Abstract
Agrobacterium tumefaciens infects its plant hosts by a mechanism of horizontal gene transfer. This capability has led to its widespread use in artificial genetic transformation. In addition to DNA, the bacterium delivers an abundant ssDNA binding protein, VirE2, whose roles in the host include protection from cytoplasmic nucleases and adaptation for nuclear import. In Agrobacterium, VirE2 is bound to its acidic chaperone VirE1. When expressed in vitro in the absence of VirE1, VirE2 is prone to oligomerization and forms disordered filamentous aggregates. These filaments adopt an ordered solenoidal form in the presence of ssDNA, which was characterized previously by electron microscopy and three-dimensional image processing. VirE2 coexpressed in vitro with VirE1 forms a soluble heterodimer. VirE1 thus prevents VirE2 oligomerization and competes with its binding to ssDNA. We present here a crystal structure of VirE2 in complex with VirE1, showing that VirE2 is composed of two independent domains presenting a novel fold, joined by a flexible linker. Electrostatic interactions with VirE1 cement the two domains of VirE2 into a locked form. Comparison with the electron microscopy structure indicates that the VirE2 domains adopt different relative orientations. We suggest that the flexible linker between the domains enables VirE2 to accommodate its different binding partners.
Keywords: DNA binding protein, genetic transformation, type IV secretion, x-ray crystallography, protein conformation
The soil bacterium Agrobacterium tumefaciens is a natural pathogen with the unique ability to genetically transform plants. Thus, the “crown gall” disease results from expression in the plant of genes that originated in the bacterium (1–4). The gene transfer mechanism of Agrobacterium has been widely adopted for genetic engineering of plants (5). Under laboratory conditions, Agrobacterium was shown to transform fungal species and mammalian tissue culture cells (6–8), suggesting potential applications in genetic therapy.
The Agrobacterium–plant interaction is a complex process involving both bacterial and host proteins. The DNA transport itself bears a strong resemblance to bacterial conjugation (9). Most of the virulence apparatus is contained on a tumor-inducing plasmid (Ti-plasmid), which encompasses distinct regions containing virulence genes (vir) and the transferred DNA (T-DNA). T-DNA is excised between specific border repeats as a single-stranded oligonucleotide, and a single VirD2 protein is left bound covalently to its 5′ end. This T strand is then secreted through a type IV secretion system channel (10).
The second major secretion substrate is the essential virulence protein VirE2. VirE2 binds ssDNA without sequence specificity (11–13) and interacts with the T strand in the plant cytoplasm (14), protecting it from cytoplasmic nuclease activity. Together with VirD2 (15), it mediates T-DNA import to the host cell nucleus via an interaction with the host protein VirE2-interacting protein 1 (VIP1) (16, 17). VirE2 has been studied widely, but often independently, for the nopaline and octopine strains of A. tumefaciens (18). In addition to multiple roles in the host cytoplasm, it has been suggested that VirE2 forms a membrane channel that assists in the transfer of the T strand through the plant plasma membrane (19, 20).
In Agrobacterium, VirE2 is coexpressed with its specific chaperone VirE1 (21–23). VirE1 is also essential for virulence, although it is not thought to be secreted to the plant. When expressed in vitro in the absence of VirE1, VirE2 is extremely prone to oligomerization and forms disordered filamentous aggregates (24). In the presence of ssDNA, VirE2 adopts a solenoidal form (e.g., a “telephone cord”) (25) whose three-dimensional structure has been characterized by using electron microscopy (EM) and a real-space helical image-processing approach (26). When VirE2 is coexpressed in vitro with VirE1, however, a soluble heterodimer is formed (20, 24). VirE1 thus prevents VirE2 oligomerization and competes with its binding to ssDNA.
To address the mechanisms of VirE2 binding to its protein and DNA partners, we determined the crystal structure of the VirE1–VirE2 complex. A unique fold was revealed independently in two structurally similar domains of VirE2 that are connected by a single interdomain linker. VirE1 is composed of a single helix situated between the two VirE2 domains, and it forms multiple interactions with both of them. A comparison of the crystal structure of VirE2 in complex with VirE1 to the EM reconstruction of VirE2 in the presence of ssDNA suggests rearrangement of the VirE2 domains. We propose that VirE2 is a dynamic structure that can accommodate its different partners because of the flexibility of its interdomain linker. These structural features reflect the physiological requirements of VirE1 as an inhibitory chaperone of VirE2 in the bacterium, and of VirE2 as a chaperone of the T strand in the plant.
Results
VirE1–VirE2 Complex: Identification of a Novel Fold in VirE2.
