Abstract
The product of the Nijmegen breakage syndrome gene (NBS1) plays crucial roles in DNA damage response through its association with many proteins, including MRE11 and RAD50. However, it remains to be determined exactly how NBS1 accumulates at or near DNA double-strand breaks. Here we report that MDC1 directly binds to NBS1 and targets NBS1 to the sites of DNA damage. The MDC1–NBS1 interaction occurs through a specific region (residues 200–420) of MDC1, which contains multiple consensus casein kinase 2 (CK2) phosphorylation sites. In addition, this interaction requires both the forkhead-associated (FHA) and tandem BRCA1 C-terminal (BRCT) domains of NBS1. Disruption of the MDC1–NBS1 interaction results in failure of NBS1 accumulation at DNA double-strand breaks and impairment of intra-S checkpoint activation. These studies provide important mechanistic insights as to how MDC1 regulates NBS1 and the intra-S-phase checkpoint in response to DNA damage.
Keywords: 53BP1, BRCA1, MRE11, casein kinase 2
The evolutionarily conserved MRE11/RAD50/NBS1 (MRN) complex plays important roles in cell cycle checkpoint signaling and DNA repair. Hypomorphic mutations in NBS1 and MRE11 cause Nijmegen breakage syndrome (NBS) and ataxia–telangiectasia-like disorder (ATLD) respectively (1–3), characterized by developmental defects, immunodeficiency, and a high incidence of cancer. Cells derived from NBS and ATLD patients are hypersensitive to radiation and display impaired intra-S-phase checkpoint activation (1–3), supporting critical roles of this complex in DNA damage response.
In response to DNA damage, the MRE11/RAD50/NBS1 complex regulates the activation of ATM, the protein kinase essential for the DNA damage signaling. The key component involved in ATM activation is believed to be NBS1, because NBS1 directly binds to ATM through a very C-terminal motif and modulates ATM autophosphorylation (4–7). Biochemical evidence also suggests that the MRN complex recruits ATM to the vicinity of DNA double-strand breaks (DSBs) and stimulates ATM activation (8). In addition to playing a role in ATM activation, NBS1 also functions downstream of ATM in regulating the intra-S-phase checkpoint (9–11). Besides regulating ATM-dependent phosphorylation of SMC proteins (12, 13), the mechanism by which NBS1 participates in this intra-S-phase checkpoint remains elusive.
The human NBS1 gene encodes a 754-aa nuclear protein with an N-terminal forkhead-associated (FHA) domain and breast cancer BRCA1 C-terminal (BRCT) domain. Recently, a second BRCT domain (termed BRCT2) has been identified adjacent to the first BRCT domain (termed BRCT1) (14). Both FHA and BRCT domains mediate protein–protein interaction by recognizing phosphoserine (pSer) and phosphothreonine (pThr)-containing motifs, and many proteins involved in the DNA damage responses contain functional FHA and BRCT domains. Previous reports indicate that retention of NBS1 at DSBs, after DNA damage, is mediated by its interaction with MDC1 (15–19). Moreover, the FHA and BRCT domains of NBS1 are required for its recruitment to DSBs (nuclear foci) after DNA damage (20–22). Despite these results, it is still unclear how exactly NBS1 is recruited to the sites of DNA damage. We hypothesize that the FHA and BRCT domains of NBS1 bind directly to MDC1, which facilitates NBS1 localization to the sites of DNA DSBs. In this study, we mapped the regions of MDC1 and NBS1 that are required for their interaction and demonstrated that MDC1 acts upstream of NBS1 and directly targets NBS1 to DNA DSBs. In addition, we showed that the interaction between MDC1 and NBS1 is required for proper control of the intra-S-phase checkpoint.
Results and Discussion
MDC1 Promotes NBS1 Accumulation at the Sites of DNA DSBs.
