Abstract
Prostaglandins (PG) are bioactive lipids produced from arachidonic acid via the action of cyclooxygenases and terminal PG synthases. Microsomal prostaglandin E synthase 1 (MPGES1) constitutes an inducible glutathione-dependent integral membrane protein that catalyzes the oxidoreduction of cyclooxygenase derived PGH2 into PGE2. MPGES1 has been implicated in a number of human diseases or pathological conditions, such as rheumatoid arthritis, fever, and pain, and is therefore regarded as a primary target for development of novel antiinflammatory drugs. To provide a structural basis for insight in the catalytic mechanism, we determined the structure of MPGES1 in complex with glutathione by electron crystallography from 2D crystals induced in the presence of phospholipids. Together with results from site-directed mutagenesis and activity measurements, we can thereby demonstrate the role of specific amino acid residues. Glutathione is found to bind in a U-shaped conformation at the interface between subunits in the protein trimer. It is exposed to a site facing the lipid bilayer, which forms the specific environment for the oxidoreduction of PGH2 to PGE2 after displacement of the cytoplasmic half of the N-terminal transmembrane helix. Hence, insight into the dynamic behavior of MPGES1 and homologous membrane proteins in inflammation and detoxification is provided.
Keywords: electron crystallography, inflammation, MAPEG, membrane protein
Microsomal prostaglandin E synthase 1 (MPGES1) is the key enzyme in pathology related production of PGE2 from cyclooxygenase (Cox) derived PGH2 (1). The protein is a member of the MAPEG protein family, which includes 5-lipoxygenase activating protein (FLAP), leukotriene C4 synthase (LTC4S), microsomal glutathione transferase (MGST)1, MGST2, and MGST3 (2, 3). MPGES1 is the most efficient PGES known and catalyzes the oxidoreduction of prostaglandin endoperoxide H2 into PGE2 with an apparent kcat/Km of 310 mM−1s−1 [supporting information (SI) Fig. S1]. The enzyme equally well catalyses the oxidoreduction of endocannabinoids into prostaglandin glycerol esters (4) and PGG2 into 15-hydroperoxy-PGE2 (5). In addition, the enzyme confers low glutathione transferase and glutathione-dependent peroxidase activities (5). The biological significance of the latter activities remains unclear but is thought to reflect the close evolutionary distance to MGST1.
MPGES1 protein expression levels are in most cases low, and proinflammatory stimuli induce its cellular expression and activity, which is prevented by corticosteroids (1, 6–8). The predominant source of PGH2 seems derived from Cox-2, although Cox-1 may also contribute (9). Studies, mainly from disruption of the MPGES1 gene in mice, indicate key roles for MPGES1-generated PGE2 in pathological conditions such as chronic inflammation, pain, fever, anorexia, atherosclerosis, stroke and tumorigenesis (10). Recently, a role for MPGES1 in regulating neonatal respiration was described in ref. 11. MPGES1 has been shown to be overexpressed in rheumatic diseases such as rheumatoid arthritis and myositis and the protein expression in this context seems resistant to anti-TNFα treatment or oral administration of corticosteroids (12, 13). Together, these findings suggest that MPGES1 is a potential target for development of therapeutic agents for the treatment of several diseases (10).
Results
Structural Determination.