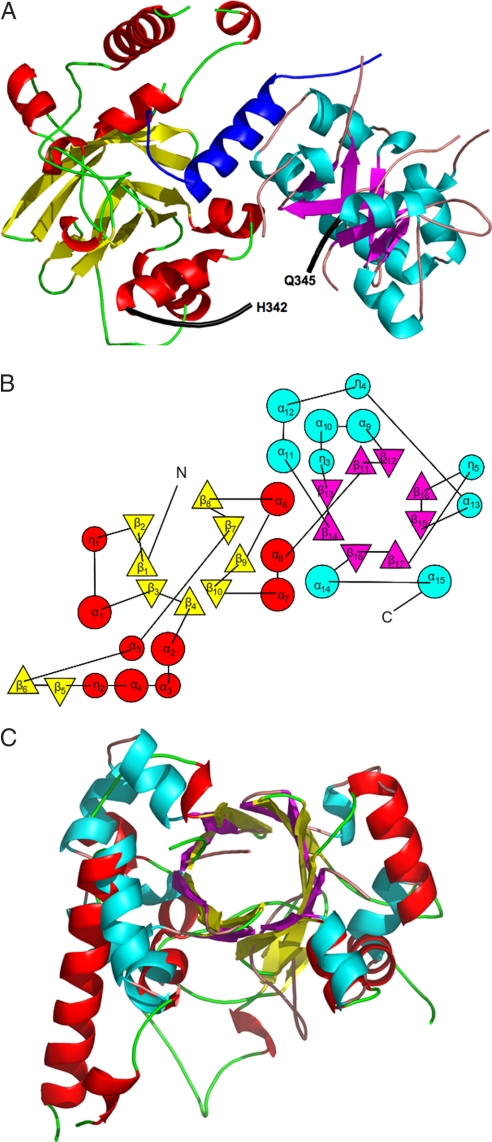
Full-length nopaline VirE2 (P08062) (residues 1–556) was coexpressed with its small chaperone VirE1 (P08063) (residues 1–63), producing crystals that contained a single heterodimeric VirE1–VirE2 complex at a 1:1 ratio in the asymmetric unit cell. This stoichiometry is consistent with prior observations by mass spectrometry (24). VirE1 exhibits electron density for residues 30–57, with a single α-helix formed by residues 38–53. The VirE2 structure has an α/β-fold and consists of two independent domains, the N-terminal domain composed of residues 112–342 and the C-terminal domain containing amino acids 345–517 (Fig. 1A). Secondary structure elements of VirE2 together with its sequence are illustrated in supporting information (SI) Fig. S1 [an alignment with the octopine VirE2 sequence (P0A3W9) is presented as well]. Both domains of VirE2 form a TIM-barrel, composed of α-helices and β-strands resulting in a donut shape with interior β-strands and exterior α-helices. The barrel is characterized by an 8-stranded β-sheet that is closed with shear number S = 8, exhibiting a unique topology (Fig. 1B). Instead of eight repeats of the αβ-motif as in a classical TIM-barrel, four ββαα-motifs are present in the C-terminal domain and variations of this motif in the N-terminal domain. The topology of the VirE2 barrel, in fact, resembles that of the dimeric enhancer of rudimentary gene (ERH domain PDB ID code 1W9G). Taken together, the combination of architecture and topology of the VirE2 domain allows its classification as a novel fold, denoted here as the VirE2 fold (A. G. Murzin, personal communication). The two VirE2 domains share the same fold with rms deviation of 3.1 Å (Fig. 1C), which is remarkable considering the low sequence homology between them (14.3%). The most significant differences in topology are the partial opening between β2 and β8 in the N-terminal barrel, and the extra secondary elements (η2, β5, and β6) compared with the C-terminal domain.
Fig. 1.
The VirE2 fold. (A) Ribbon representation of VirE1–VirE2 complex [created with PyMol (51)]. The helix of VirE1 is shown in blue. The N-terminal domain of VirE2 is depicted in red for α-helices and yellow for β-strands, and the C-terminal domain is shown in cyan for α-helices and magenta for β strands. Both folded domains construct TIM barrels with a unique topology defining the VirE2 fold. The interdomain linker (residues 337–346) is shown as a thick black line for which no electron density was observed for residues N343 and K344. (B) A topology representation [obtained by using TOPS (52)] of the VirE2 fold; β-strands are represented by triangles and α-helices by circles, with colors as in A. (C) Superposition of the N- and C-terminal domains of VirE2 showing that they have the same fold despite their low sequence homology.
