMAL-S5 Fusion Protein.
For autolysis assay, 20 μl purified pMAL-S5 (10 μg) were incubated with 0.5 volume of each pH buffer described above to allow autolysis overnight at 37°C, with the one adding column buffer as the control. The autolytic products were displayed in a 12% SDS-PAGE gel, using the minigel system (Bio-Rad) according to the manufacturer's instructions.
For immunoblot, the protein samples were blotted onto a polyvinylidene difluoride membrane (Millipore) and probed with the antibodies according to standard protocols (42).
Immunolocalization of S5 by TEM.
Ovaries at the stage of megasporocytes were collected and fixed in a mixture of 4% paraformaldehyde and 0.5% glutaraldehyde in 0.01 M phosphate buffer (pH 7.2). Immunolocalization of S5 was performed essentially as previously described (43). Samples were dehydrated, infiltrated, and finally embedded in Lowicryl K4M (Electron Microscopy Sciences, Catalog 14330). Ultrathin sections cut on a Sorvall MT-6000 ultratome and mounted on formvar-coated nickel grids were first incubated in PBST [0.1 M phosphate buffer (pH 7.2) containing 0.1% Tween-20] containing 0.2 M glycine and 1% BSA for 20 min in order to block unspecific binding, and then in 1:20 diluted anti-NS5 for 5 h at 37°C. After rinse, sections were incubated in 1:10 diluted goat anti-rabbit IgG [labeled with colloidal gold (diameter 10 nm); Sigma, Catalog G7402] for 2 h at 37°C, washed and stained with saturated uranyl acetate, and examined and photographed on a JEM-100CX/II transmission electron microscope. Control experiments were performed simultaneously with omission of anti-NS5.
Supplementary Material
Acknowledgments.
DNA sequences of the three alleles of S5 and their predicted amino acids have been deposited in GenBank under accession no. EU889293(S5-n), EU889294(S5-j), and EU889295(S5-i). This work was supported by the Ministry of Science and Technology and the National Natural Science Foundation of China.
Footnotes
Conflict of interest statement: A patent based on this work was filed.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU889293(S5-n), EU889294(S5-j), and EU889295(S5-i)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0804761105/DCSupplemental.
References
- 1.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia Univ Press; 1937. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 3.Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 4.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 5.Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- 6.Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 7.Masly JP, Jones CD, Noor MA, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 8.Escobar-Restrepo J-M, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 9.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato S, Kosaka H, Hara S. On the affinity of rice varieties as shown by fertility of hybrid plants. Bull Sci Fac Agric Kyushu Univ. 1928;3:132–147. [Google Scholar]
- 11.Ikehashi H, Araki H. Variety screening of compatibility types revealed in F1 fertility of distant cross in rice. Jpn J Breed. 1984;34:304–313. [Google Scholar]
- 12.Liu KD, Zhou ZQ, Xu CG, Zhang Q, Saghai Maroof MA. An analysis of hybrid sterility in rice using a diallel cross of 21 parents involving indica, japonica and wide compatibility varieties. Euphytica. 1996;90:275–280. [Google Scholar]
- 13.Ikehashi H, Araki H IRRI, editor. Rice Genetics. Manila: IRRI; 1986. Genetics of F1 sterility in remote crosses of rice; pp. 119–130. [Google Scholar]
- 14.Liu YS, Zhu LH, Sun JS, Chen Y. Mapping QTLs for defective female gametophyte development in an inter-subspecific cross in Oryza sativa L. Theor Appl Genet. 2001;102:1243–1251. [Google Scholar]
- 15.Li D, et al. Fine mapping of S32(t), a new gene causing hybrid embryo sac sterility in a Chinese landrace rice (Oryza sativa L.) Theor Appl Genet. 2007;114:515–524. doi: 10.1007/s00122-006-0450-8. [DOI] [PubMed] [Google Scholar]
- 16.Song X, Qiu SQ, Xu CG, Li XH, Zhang Q. Genetic dissection of embryo sac fertility, pollen fertility, and their contributions to spikelet fertility of intersubspecific hybrids in rice. Theor Appl Genet. 2005;110:205–211. doi: 10.1007/s00122-004-1798-2. [DOI] [PubMed] [Google Scholar]