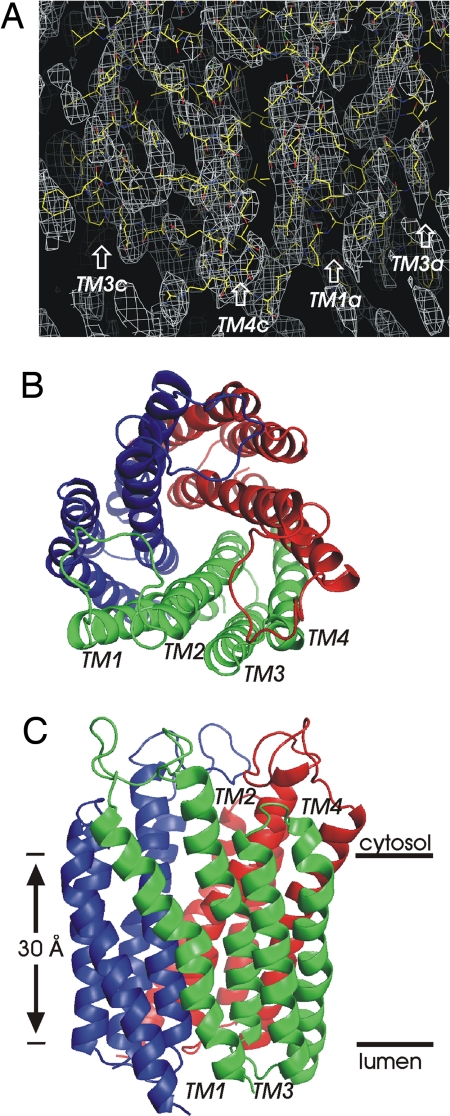
We have determined the structure of human MPGES1 in complex with the tripeptide γ-l-glutamyl-l-cysteinyl-glycine, glutathione (GSH) at 3.5 Å in-plane resolution using electron crystallography (Table S1). The protein, fused to an N-terminal His6-tag, was overexpressed in Escherichia coli and purified in a Triton X-100 solubilized form. 2D crystals were grown by dialysis in the presence of added phospholipids at a low lipid to protein ratio. The crystals often appeared as rounded rectangular sheets extending several μm in the long dimension and ≈1 μm across (Fig. S2A). In the crystalline sheets, the protein molecules were tightly packed and tilted 20° relative to the normal of the layer (Fig. S3). This arrangement explained an earlier observation that no end on views revealing the positions of transmembrane helices were observed from 0° projection maps (Fig. S2A) as had been demonstrated earlier for the MAPEG members MGST1 (14, 15) and LTC4S (16). Polar interactions within one unit cell involving arginine residues with exposed side chains at either the lumenal or the cytoplasmic face of the protein were found to play pertinent roles for crystal contacts (Fig. S3).
Overall Structure.
The subunits of MPGES1 form a homotrimer (Fig. 1) in a similar way as for the other structurally characterized MAPEG members, MGST1 (17), FLAP (18), and LTC4S (19, 20) and in agreement with earlier low resolution and hydrodynamic data on MPGES1 (5). Our result thus supports the suggestion that a trimeric arrangement is common to all MAPEG proteins (21). Because the 2D crystals of MPGES1 are orthorhombic the symmetry component within the trimer is a local pseudo threefold axis. The variations in the immediate surroundings of the subunits in the asymmetric units of the unit cell suggest local structural variations at least in the peripheral parts of the trimers.
Fig. 1.
Overall structure of MPGES1. (A) 2Fo − Fc map contoured at 1.2 σ as seen from the side at the interface between two subunits. (B and C) The trimer subunits have been colored in ribbon representation. (B) Top view from the face corresponding to the cytoplasm when the protein is positioned in its natural membrane environment. (C) Side view with the cytoplasmic side up. The transmembrane helices of one subunit have been labeled at their C-terminal ends.
The His6-tagged N terminus located on the lumenal side of the membrane embedded protein is flexible and the determined model starts at Pro-11. The model begins with a long transmembrane (TM) helix ending at Lys-41. It is followed by a relatively large cytoplasmic loop region. Sequence alignment of MAPEG members (Fig. S4) shows that this domain is highly variable and it is thus unlikely to be crucial for the catalytic functions of the MAPEG enzymes. The second TM helix, similar in length as the first one, spans residues 61–90. A proline residue at position 81 gives rise to a slight kink located in the lumenal half of this helix. This proline is highly conserved throughout the MAPEG family but a shift in position between MPGES1/MGST1 and LTC4S/FLAP (Fig. S4) gives rise to differences in helical arrangements. A short loop on the lumenal side connects TM2 with TM3. Both TM3 (Pro-96-Gly-119) and TM4 (Pro-124-His-151), connected by a four-residue loop, are less exposed on the cytoplasmic side of the membrane than the N-terminal helices. In the trimer the N terminus of one subunit is in close proximity to the C terminus of one of its neighbors. The three TM2s of the trimer form an inner core structure, which narrows down into complete closure toward the lumenal side by aromatic side chains. The funnel shaped opening toward the opposite cytoplasmic side is partly covered by the three large connections between TM1s and TM2s. These domains protrude toward the loops between TM3 and TM4 of neighboring subunits.