The two VirE2 domains are linked by residues 337–346. No electron density was observed for residues 343 and 344, a finding that could be attributed to the flexible nature of the unstructured loop (Fig. 1A). The only direct interdomain contacts involve interactions between residues Y319 and Y373 and between S322 and N377. Consequently, the two VirE2 domains are held together primarily by tight bilateral interactions with the single helix of VirE1. The interdomain linker maintains continuity of the protein but cannot constrain the relative orientations of the two domains. In this sense, the two domains form an interlocked pseudoheterodimer that shares their active site, the VirE1 binding site.
Interaction Between VirE1 and VirE2.
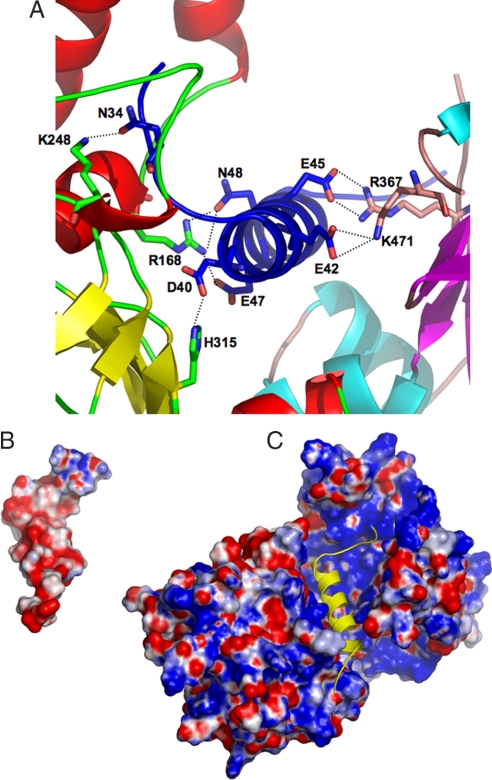
In the heterodimer structure there are 44 contacts (up to 3.5Å), between VirE1 and VirE2, predominantly of electrostatic nature. These are contributed by 16 residues from VirE1 (of 27) and 18 residues from the two domains of VirE2. Of these residues, 11 are in the N-terminal domain of VirE2 and 7 are in its C-terminal domain (see Table S1). VirE2 contains clusters of aromatic residues with side chains pointing toward the VirE1 binding site, which are involved in VirE1 binding. These include Y319 and W323 from the N-terminal and W383 from the C-terminal domain. Most of the VirE1–VirE2 interactions are electrostatic, involving salt bridges between residues R168, K248, H315, R367, and K471 from VirE2 and N34, D40, E42, E45, E47, and N48 from VirE1 (Fig. 2A). The latter acidic residues cause the VirE1 to display a strong electronegative surface (Fig. 2B). The VirE2 surface displays both electronegative and electropositive patches with its VirE1 binding pocket being predominantly electropositive (Fig. 2C). The electrostatic complementarity cements the two domains of VirE2 around the VirE1 helix in a “locked” conformation, covering most of its surface area.
Fig. 2.
Electrostatic interactions between VirE1 and VirE2. (A) Residues from both domains of VirE2 form electrostatic interactions (dashed lines) with acidic residues from VirE1 (colors as in Fig. 1A). (B) A representation of the electrostatic surface potential of VirE1 showing an overall negative charge (red). The orientation of VirE1 is as in C [Figure created with PyMol (51)]. (C) A representation of the electrostatic surface potential of VirE2 is shown, calculated in the absence of VirE1. Although the VirE2 surface potential has both positive and negative regions, the VirE1 binding pocket is predominantly electropositive (blue). The single helix in VirE1 is depicted as a yellow diagram in the same orientation as in B.
Two large portions of the VirE2 molecule are not observed in the crystal structure. The experimental maps contained no electron density at the C terminus (last 39 residues), which suggests a flexible and possibly unstructured form. Notably, this region largely coincides with the C-terminal, type IV secretion signal of VirE2 (27, 28). A larger segment at the N terminus (111 residues) also lacks electron density. No electron density was observed for these regions in a second crystal form that was obtained under different conditions and crystallized in a different space group (P21) (data not shown). Repeated attempts to analyze the protein content within the crystals by mass spectrometry were unsuccessful because of their small and fragile nature. These findings suggest that the terminal regions of VirE2 are flexible or intrinsically unstructured, although partial proteolysis may also have occurred during crystallization. Note, however, that the crystallographic refinement parameters including the Rfree and Rwork values (25.3% and 21.0%, respectively) indicate that most of the density is accounted for in the structure. Thus, we conclude that core residues of VirE2 are responsible for its binding to VirE1.
Structural Rearrangement of VirE2 Domains in the Solenoidal Filament.