- 17.Li WT, Zeng RZ, Zhang ZM, Zhang GQ. Mapping of S-b locus for F1 pollen sterility in cultivated rice using PCR based markers. Acta Bot Sin. 2002;44:463–467. [Google Scholar]
- 18.Zhuang CX, Fu Y, Zhang GQ, Mei MT, Lu YG. Molecular mapping of S-c, an F1 pollen sterility gene in cultivated rice. Euphytica. 2002;127:133–138. [Google Scholar]
- 19.Wang GW, He YQ, Xu CG, Zhang Q. Identification and confirmation of three neutral alleles conferring wide-compatibility in inter-subspecific hybrids of rice (Oryza sativa L.) using near isogenic lines. Theor Appl Genet. 2005;111:702–710. doi: 10.1007/s00122-005-2055-z. [DOI] [PubMed] [Google Scholar]
- 20.Yanagihara S, et al. Molecular analysis of the inheritance of the S-5 locus, conferring wide compatibility in indica/japonica hybrids of rice (O. sativa L) Theor Appl Genet. 1995;90:182–188. doi: 10.1007/BF00222200. [DOI] [PubMed] [Google Scholar]
- 21.Liu KD, et al. A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor Appl Genet. 1997;95:809–814. [Google Scholar]
- 22.Qiu SQ, et al. Delimitation of the rice wide compatibility gene S5n to a 40-kb DNA fragment. Theor Appl Genet. 2005;111:1080–1086. doi: 10.1007/s00122-005-0033-0. [DOI] [PubMed] [Google Scholar]
- 23.Ji Q, Lu J, Chao Q, Gu M, Xu M. Delimiting a rice wide-compatibility gene S5n to a 50 kb region. Theor Appl Genet. 2005;111:1495–1503. doi: 10.1007/s00122-005-0078-0. [DOI] [PubMed] [Google Scholar]
- 24.Davies DR. The structure and function of aspartic proteinases. Annu Rev Biophys Biophys Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings ND, Barrett AJ. Families of aspartic peptidases, and those of unknown catalytic mechanism. Methods Enzymol. 1995;248:105–120. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- 26.Yan R, et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 27.Kervinen J, Wlodawer A, Zdanov A. 17. Phytepsin. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 2nd Ed. Amsterdam: Elsevier/Academic; 2004. pp. 77–84. [Google Scholar]
- 28.Bi X, Khush GS, Bennett J. The rice nucellin gene ortholog OsAsp1 encodes an active aspartic protease without a plant-specific insert and is strongly expressed in early embryo. Plant Cell Physiol. 2005;46:87–98. doi: 10.1093/pcp/pci002. [DOI] [PubMed] [Google Scholar]
- 29.Xia Y, et al. An extracellular aspartic protease functions in Arabidopsis disease resistance signalling. EMBO J. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge X, et al. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep. 2005;6:282–288. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 32.Glathe S, et al. Transport and activation of the vacuolar aspartic proteinase phytepsin in barley (Hordeum vulgare L.) J Biol Chem. 1998;273:31230–31236. doi: 10.1074/jbc.273.47.31230. [DOI] [PubMed] [Google Scholar]
- 33.Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, Saghai Maroof MA, Lu TY, Shen BZ. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor Appl Genet. 1992;83:495–499. doi: 10.1007/BF00226539. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q, et al. Molecular marker diversity and hybrid sterility in indica-japonica rice crosses. Theor Appl Genet. 1997;95:112–118. [Google Scholar]
- 36.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Fujinaga M, Chernaia MM, Tarasova NI, Mosimann SC, James MN. Crystal structure of human pepsin and its complex with pepstatin. Protein Sci. 1995;4:960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YJ, Zhang Q. Optimizing the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- 39.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 40.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 42.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. [Google Scholar]
- 43.Qin Y, Zhao J. Localization of arabinogalactan proteins in egg cells, zygotes, and two-celled proembryos and effects of beta-D-glucosyl Yariv reagent on egg cell fertilization and zygote division in Nicotiana tabacum L. J Exp Bot. 2006;57:2061–2074. doi: 10.1093/jxb/erj159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.