Essentially no charged side chains are found exposed toward the phospholipid bilayer for MPGES1 (Fig. S5). The side chain of Lys-26 in the center of TM1 points toward the interior of the structure and makes an ion pair with Asp-75 of TM2 in the same subunit. This interaction gives rise to a distortion and bending of TM1. Arg-110, located in the center of TM3, is not exposed to the lipid bilayer but rather involved in intermolecular contacts. Compared with MPGES1, the crystal structure of LTC4S has more exposed charged side chains (Fig. S5).
Mutagenesis.
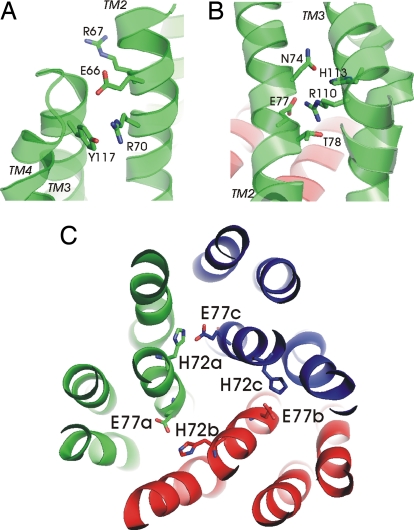
Our present structure and effects of site-directed mutagenesis on activity have identified several important intra- and intermolecular contacts (Fig. 2 and Table 1). The cytoplasmic face of TM2 is crucial for GSH binding and is stabilized toward TM3 of the same subunit close to its loop connection to TM4 (Fig. 2A). The polar side chain residues involved in this interaction Glu-66, Arg-67, Arg-70, and Tyr-117 are highly conserved in MAPEG (Fig. S4). Among the human members MGST3 has substitutions to asparagine and cysteine at positions corresponding to 66 and 67, respectively, whereas the noncatalytic FLAP has a threonine instead of arginine at position 70. Mutating Arg-67 to alanine in MPGES1 completely abolishes activity (Table 1). Further stabilization toward the center of the structure is achieved through side chains interactions from Asn-74, Glu-77 and Thr-78 of TM2 to Arg-110 and His-113 of TM3 (Fig. 2B). The arginine residue at position 110 is conserved among all MAPEG members. Both Glu-77 and Arg-110 are essential for MPGES1 catalysis, whereas the His72Ala mutant shows some activity (Table 1). Toward the lumenal side of the trimer phenylalanines, tyrosines, and methionines are in close proximity contributing to aromatic side chain interactions within and between subunits. His-72 in one subunit forms a salt bridge to Glu-77 in the neighboring subunit (Fig. 2C). This histidine residue in MGST1/MPGES1 is substituted by a glutamine in LTC4S and MGST2, whereas position 77 is a glutamic or an aspartic acid in all MAPEGs.
Fig. 2.
Intra- and intersubunit interactions in MPGES1. (A) Stabilization of the cytoplasmic face of transmembrane helices 2 and 3. (B) Interactions between the same helices close to the GSH binding site. (C) Stabilization of the trimer through the His-72–Glu-77 ion pair.
Table 1.
Site-directed mutagenesis for MPGES1
Glutathione Binding.