The structure of VirE2 in complex with ssDNA has been characterized by EM and three-dimensional image reconstruction, revealing a solenoid form with interacting VirE2 units arranged in a spiral around a hollow core (26). There were 4.25 protein units per helical turn, and a rise of 5.1 nm per turn. Although ssDNA could not be observed in the low-resolution EM structure, the geometrical contraction of a circular DNA substrate suggested that the oligonucleotide wraps along the inner circumference of the protein shell. This is further supported by a recent study of the VirE2–ssDNA interaction in which optical tweezers were used to follow the complex formation mechanically (29).
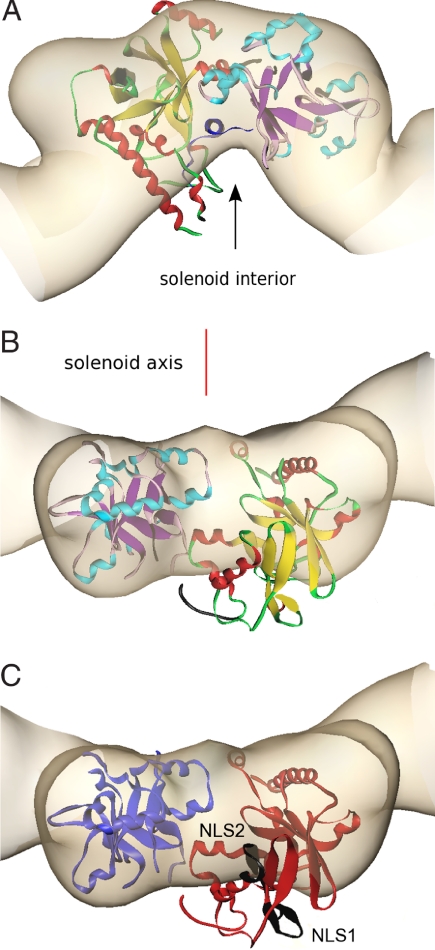
To gain insight into the manner by which VirE2 accommodates each of its partners, we tried to align the crystal structure of VirE1–VirE2 manually into the molecular envelope of VirE2 as determined by EM. We constrained the face that binds VirE1, and presumably ssDNA, to align with the near-vertical groove facing the interior of the solenoid (Fig. 3A). This orientation places the two nuclear localization signals (NLS) in VirE2, 228KLR … KYGRR244 and 296KTK … KLKSK310 (30), facing the exterior of the solenoid assembly (Fig. 3C). The alternative orientation with the interdomain cleft matching the deep grove at the exterior of the complex is unfavored because it would place the nuclear localization signals on the interior, hidden from protein interaction in the plant, and the basic VirE1 binding residues facing outward. It was immediately clear that fitting the heterodimer of VirE1–VirE2 as a single rigid body into the density map obtained by EM was not possible. Either domain could be fit neatly, causing the other to protrude from the EM envelope (Fig. 3B). We can envision a more reasonable fit by permitting rearrangement between the two VirE2 domains. This is structurally feasible because of the presence of the extended linker (residues 337–346) that bridges the two domains. Minor conformational changes within secondary elements of the domains are likely to take place as well. Therefore, it appears that the two domains of VirE2 acquire a different relative orientation on binding of VirE1 versus ssDNA. In the presence of VirE1, the two domains of VirE2 are locked in an orientation that is unfavorable for the VirE2–VirE2 homodimeric interactions required for assembly of both the solenoids and the irregular filaments, preventing as well the binding to ssDNA.
Fig. 3.
Alignment of the crystal structure of VirE1–VirE2 complex with the VirE2 envelope obtained by EM in the presence of ssDNA, represented at the same length scales. The crystal structure of VirE2 (colors as in Fig. 2A), treated as a single rigid body, was manually aligned into the envelope of one repeat of VirE2 in the solenoid as determined by EM (beige) and subject to constraints as described in the text. [Figures created with Amira (Mercury Computer Systems)]. (A) The view down the solenoid axis. For alignment purposes, VirE1 (dark blue) was introduced as shown facing the solenoid interior (black arrow), although it is in fact absent from the VirE2 complex with ssDNA. (B) Rotation of view A by 90° perpendicular to the solenoid axis (red line), showing one VirE2 repeat viewed from the exterior of the solenoid. It is clear that, although one domain of VirE2 neatly fits the envelope, the other protrudes from it (the interdomain flexible linker is shown in black). (C) Indication of the two NLS sequences (black) facing the exterior of the solenoid. The orientation is as in B, and the color scheme is simplified for clarity (N-terminal domain, red; C-terminal domain, blue).