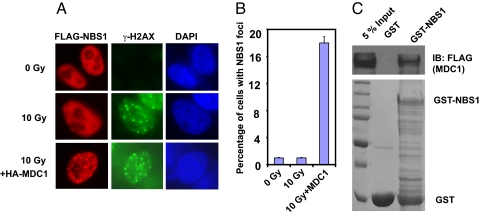
We noticed that in contrast to robust MDC1 foci formation after DNA damage, the formation of NBS1 foci in cells expressing exogenous NBS1 was not readily detectable (Fig. 1A). We speculated that this failure of proper NBS1 localization could be due to the limited expression of an endogenous protein or proteins required for NBS1 foci formation in vivo. We and others have shown that MDC1 is required for NBS1 foci formation, and MDC1 interacts with the MRN complex (15–18, 23). Thus, MDC1 is the likely candidate that directs NBS1 localization. Indeed, we found that cotransfection of MDC1 with NBS1 significantly enhanced NBS1 foci formation (Fig. 1A), which colocalized with the DSB marker γ-H2AX in the cell. Quantitatively, ≈20% of cells expressing exogenous NBS1 show NBS1 foci formation in cotransfected cells, whereas <1% of cells show NBS1 foci in cells transfected with exogenous NBS1 alone (Fig. 1B). In addition, we found that bacterially expressed GST-NBS1 could retrieve MDC1 from cell lysates in a pull-down assay (Fig. 1C). Together, these results indicate that NBS1 interacts with MDC1, and this interaction recruits NBS1 to DNA DSBs after DNA damage.
Fig. 1.
MDC1 promotes the recruitment of NBS1 to DSBs after DNA damage. (A) A plasmid encoding FLAG-tagged full-length NBS1 was used to transiently transfect HeLa cells with or without HA-tagged full-length MDC1. Thirty-six hours later, cells were left untreated or exposed to ionizing radiation (10 Gy) and immunostained with anti-FLAG and anti-γ-H2AX antibodies. (B) Quantification of cells described in A with NBS1 foci. The results represent the average of two independent experiments. (C) The interaction between NBS1 and MDC1. GST or GST-NBS1 was incubated with cell lysates containing exogenously expressed FLAG-tagged full-length MDC1. (Upper) Bound MDC1 was analyzed by immunoblotting with anti-FLAG antibody. (Lower) Input GST or GST-NBS1 proteins.
A Region Comprising Residues 200–420 of MDC1 Is Required for NBS1 Foci Formation.
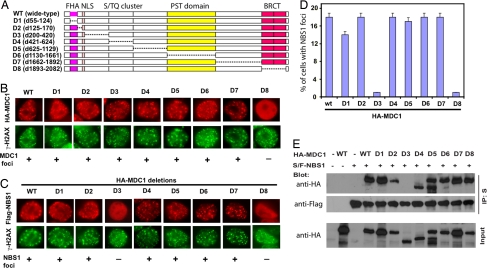
We first attempted to determine which region of MDC1 is required for recruiting NBS1 to DNA damage foci. A series of HA-tagged MDC1 deletion mutants were generated (Fig. 2A). In all of these deletion mutants, the nuclear localization sequence (residues 170–200) was kept intact to ensure that all mutants of MDC1 localize normally in nuclei (Fig. 2A). All except one deletion mutant of MDC1 localized to DSB sites as indicated by their colocalization with γ-H2AX (Fig. 2B). The only MDC1 deletion mutant that could not form foci is the BRCT domain deletion mutant (D8), because this mutant could not bind to phosphorylated H2AX as reported (15, 24) and thus failed to localize to the sites of DNA damage.
Fig. 2.
A specific region of MDC1 (residues 200–420) is required for NBS1 recruitment to DSB sites after DNA damage. (A–C) Plasmids encoding HA-tagged full-length or deletion mutants of MDC1 (A) were transfected alone (B) or cotransfected with Flag-tagged full-length NBS1 (C) into HeLa cells. After exposure to ionizing radiation, cells were immunostained with antibodies as indicated. (D) Percentages of cells with NBS1 foci described in C were quantified. The results represent the average of two independent experiments. (E) Plasmids encoding S/FLAG (S/F)-tagged NBS1 were cotransfected with constructs encoding HA-tagged full-length or deletion mutants of MDC1 as indicated. Cell lysates were then subjected to pull-down with S-Sepharose and immunoblotted with anti-FLAG (NBS1) or anti-HA (MDC1) antibodies.