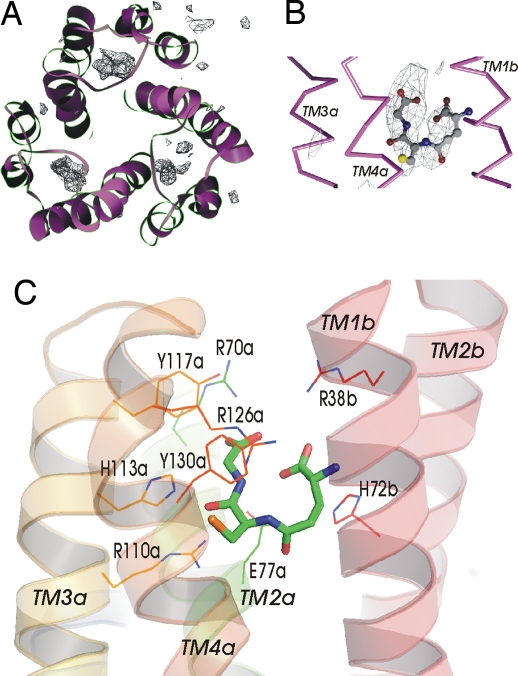
We have grown the crystals of MPGES1 in the presence of GSH. An omit map calculated between the observed electron diffraction amplitudes and amplitudes calculated from the protein model showed three distinct densities in more or less identical positions relative to each protein monomer (Fig. 3A). These regions were U-shaped (Fig. 3B) similar to what has been observed for GSH in LTC4S (19, 20) but located deeper and at a different angle. In the vicinity of the GSH binding pocket a number of conserved residues are found that are involved in GSH binding (Fig. 3C). One GSH molecule is fixed by interacting with TM1 and 2 from one subunit and with TM2, 3 and 4 from its neighbor. The two arginine/tyrosine pairs Arg-70/Tyr-117 and Arg-126/Tyr-130 are in close contact to the glycine and cysteine moieties of GSH, respectively. Arg-70 and Arg-38 make salt bridges to the carboxylates at either end of the bent GSH molecule. Site-directed mutagenesis showed that substitution of Arg-70 for an alanine or a serine preserves some enzymatic activity, whereas the Arg70Ala/Tyr117Ala double mutant lost almost all activity (Table 1). The thiol group of GSH is stabilized by Arg-126 (3.8 Å) and directed toward the membrane between helices 1 and 4 of neighboring subunits.
Fig. 3.
Binding of GSH. (A) Top view of the MPGES1 trimer from the cytoplasmic side with the omit map contoured at 2.5 σ, showing the positions of GSH molecules. (B) The U-shaped GSH molecule in ball and stick representation with the omit map density as seen from the side at the interface between transmembrane helices 1 and 4 from two different subunits. (C) Side chains involved in formation of the binding pocket for GSH.
Discussion
Open/Closed Conformation of MPGES1.
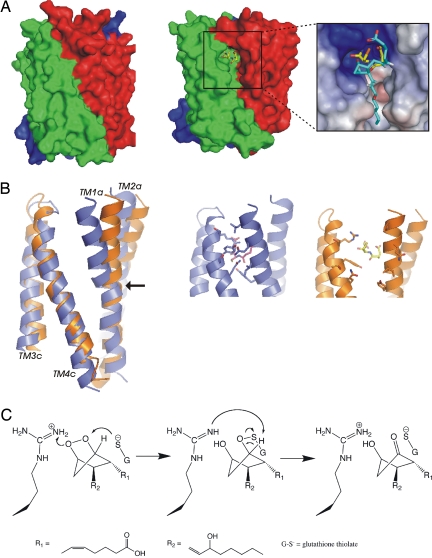
In LTC4S an opening between helix 1 in one subunit and helix 4 in a neighboring subunit forms a V-shaped cleft that allows access to GSH from the interior of the membrane (19, 20). This represents a natural entry point for labile hydrophobic substrates, such as PGH2 and LTA4 that, by way of membrane entry, are protected from hydrolytic degradation. In addition, PGH2 is produced by cyclooxygenases located on the lumenal side of the endoplasmatic reticulum and diffuses through the membrane to the active site of MPGES1 located on the cytoplasmic side. In MPGES1, the corresponding helices 1 and 4 are closer together and do not allow access to GSH. Therefore, we conclude that our MPGES1 structure represents a closed conformation of the enzyme. Because no alternate access path to GSH is evident it follows that dynamic opening and closing of the active site involving helices 1 and 4 is required to accommodate PGH2 (Fig. 4). Hydrogen/deuterium exchange dynamics of the closely related MGST1 supports dynamic flexibility in helix 1 (22) that is decreased when a hydrophobic ligand is present (23). Furthermore, in the structures of LTC4 synthase a detergent molecule occupies the V-shaped cleft between subunits, which was proposed to be the substrate binding site (19, 20). To generate a model (24) of an open structure for MPGES1, we used LTC4S as a template (Fig. 4 A and B). It is apparent that helix 1 occludes the active site in the closed conformation as it clashes with PGH2 modeled to contact GSH whereas the open conformation allows access (Fig. 4A Inset). The salt bridge between Lys-26 and Asp-75 appears to act as a hinge for the displacement