Discussion
We have presented here the structure of nopaline VirE2 in complex with VirE1. VirE2 in this complex exhibits two structurally similar domains, each of which presents a combination of TIM barrel architecture with a ββαα topology. This combination defines the VirE2 fold. The structural similarity between the domains is remarkable considering the very low sequence homology between them. Also notable is that the two domains are connected by a single, extended linker that appears to be flexible, with very few direct contacts between the VirE2 domains. In the C-terminal domain, there are four repeats of a (consensus) ββαα motif, related by the pseudo tetrahedral symmetry. The VirE2 fold could have evolved by a series of duplication events of this motif. The motif assembly into the barrel is likely facilitated by the subunit β-hairpins (A.G. Murzin, personal communication). The structural similarity of the two domains is suggestive of an evolutionary selection for the VirE2 fold as an efficient scaffold for sequestering VirE1, ssDNA, and its other partners. Octopine VirE2 shares a high sequence homology of 68% with the nopaline protein (Fig. S1). This strongly suggests that the two proteins adopt very similar folds. The VirE2 fold is distinct from the previously described OB fold (31) used by a variety of proteins that are involved in protein–nucleic acid, protein–polysaccharide, and protein–protein interactions. The VirE2 fold also differs from those observed in other ssDNA binding proteins such as RecA (32) and Adenovirus DBP (33), which do not adopt an OB fold.
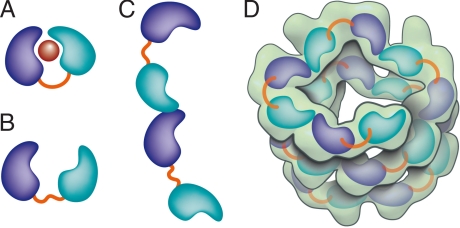
In the heterodimer, the two folded domains of VirE2 clamp tightly around the single α-helix of VirE1 by using only core residues of VirE2. Prior analysis by analytical ultracentrifugation and mass spectrometry showed that the soluble VirE1–VirE2 complex is monomeric and has a 1:1 stoichiometry (24). Therefore, the unobserved segments at the N and C termini of VirE2 are not likely to participate in VirE1 binding. An earlier deletion analysis (34) identified a VirE1 binding region between residues 189 and 391 of VirE2 (octopine). These residues span both VirE2 domains in our structure (corresponding nopaline residues 201–413) and include most of the VirE2-interacting residues (Table S1). Both electrostatic and hydrophobic interactions with VirE1 cement the two domains of VirE2 into a locked conformation (depicted schematically in Fig. 4A) where the flexible linker joining these two independent VirE2 domains does not constrain their relative orientation (Fig. 4B). This conformation bears an intriguing relation to the structure of the flagellin protein FliC in complex with its chaperone FliS, although its core domains are simple α-helices (35). Another intriguing similarity is to the type III secretion system translocon protein EspA, which forms extracellular filaments but whose polymerization in the bacterioplasm is inhibited by association with its chaperone CesA (36). Although the sequences and folds bear no relation to VirE2, oligomerization of the latter seems to be regulated in a similar manner by VirE1.
Fig. 4.
Schematic representation of VirE2 showing how its interdomain flexible linker permits structural rearrangements in complex with its different partners. The two domains of VirE2 are shown in purple and cyan linked by their interdomain flexible linker shown in orange. (A) In the presence of VirE1 (red), the two VirE2 domains are locked by their interaction with VirE1. (B) In the absence of VirE1, the domains of VirE2 are unlocked and free to rotate around the flexible interdomain linker. (C) In the unlocked form, VirE2 has a strong tendency to self-assemble forming N- to C-terminal interactions. Because of the flexible linker, the two domains of VirE2 can adopt a range of orientations resulting in irregular filaments. (D) On addition of ssDNA to the filaments, an ordered solenoid assembly is formed (gray envelope: a tracing of the EM model with 4.25 VirE2 units per turn (26). The ssDNA should wrap along the inner wall of the protein structure, limiting the degree of freedom in the linker and thereby imposing a favorable VirE2–VirE2 arrangement.
VirE2 can bind alternatively to VirE1 or to ssDNA, and ssDNA can displace VirE1 from VirE2 (20, 24). The acidic nature common to both substrates suggests that they bind via electrostatic interactions to a common region of VirE2. Results of insertion mutations and yeast two-hybrid analysis indicate an overlap of VirE1 and ssDNA binding domains (37, 38). Our structure reveals that the overall positive charge on VirE2 is not completely neutralized by VirE1 because of the presence of additional basic residues that do not participate in VirE1 binding. This may explain a residual affinity of VirE2 in the heterodimer for ssDNA (20, 24). The two VirE2 domains also contain clusters of aromatic residues with side chains pointing toward the VirE1 binding site forming hydrophobic pockets. These include Y160, Y194, Y373, Y319, W323, and W383, with the last three being involved in VirE1 interactions (Table S1). These planar side chains may form additional stacking interactions with the nucleotide bases of DNA. Our data do not rule out additional interactions between ssDNA and parts of VirE2 that do not interact with VirE1. It was reported for octopine VirE2 that the C-terminal 37 residues are necessary, but not sufficient, for ssDNA binding (12), and that amino acid insertions at many sites throughout the primary sequence strongly reduced ssDNA binding (40).