These deletion mutants of MDC1 were then cotransfected with FLAG-tagged NBS1, and their effects on NBS1 foci formation were determined. Strikingly, we found that the deletion mutant D3 (deletion of residues 200–420), which still harbors an intact FHA domain, failed to promote NBS1 foci formation after DNA damage (Fig. 2C). The quantification of the effect of these MDC1 deletion mutants on NBS1 foci formation is summarized in Fig. 2D.
We further performed coimmunoprecipitation experiments to examine the interaction between NBS1 and these MDC1 deletion mutants. Although WT MDC1 or MDC1 mutants with the deletion of either FHA or BRCT domain could still interact with NBS1, the deletion of residues 200–420 specifically abolished the MDC1–NBS1 interaction (Fig. 2E). Together, these data suggest that the region consisting of residues 200–420 of MDC1 is required for its interaction with NBS1 and support the idea that the interaction between MDC1 and NBS1 is critical for NBS1 foci formation.
Phosphorylation of MDC1 on CK2 Consensus Sites Mediates the MDC1–NBS1 Interaction.
We tried to generate smaller deletion mutants within this region (residues 200–420) of MDC1 and further map the minimal domain required for NBS1 localization to the sites of DNA damage. However, those smaller deletion mutants (deletions of residues 200–306, 307–383, 384–420, 307–420, 307–397, or 360–397) could, at best, partially reduce NBS1 foci formation (data not shown), raising the possibility that there may be functionally redundant sequences or motifs within this region of MDC1.
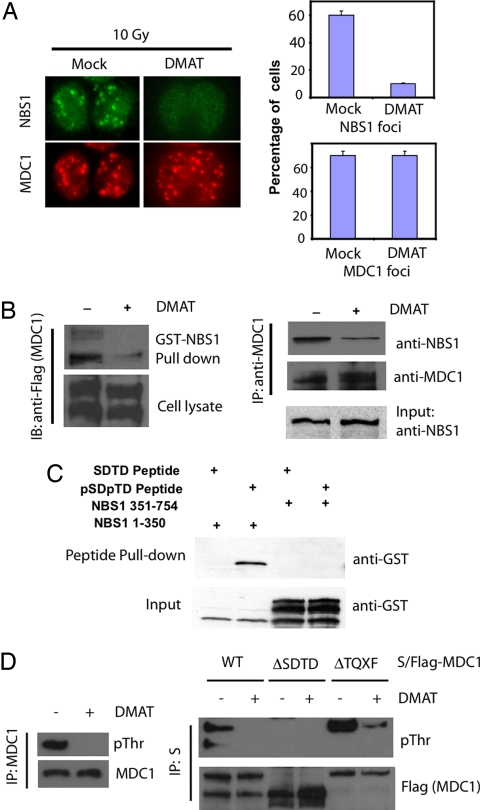
This particular region of MDC1 (residues 200–420) contains six conserved SDTDXD/E clusters, fulfilling the criteria for CK2 phosphorylation site S/TXXD/E (25). The six SDTDXD/E motifs are also highly conserved across species (data not shown), suggesting a conserved function for these motifs. We suspected that CK2 might phosphorylate both Ser and Thr residues at these SDTDXD/E clusters (6 Ser and 6 Thr, a total of 12 residues) and therefore generate docking sites for NBS1. To directly test this possibility, we pretreated cells with 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT), a CK2 inhibitor (26), before irradiation and then examined endogenous MDC1 and NBS1 foci formation. While the MDC1 foci formation was not affected, NBS1 foci were significantly reduced after DMAT treatment (Fig. 3A). We also examined how the inhibition of CK2 activity would affect the MDC1–NBS1 interaction. Corroborating the above data of NBS1 foci formation, DMAT treatment significantly decreased the binding of MDC1 to GST-NBS1 (Fig. 3B Left) or endogenous NBS1 (Fig. 3B Right). To validate that CK2 phosphorylation on MDC1 generates docking sites for NBS1, we synthesized biotinylated phosphopeptides containing the consensus (p)SD(p)TDXE motif of MDC1 and control unphosphorylated peptides with the identical sequence. These peptides were coupled to streptavidin-Sepharose beads and incubated with a bacterially expressed GST-NBS1 fragment containing the FHA and tandem BRCT domains (residues 1–350). As shown in Fig. 3C, phospho-MDC1 peptides specifically interacted with the N terminus of NBS1 (residues 1–350) containing NBS1 FHA and BRCT domains, but not with a C-terminal fragment of NBS1 (residues 351–754).