of the cytoplasmic half of TM1, which in that part does not have any strong interactions with neighboring helices. The open form of MPGES1, resembling LTC4S regarding substrate access, thus constitutes a model for the productive enzyme. We believe that the closed state is in rapid equilibrium with an open state that is catalytically competent and very efficiently pulls the equilibrium in favor of product formation. In the resting enzyme the presumed GSH thiolate could in this way be protected from oxidation or unwanted side reactions. (The enzyme does display low glutathione transferase and peroxidase activity.)
Fig. 4.
Conformational change allowing PGH2 access and schematic chemical mechanism for MPGES1. (A Left) Surface representation of the MPGES1 trimer structure as determined from the 2D crystals, showing a closed conformation. The three subunits are green, red, and blue, respectively. (A Right) Open conformation based on homology modeling from the structure of LTC4S (PDB entry 2UUH) exposes the GSH molecules. (Inset) Enlargement of the cleft between subunits where PGH2 is expected to bind. GSH is shown in yellow and a manually fitted PGH2 molecule shown in cyan. The surface rendering shows electrostatic potential of the open homology model. Blue represents positively charged, red represents negatively charged, and white represents a nonpolar surface. (B) (Left) The largest difference between the closed and open conformation is a bending of the cytoplasmic half of TM1 about a hinge fixed by the Lys-26/Asp-75 salt bridge (arrow). (Center) The closed conformation with Ser-127, Tyr-130, Thr-131, and Gln-134 onTM4 pointing toward residues Ile-32, Gln-34, and Leu-39 in TM1 in the next subunit. Pointing inwards binding to the GSH in magenta are Arg-126 from TM4 and Tyr-28 from TM1. (Right) The open conformation model with the same residues shown and with GSH in yellow. (C) Suggested chemical mechanism for the MPGES1 catalyzed PGE2 synthesis involves an attack of the GSH thiolate on the O9 of the endoperoxide bridge followed by proton donation to O11 via Arg-126. Arg-126 then abstracts a proton from C9 where a carbonyl forms as the oxygen sulfur bond is broken. The leaving GSH thiolate is stabilized by Arg-126.
Mechanism.
Two chemical mechanisms have been suggested for PGE2 synthesis (25) where one involves attack of a thiol on the O9 of the endoperoxide bridge in a glutathione peroxidase like mechanism. Because MPGES1 also catalyses GSH-dependent peroxidase activity toward other (hydro)peroxides, we favor this mechanism that requires attack of the GSH thiolate, stabilized by Arg-126 on the O9 oxygen of PGH2 (Fig. 4C). Strict restrictions are put on binding of PGH2 so that the correct oxygen is attacked. A proton donor and acceptor are then required to assist in the chemical conversion (Fig. 4C) and in a radius of 8 Å around the GSH sulfur only Arg-126 and tyrosines 28 and 130 are reasonable candidates for protonation of O11 and proton abstraction from C9. Arg-126 could stabilize the evolving oxyanion O11 and donate a proton as the endoperoxide is opened. This would allow Arg-126 to abstract a proton (26) from C9 forming the carbonyl as the oxygen sulfur bond is broken regenerating the GSH thiolate (Fig. 4C). This mechanism is consistent with in vitro mutagenesis data supporting the GSH binding site location in MPGES1 (Table 1). Alternatively, tyrosine(s) alone or in combination with arginine could catalyze the chemistry.