VirE2, in the absence of VirE1, has a strong tendency to oligomerize into poorly soluble, irregular filaments. In the presence of ssDNA, these VirE2 filaments take an ordered solenoidal form dominated by head-to-tail VirE2–VirE2 interactions (26). Both forms are consistent with repeating contacts in the VirE2 self-assemblies involving both N and C termini (24). We suggest that the ordered and disordered forms differ primarily in the relative orientation between the two domains of VirE2, based on the rotational freedom permitted by the extended interdomain linker. Specifically, in the absence of VirE1, the domains would be “unlocked” and free to rotate (Fig. 4B). N- to C-terminal interactions then naturally lead to irregular filaments (Fig. 4C). ssDNA, however, may limit the degree of freedom in the linker and thereby impose a favorable VirE2 arrangement for ordered solenoid assembly (Fig. 4D). The EM structure affords a view of VirE2 in the presence of ssDNA. In this protein–DNA complex that lacks VirE1, we expect to see a different constrained configuration of VirE2. The low resolution of the EM reconstruction envelope, as well as the missing N- and C-terminal segments of VirE2 in the crystal structure, leave too much freedom to perform a quantitative docking. However, with the constraints of DNA binding on the interior and NLS on the exterior of the solenoid, only relatively minor adjustments are possible. It is then clear that the locked form is too globular to fit the protein units in the EM map, where VirE2 adopts a more elongated, extended conformation. Thus, it appears that ssDNA binding to VirE2 is associated with rearrangements between the domains and possibly minor conformational changes.
Structural rearrangements on DNA binding are a common theme among ssDNA binding proteins. RecA is a well known example, where the protein subunits in complex with ssDNA were rotated with respect to those in a crystal structure obtained in the absence of ssDNA (32). In the structure of human replication protein A (RPA), large conformational changes within individual OB folds are induced in the presence of ssDNA as well (39). RPA demonstrates a number of other similarities with VirE2. Specifically, RPA possesses two independent functional OB motifs, with low sequence identity between them, connected by a flexible linker of 10 aa. The crystal structure of RPA was determined with and without ssDNA. There too, a major rearrangement involving a flexible linker was observed on binding of ssDNA, along with conformational changes within the OB domains (39). Although this holds true for many OB proteins, others appear to have preformed surfaces for ssDNA binding as in the case of the major cold shock proteins from Bacillus (40). Although the protein folds may be quite variable, the chemical basis of ssDNA recognition remains a conserved process by using non-sequence-specific electrostatic interactions, hydrogen bonds, and stacking interactions of the nucleotide bases with aromatic residues (41, 42).
Based on the structure of VirE1–VirE2 and prior biochemical evidence, we can suggest a model for the physiological functions of these proteins. In the bacterium, VirE1 maintains VirE2 in a globular heterodimeric form that is resistant to disruption by ssDNA. Because VirE1 binds to core residues, the termini of VirE2 remain available for other protein interactions. Most notably the C-terminal secretion signal can interact with the VirD4-coupling protein for secretion. In an oligomerized form, with or without ssDNA, the C terminus would likely be blocked and consequently unavailable for the interaction with other partners such as VirD4. Consistently, virE1 null bacteria are avirulent (22). The process of delivery to the plant cytoplasm is thought to involve removal of the VirE1. One possibility is that VirE2 is delivered simultaneously with the T-DNA. This would permit formation of the T-complex (VirE2 bound to the T-strand) at or near the channel, whose inherently crowded environment might promote the exchange of VirE2 binding partners, that is, from VirE1 to ssDNA, as observed in vitro (24). A second possibility is that the VirE1 is stripped from VirE2 in the secretion channel independently of DNA, perhaps by mechanical action in bending the domains and rupturing the stabilizing interactions. This is supported by complementation assays where VirE2 and T-DNA arrive from different sources (30, 43). In either case, our observations in vitro suggest that VirE2 lacking VirE1 would form filamentous aggregates in the plant, which would be ready to bind the incoming T-strand. This T-complex could then safely move to the nucleus via its interaction with the host cell nuclear import machinery. In this multistep process, VirE2 interacts with diverse partners. The crystal structure of VirE2 with VirE1 points to a flexible interdomain linker permitting the necessary versatility of molecular interaction.