Fig. 3.
Phosphorylation of MDC1 on CK2 consensus sites mediates the MDC1–NBS1 interaction. (A) U2OS cells were mock treated or pretreated with CK2-specific inhibitor DMAT (20 μM) overnight before exposure to ionizing radiation. Cells were then fixed and immunostained with the indicated antibodies. The percentages of cells with NBS1 foci and MDC1 foci were quantified. The results represent the average of two independent experiments. (B) 293T cells were pretreated with DMAT (20 μM) for 6 h, and cell lysates were incubated with GST-NBS1 coupled to GSH-Sepharose (Left) or subjected to immunoprecipitation with anti-MDC1 antibodies (Right). Proteins retained on beads were then blotted with indicated antibodies. (C) Phosphorylated or control MDC1 peptides mimicking CK2 phosphorylation were incubated with purified GST-NBS1 N-terminal fragment containing the FHA-BRCT1-BRCT2 domain (residues 1–350) or GST-NBS1 C-terminal fragment (residues 350–754). GST-fusion proteins associated with peptides were detected by immunoblotting with anti-GST antibodies. (D) 293T cells were left untreated or pretreated with DMAT (20 μM) for 8 h. Cell lysates were prepared and subjected to immunoprecipitation using anti-MDC1 antibodies, and then immunoblotted with indicated antibodies (Left). 293T cells were transfected with WT or deletion mutants of S/Flag-tagged MDC1. Cells were left untreated or pretreated with DMAT before harvesting. Immunoprecipitation was carried out by using S-protein beads followed by immunoblotting using the indicated antibodies (Right). ΔSDTD, deletion of residues 200–420 of MDC1; ΔTQXF, deletion of residues 698–768 of MDC1.
To confirm that these SDTD motifs of MDC1 are phosphorylated in vivo, we used a pThr antibody. As shown in Fig. 3D, endogenous MDC1 was phosphorylated as indicated by the anti-pThr blotting, and this phosphorylation was drastically reduced by the pretreatment of cells with the CK2 inhibitor DMAT (Fig. 3D Left). Although phosphorylation of WT and the ΔTQXF (del 698–768) mutant of MDC1 could be inhibited by the DMAT treatment, the ΔSDTD (del 200–420) mutant of MDC1 was not appreciably phosphorylated even without DMAT treatment (Fig. 3D Right). Moreover, we have performed mass spectrometry analysis for the identification of in vivo phosphorylation sites of MDC1 purified from 293T cells stably expressing tagged MDC1. As shown in Table 1, we isolated phosphopeptides corresponding to each of the six SDTD motifs. In fact, some of these phosphorylation sites have been reported (27, 28). Collectively, these in vivo and in vitro results suggest that the SDTD sites on MDC1 are indeed phosphorylated in vivo in a CK2-dependent manner, and these phosphorylation sites directly mediate the interaction with NBS1.
Table 1.
MDC1 phosphopeptides identified by mass spectrometry analysis
| Phosphopeptide | Putative CK2 sites |
|---|---|
| TTSSSVIVPEpSDEEGHpSPVLGGLGPPFAFNLNpSDpTDVEEGQQPATEEASSAAR | S218, T220 |
| SQPPGEDpSDpTDVDDDSRPPGRPAEVHLER | S299, T301 |
| AQPFGFIDpSDpTDAEEER | S329, T331 |
| PGAPGLAHLQEpSQAGpSDpTDVEEGKAPQAVPLEK | S376, T378 |
| SQASMVINpSDpTDDEEEVSAALTLAHLK | S402, T404 |
| SQTTTERDpSDpTDVEEEELPVENR | S453, T455 |
Separate Phosphorylated Regions of MDC1 Participate in Distinct Mediator Functions.