The structure of MPGES1 is an attractive target for developing drugs that could stabilize the closed form and thereby efficiently inhibit catalysis. In addition, LTC4S could also exist in a closed conformation that was not experimentally accessible as the detergent used in crystallization behaved as a substrate analogue (19, 20). The position of the hydrophobic substrate in LTC4S and the similar location of GSH in MPGES1/LTC4S show that PGH2 binds to the cleft between TM1 and TM4. We have modeled a tentative binding mode, using the molecular ruler concept that places the omega-end of PGH2 toward the middle of the bilayer, lets the endoperoxide reside in proximity to GSH, and allows the carboxylate to reside in the hydrophilic protein water interface (Fig. 4A Inset). This tentative model suggests reasonable hydrogen bonding and polar interactions allowing the hydrocarbon part to be bound along the cleft without steric clashes, which should be confirmed by the crystal structure of MPGES1 with a ligand.
Materials and Methods
Human MPGES1 containing a His6-tag at the N terminus was overexpressed in E. coli and purified as published in ref. 5 and finally stored at a concentration of 1 mg/ml in a buffer containing 50 mM NaPi (pH 7.5), 50 mM NaCl, 10% glycerol, 1 mM GSH, 1% Triton X-100, and 0.1 mM EDTA.
For 2D crystallization, we mixed aliquots of typically 50–100 μl of His6-MPGES1 with bovine liver lecithin in Triton X-100 at a molar lipid to protein ratio of 9. The mixture was subjected to dialysis against the same buffer as the one used for protein storage but with 20% glycerol and lacking the detergent. Dialysis, using membranes with a cut-off molecular mass of 12–14 kDa, was performed for at least 1 week at room temperature.
For electron microscopy, we prepared grids with the inverted technique (27), using conventional 400 mesh grids, thin carbon layers, typically 7% trehalose and 2 μl of the crystallization suspension. Grids were placed in liquid nitrogen cooled Gatan 626 or 914 cryoholders before insertion into a JEOL 2100F electron microscope equipped with a TemCam-F415 4k × 4k CCD camera (Tietz Video and Image Processing Systems).
Electron diffraction patterns were recorded on the CCD with two times binning at a camera length of 200 cm (Fig. S2B). For tilted diffraction patterns, pairwise acquisitions were always made in such a way that the first tilted recording was followed by a 0° pattern that was used entirely for classification and quality assessment. By applying this approach of nontilted diffraction pattern based sorting, we solved the problem of substantial crystalline heterogeneity and acquired a final set of 100 diffraction patterns showing good merging statistics (Table S1). Background corrected amplitudes were extracted using the MRC software (28). The diffraction dataset was subjected to merging using DIFFMERG, and, subsequently, LATLINE was used for lattice line fitting.
We used the trimeric model of microsomal glutathione transferase 1 (MGST1) (17) for a molecular replacement search, because the sequence identity is high (≈40%), and our studies have shown that MPGES1 also forms a trimer in 2D crystals (5). One of the top score solutions from AMoRe (29) was unique, because it did not present severe clashes between trimers and it fitted a low resolution projection map. Model building in O (30) started from a polyalanine model of the protein at the determined position and orientation. Numerous rounds of rigid body and restrained refinement, using tight geometry restraints and medium noncrystallographic symmetry in REFMAC5 in combination with geometry idealization and manual rebuilding in O, resulted in a final structure of the MPGES1 trimer in which all residues except the His-tags and the first 10 N-terminal residues were included. At the present resolution, the difference between using electron- or x-ray form factors are negligible; thus, x-ray values were used throughout the refinement. 2Fo − Fc and Fo − Fc maps were continuously generated to evaluate the accuracy and quality of the refined model. Reliable Rfree values were obtained by using a final fraction of ≈5% of the observed structure factor amplitudes. PROCHECK (31) was used to examine the molecular geometry. For analysis and illustrations, we used PyMOL (32) and CHIMERA (33).