Materials and Methods
Coexpression and Purification of Selenomethionine Labeled VirE1–VirE2.
VirE1 (P08063) (1–63) and VirE2 (P08062) (1–556) (Nopaline) genes were cloned into pACYCDuet-1 (Novagen). VirE1 was fused at its N terminus to a 6-His tag separated by a TEV cleavage site (ENLYFQG), whereas VirE2 was expressed in its native form. BL21(DE3) bacteria coexpressing VirE1 and VirE2 were grown at 37°C in M9 minimal medium containing glucose (0.4 wt/vol) and chloramphenicol (34 mg/ml). When cultures reached A600 = 0.6, the cultures were supplemented as follows (reagents were added as solids): selenomethionine (50 mg/liter culture), along with lysine hydrochloride (100 mg), threonine (100 mg), phenylalanine (100 mg), leucine (50 mg), isoleucine (50 mg), and valine (50 mg). Protein expression was induced with 50 μM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at 15°C for 24 h. Purification of the VirE1–VirE2 complex was performed as described in ref. 24. The purified protein in final buffer containing 20 mM Tris (pH = 8), 50 mM NaCl, and 1 mM DTT was concentrated to 10.5 mg/ml for crystallization trials.
Crystallization, Data Collection, and Refinement.
Plate-like fragile crystals of the VirE1–VirE2 complex were obtained at 19C° by the microbatch method (44), under oil by using the Oryx6 robot (Douglas Instruments). Selenomethionine VirE1 and VirE2 crystals were grown from a precipitating solution of 100 mM Hepes (pH = 7), 20% PEG 6000, 0.2M NH4Cl, and 1% Triton X-405. Crystals formed in a space group P212121, with cell constants a = 51.02 Å, b = 96.26 Å, c = 112.48 Å, and contained a single monomer in each asymmetric unit cell with Vm of 2.16 Å3/Da. Single-wavelength anomalous diffraction (SAD) data from a single crystal were collected at the European Synchrotron Radiation Facility (ESRF) beam line, ID14-4. Data to 2.3-Å resolution were collected at the peak wavelength. The diffraction images were indexed and integrated by using the program HKL2000 (45). The integrated reflections were scaled by using the program SCALEPACK (45). Structure factor amplitudes were calculated by using TRUNCATE from the CCP4 program suite (46). Details of the data collection are described in Table S2. Selenium sites were identified with the program, SHELX (47). Refinement were carried out with the program CCP4/Refmac5 (48). The model was built into 2Fobs − Fcalc, and Fobs − Fcalc maps by using the program COOT (49). In later rounds of refinement, water molecules were built into peaks >3σ in Fobs − Fcalc maps. The final model includes residues 112–179, 183–342, 345–356, 364–439, 445–473, and 476–517 of VirE2 and residues 30–57 of VirE1, 68 water molecules and one molecule of PEG. The Rfree value is 25.3% (for the 5% of reflections not used in the refinement), and the Rwork value is 21.0% for all data to 2.3 Å. The VirE1–VirE2 model was evaluated with the program PROCHECK (50). Details of the refinement statistics of VirE1–VirE2 structure are described in Table S2. The coordinates and structure factors for VirE1–VirE2 have been deposited in the RCSB Protein Data Bank under the ID code 3BTP.
Supplementary Material
Acknowledgments.
We thank Prof. Joel L. Sussman for helpful discussions, Prof. Alexey G. Murzin for confirming the VirE2 fold, and Dr. Gordon Leonard, at the European Synchrotron Radiation Facility (ESRF) beam line, ID14–4, whose outstanding efforts have made these experiments possible. M.E. thanks Prof. Tzvi Tzfira for illuminating discussions about VirE2. The structure was determined in collaboration with the Israel Structural Proteomics Center (ISPC), supported by The Israel Ministry of Science, Culture, and Sport, the Divadol Foundation, the Neuman Foundation and the European Commission Sixth Framework Research and Technological Development Program ‘SPINE2-COMPLEXES’ Project Contract No. 031220. This work was supported in part by a grant from the Human Frontiers Science Program, by the Gerhardt M.J. Schmidt Minerva Center for Supramolecular Architecture, and by the historic generosity of the Harold Perlman Family.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BTP). An animated wiki page appears at http://www.proteopedia.org/wiki/index.php/3btp
This article contains supporting information online at www.pnas.org/cgi/content/full/0801525105/DCSupplemental.