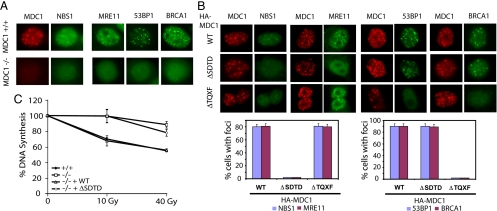
The region of MDC1 (residues 200–420) involved in its interaction with NBS1 is distinct from the region recently identified for RNF8 binding (29–32), suggesting that MDC1 may use two separate domains in the recruitment of different downstream effectors. Because the MDC1-mediated RNF8 recruitment is required for the accumulation of BRCA1 and 53BP1 to the sites of DNA damage, we decided to examine the focus localization of NBS1 complex, BRCA1, and 53BP1 in MDC1−/− mouse embryonic fibroblasts (MEFs), after transient expression of WT or two deletion mutants of MDC1 (ΔSDTD, deletion of residues 200–420, which contains SDTDxD/E motifs required for NBS1 binding; ΔTQXF, deletion of residues 698–768, which contains TQXF motifs required for RNF8 binding).
We first confirmed that IR-induced foci formation for NBS1, MRE11, 53BP1, and BRCA1 were abolished in MDC1−/− MEF cells, whereas all these proteins accumulate to DSBs and colocalize with endogenous MDC1 in MDC1+/+ MEF cells (Fig. 4A and data not shown). As shown in Fig. 4B, the expression of WT MDC1 in MDC1−/− cells fully restored focus formation of NBS1 complex, BRCA1, and 53BP1. As expected, the expression of MDC1 ΔTQXF mutant rescued focus formation of NBS1 and MRE11, but failed to do so for either BRCA1 or 53BP1 (Fig. 4B). On the other hand, expression of the MDC1 ΔSDTD mutant with the deletion of NBS1-binding domain could not rescue NBS1 or MRE11 foci formation, but restored BRCA1 and 53BP1 foci formation (Fig. 4B). These data support the hypothesis that two independent domains of MDC1 are involved in the accumulation of different downstream checkpoint proteins to the sites of DNA damage.
Fig. 4.
Separate phosphorylated regions of MDC1 participate in distinct mediator functions. (A) MDC1+/+ and MDC1−/− MEF cells were exposed to ionizing radiation (10 Gy), fixed, and stained with the indicated antibodies. (B Upper) MDC1−/− MEF cells transfected with WT or deletion mutants of HA-tagged MDC1 were exposed to ionizing radiation, fixed, and stained with the indicated antibodies. (Lower) Percentages of cells containing NBS1, MRE11, 53BP1, and BRCA1 foci in cells respectively expressing WT, ΔSDTD mutant, or ΔTQXF mutant of MDC1 were determined. (C) MDC1+/+ MEF cells and MDC1−/− MEF cells stably expressing WT or a deletion mutant of MDC1 were left untreated or exposed to ionizing radiation, and DNA synthesis was measured as indicated in Materials and Methods.
We also established MDC1−/− derivative cell lines stably expressing WT or the ΔSDTD mutant of MDC1. Whereas WT MDC1 fully restored ionizing radiation-induced intra-S-phase checkpoint control, the ΔSDTD mutant of MDC1 failed to do so, implying that the interaction between MDC1 and NBS1 is important for this checkpoint function.
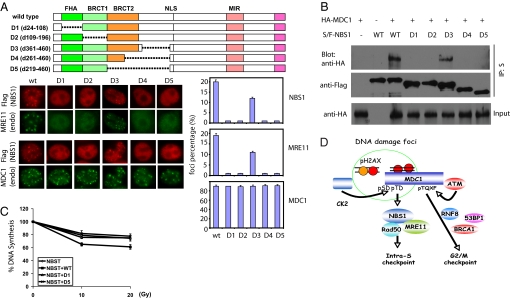
The FHA and Tandem BRCT Domains of NBS1 Promote Foci Formation Through Interaction with MDC1.