Sequence alignment was performed with ClustalW (34) followed by structural based manual adjustments. MODELLER 8, Version 2 (24) was used to calculate homology models.
The plasmid pSP19T7LT containing wild-type MPGES1 with an N-terminal His6-tag (5) was subjected to site-directed mutagenesis using GeneTailor site-directed mutagenesis system (Invitrogen) and specific mutation primers. Mutant protein was expressed in BL21 Star (DE) E. coli cells grown at 37°C in terrific broth. Subsequently, we isolated the membrane fractions by differential centrifugation and quantified the amount of mutant MPGES1 by comparing the signals with known amounts of purified MPGES1 in Western blot analysis. For activity measurements, we used accordingly 400 ng of mutant or wild-type MPGES1 in the assay described in ref. 6.
Supplementary Material
Acknowledgments.
This work was supported by the Swedish Research Council, Vinnova (Swedish Governmental Agency for Innovation Systems), the Japan Science and Technology Agency, the Swedish Cancer Society, and funds from Karolinska Institutet.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3DWW).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802894105/DCSupplemental.
References
- 1.Jakobsson P-J, Thoren S, Morgenstern R, Samuelsson B. Identification of human prostaglandin E synthase: A microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci USA. 1999;96:7220–7225. doi: 10.1073/pnas.96.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jakobsson P-J, Morgenstern R, Mancini J, Ford-Hutchinson A, Persson B. Common structural features of MAPEG—a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein Sci. 1999;8:689–692. doi: 10.1110/ps.8.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresell A, et al. Bioinformatic and enzymatic characterization of the MAPEG superfamily. FEBS J. 2005;272:1688–1703. doi: 10.1111/j.1742-4658.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 4.Kozak KR, et al. Metabolism of the endocannabinoids, 2-arachidonylglycerol and anandamide, into prostaglandin, thromboxane, and prostacyclin glycerol esters and ethanolamides. J Biol Chem. 2002;277:44877–44885. doi: 10.1074/jbc.M206788200. [DOI] [PubMed] [Google Scholar]
- 5.Thoren S, et al. Human microsomal prostaglandin E synthase-1: Purification, functional characterization, and projection structure determination. J Biol Chem. 2003;278:22199–22209. doi: 10.1074/jbc.M303227200. [DOI] [PubMed] [Google Scholar]
- 6.Thoren S, Jakobsson P-J. Coordinate up- and down-regulation of glutathione-dependent prostaglandin E synthase and cyclooxygenase-2 in A549 cells. Inhibition by NS-398 and leukotriene C4. Eur J Biochem. 2000;267:6428–6434. doi: 10.1046/j.1432-1327.2000.01735.x. [DOI] [PubMed] [Google Scholar]
- 7.Murakami M, et al. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J Biol Chem. 2000;275:32783–32792. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 8.Stichtenoth DO, et al. Microsomal prostaglandin E synthase is regulated by proinflammatory cytokines and glucocorticoids in primary rheumatoid synovial cells. J Immunol. 2001;167:469–474. doi: 10.4049/jimmunol.167.1.469. [DOI] [PubMed] [Google Scholar]
- 9.Ueno N, Takegoshi Y, Kamei D, Kudo I, Murakami M. Coupling between cyclooxygenases and terminal prostanoid synthases. Biochem Biophys Res Commun. 2005;338:70–76. doi: 10.1016/j.bbrc.2005.08.152. [DOI] [PubMed] [Google Scholar]
- 10.Samuelsson B, Morgenstern R, Jakobsson P-J. Membrane prostaglandin E synthase-1: A novel therapeutic target. Pharmacol Rev. 2007;59:207–224. doi: 10.1124/pr.59.3.1. [DOI] [PubMed] [Google Scholar]