References
- 1.Chilton MD, et al. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- 2.Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: A feast of fundamental insights. Plant J. 2000;23:11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]
- 3.Gelvin SB. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, et al. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr Opin Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 8.Kunik T, et al. Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci USA. 2001;98:1871–1876. doi: 10.1073/pnas.041327598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie PJ. Type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie PJ, Ward JE, Winans SC, Nester EW. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen P, Pazour GJ, Anderson D, Das A. Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J Bacteriol. 1989;171:2573–2580. doi: 10.1128/jb.171.5.2573-2580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Citovsky V, Wong ML, Zambryski P. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: Implications for the T-DNA transfer process. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelvin SB. Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J Bacteriol. 1998;180:4300–4302. doi: 10.1128/jb.180.16.4300-4302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi L, Hohn B, Tinland B. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 16.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 18.Tzfira T, Citovsky V. Comparison between nuclear localization of nopaline- and octopine-specific Agrobacterium VirE2 proteins in plant, yeast and mammalian cell. Mol Plant Pathol. 2001;2:171–176. doi: 10.1046/j.1364-3703.2001.00065.x. [DOI] [PubMed] [Google Scholar]
- 19.Dumas F, Duckely M, Pelczar P, Van Gelder P, Hohn B. An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc Natl Acad Sci USA. 2001;98:485–490. doi: 10.1073/pnas.011477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duckely M, et al. The VirE1VirE2 complex of Agrobacterium tumefaciens interacts with single-stranded DNA and forms channels. Mol Microbiol. 2005;58:1130–1142. doi: 10.1111/j.1365-2958.2005.04894.x. [DOI] [PubMed] [Google Scholar]
- 21.Sundberg C, Meek L, Carroll K, Das A, Ream W. VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1996;178:1207–1212. doi: 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, Sagulenko E, Ding Z, Christie PJ. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng W, et al. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 24.Frenkiel-Krispin D, et al. Plant transformation by Agrobacterium tumefaciens: modulation of single-stranded DNA-VirE2 complex assembly by VirE1. J Biol Chem. 2007;282:3458–3464. doi: 10.1074/jbc.M605270200. [DOI] [PubMed] [Google Scholar]
- 25.Citovsky V, Guralnick B, Simon MN, Wall JS. The molecular structure of agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J Mol Biol. 1997;271:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Arish A, et al. Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J Biol Chem. 2004;279:25359–25363. doi: 10.1074/jbc.M401804200. [DOI] [PubMed] [Google Scholar]
- 27.Vergunst AC, van Lier MC, den Dulk-Ras A, Hooykaas PJ. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 2003;133:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simone M, McCullen CA, Stahl LE, Binns AN. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 29.Grange W, et al. VirE2: A unique ssDNA-compacting molecular machine. PLoS Biol. 2008;6:e44. doi: 10.1371/journal.pbio.0060044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Citovsky V, Zupan J, Warnick D, Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 31.Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanLoock MS, et al. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 33.Tucker PA, et al. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 1994;13:2994–3002. doi: 10.1002/j.1460-2075.1994.tb06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou XR, Christie PJ. Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evdokimov AG, et al. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 36.Yip CK, Finlay BB, Strynadka NC. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 37.Dombek P, Ream W. Functional domains of Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J Bacteriol. 1997;179:1165–1173. doi: 10.1128/jb.179.4.1165-1173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundberg CD, Ream W. The Agrobacterium tumefaciens chaperone-Like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J Mol Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 41.Kur J, Olszewski M, Dlugolecka A, Filipkowski P. Single-stranded DNA-binding proteins (SSBs)—Sources and applications in molecular biology. Acta Biochim Pol. 2005;52:569–574. [PubMed] [Google Scholar]
- 42.Shamoo Y. Encyclopedia of Life Sci. London: Macmillan Publishers, Nature Publishing Group; 2002. pp. 1–7. [Google Scholar]
- 43.Otten L, et al. Restoration of virulence of Vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 44.Chayen NE, Shaw Stewart PD, Blow DM. Microbatch crystallization under oil—A new technique allowing many small-volume crystallization trials. J Cryst Growth. 1992;122:176–180. [Google Scholar]
- 45.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.French GS, Wilson KS. On the treatment of negative intensity observations. Acta Crystallogr A. 1978;34:517–552. [Google Scholar]
- 47.Sheldrick GM. In: Direct Methods for Solving Macromolecular Structures. Forteir S, editor. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 51.DeLano WL. The PyMOL Molecular Graphics System. [Accessed May 28, 2008];2002 Available at: http://www.pymol.org.
- 52.Michalopoulos I, Torrance GM, Gilbert DR, Westhead DR. TOPS: An enhanced database of protein structural topology. Nucleic Acids Res. 2004;32:D251–D254. doi: 10.1093/nar/gkh060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.