- 11.Hofstetter AO, Saha S, Siljehav V, Jakobsson P-J, Herlenius E. The induced prostaglandin E2 pathway is a key regulator of the respiratory response to infection and hypoxia in neonates. Proc Natl Acad Sci USA. 2007;104:9894–9899. doi: 10.1073/pnas.0611468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korotkova M, et al. Effects of immunosuppressive treatment on microsomal PGE synthase 1 and cyclooxygenases expression in muscle tissue of patients with polymyositis or dermatomyositis. Ann Rheum Dis. 2007 Dec 18; doi: 10.1136/ard.2007.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korotkova M, et al. Effects of antirheumatic treatments on the prostaglandin E2 biosynthetic pathway. Arthritis Rheum. 2005;52:3439–3447. doi: 10.1002/art.21390. [DOI] [PubMed] [Google Scholar]
- 14.Hebert H, et al. The 3.0 Å projection structure of microsomal glutathione transferase as determined by electron crystallography of P21212 two-dimensional crystals. J Mol Biol. 1997;271:751–758. doi: 10.1006/jmbi.1997.1216. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt-Krey I, et al. The projection structure of the membrane protein microsomal glutathione transferase at 3 Å resolution as determined from two-dimensional hexagonal crystals. J Mol Biol. 1999;288:243–253. doi: 10.1006/jmbi.1999.2683. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt-Krey I, et al. Human leukotriene C4 synthase at 4.5 Å resolution in projection. Structure. 2004;12:2009–2014. doi: 10.1016/j.str.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Holm PJ, et al. Structural basis for detoxification and oxidative stress protection in membranes. J Mol Biol. 2006;360:934–945. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson AD, et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 19.Ago H, et al. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature. 2007;448:609–612. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Molina D, et al. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 21.Hebert H, Jegerschöld C. The structure of membrane associated proteins in eicosanoid and glutathione metabolism as determined by electron crystallography. Curr Opin Struct Biol. 2007;17:396–404. doi: 10.1016/j.sbi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Busenlehner LS, et al. Stress sensor triggers conformational response of the integral membrane protein microsomal glutathione transferase 1. Biochemistry. 2004;43:11145–11152. doi: 10.1021/bi048716k. [DOI] [PubMed] [Google Scholar]
- 23.Busenlehner LS, et al. Location of substrate binding sites within the integral membrane protein microsomal glutathione transferase-1. Biochemistry. 2007;46:2812–2822. doi: 10.1021/bi6023385. [DOI] [PubMed] [Google Scholar]
- 24.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 25.Yamada T, Komoto J, Watanabe K, Ohmiya Y, Takusagawa F. Crystal structure and possible catalytic mechanism of microsomal prostaglandin E synthase type 2 (mPGES-2) J Mol Biol. 2005;348:1163–1176. doi: 10.1016/j.jmb.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 26.Guillen-Schlippe YV, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Hirai T, Murata K, Mitsuoka K, Kimura Y, Fujiyoshi Y. Trehalose embedding technique for high-resolution electron crystallography: Application to structural study on bacteriorhodopsin. J Electron Microsc. 1999;48:653–658. doi: 10.1093/oxfordjournals.jmicro.a023731. [DOI] [PubMed] [Google Scholar]
- 28.Crowther RA, Henderson R, Smith JM. MRC Image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 29.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 32.De Lano WL. The PyMOL Molecular Graphics System. [Accessed September 1, 2007];2002 Available at www.pymol.org.
- 33.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, et al. Structural and functional characterization of human microsomal prostaglandin E synthase-1 by computational modeling and site-directed mutagenesis. Bioorg Med Chem. 2006;14:3553–3562. doi: 10.1016/j.bmc.2006.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.