NBS1 contains a FHA domain and a BRCT domain in its N terminus. Recently, the BRCT2 domain was identified adjacent to domain BRCT1 (Fig. 5A) (14). The FHA and BRCT1 domains of NBS1 have been shown to be required for NBS1 foci formation after DNA damage (20–22). However, it was not clear whether this newly identified BRCT2 would also be required for NBS1 foci formation. To address this question, a series of NBS1 deletion mutants were generated (Fig. 5A) and stably expressed in NBS1-deficient NBST cells. These cells were examined for their abilities to form NBS1 and endogenous MRE11 foci without cotransfection of MDC1 (Fig. 5A). Consistent with previous reports, deletion of NBS1 FHA or BRCT1 domain abolished NBS1 and MRE11 foci formation after DNA damage (Fig. 5A D1 and D2). Intriguingly, disruption of the BRCT2 domain also abolished NBS1 and MRE11 foci formation (Fig. 5A D4 and D5). Importantly, a deletion mutant that does not disrupt any of these domains still displayed NBS1 and MRE11 foci after DNA damage (Fig. 5A D3). As a control, we showed that all of these cells have normal ionizing radiation-induced endogenous MDC1 foci formation (Fig. 5A). The quantification of foci formation for NBS1, MRE11, and MDC1 in these cells is summarized in Fig. 5A Lower Right.
Fig. 5.
The FHA and BRCT domains of NBS1 are required for MDC1-mediated foci formation and interaction. (A) Diagrams of plasmids encoding FLAG-tagged full-length or deletion mutants of NBS1 are shown. NLS, nuclear localization sequence; MIR, MRE11-interacting region. NBST cells stably expressing WT or deletion mutants of Flag-tagged NBS1 were exposed to ionizing radiation, fixed, and stained with antibodies to Flag and MRE11, or Flag and MDC1 as indicated. The percentages of cells containing foci of NBS1, MRE11, and MDC1 were quantified and summarized. The results represent the average of two independent experiments. (B) Plasmids encoding S- and FLAG-tagged full-length or deletion mutants of NBS1 were cotransfected into 293T cells along with plasmids encoding HA-tagged full-length MDC1. Cell lysates were then subjected to pull-down with S-Sepharose and immunoblotted with anti-FLAG (NBS1) or anti-HA (MDC1) antibodies. (C) NBS1-deficient NBST cells stably expressing full-length or deletion mutants of NBS1 were left untreated or exposed to ionizing radiation, and DNA synthesis was measured as indicated in Materials and Methods. (D) Diagram of the mediator function of MDC1 in recruiting different DNA damage checkpoint proteins through distinct motifs.
The failure of foci formation for these NBS1 deletion mutants may reflect their inability to interact with MDC1. To test this possibility, we performed MDC1–NBS1 coimmunoprecipitation experiments. As expected, disruption of FHA, BRCT1, or BRCT2 domain of NBS1 abolished its interaction with MDC1 (Fig. 5B D1, D2, D4, and D5). In contrast, the deletion mutant that does not disrupt any of these domains still interacted with MDC1 (Fig. 5B D3). Collectively, these results suggest that the FHA and tandem BRCT domains of NBS1 are required for its interaction with MDC1 and its subsequent accumulation at sites of DNA DSBs.
The MDC1-NBS1 Pathway Is Required for Intra-S-Phase Checkpoint Control After DNA Damage.
MDC1 is known to participate in the intra-S-phase checkpoint (16, 17, 33). On the other hand, NBS1 deficiency or mutation of NBS1 also leads to impaired intra-S-phase checkpoint after DNA damage (1–3). As shown in Fig. 4C, reconstitution of MDC1−/− MEF cells with the NBS1-binding defective ΔSDTD mutant of MDC1 was unable to restore the intra-S-phase defect. To further determine whether the interaction between NBS1 and MDC1 is important for this checkpoint function, we stably reconstituted NBS1-deficient NBST cells with WT and various deletion mutants of NBS1 and determined the rate of DNA synthesis after DNA damage. Indeed, disruption of the FHA (D1) or BRCT2 (D5) domain of NBS1 compromised the intra-S-phase checkpoint (Fig. 5C), supporting the conclusion that the MDC1-NBS1 interaction is required for ionizing radiation-induced intra-S-phase checkpoint.
In this study, we demonstrated that MDC1 recruits NBS1 to DSBs through a direct binding between MDC1 and NBS1. The FHA/BRCT domain of NBS1 and a region of MDC1 containing six SDTDXD/E clusters corresponding to consensus CK2 phosphorylation sites are required for the MDC1–NBS1 interaction. Our results suggest that CK2 phosphorylates Ser and Thr residues at the SDTDXD/E clusters of MDC1 and therefore generates binding sites for the FHA and BRCT domains of NBS1. We provide several lines of evidence here to support this scenario. First, deletion of the region containing SDTDXD/E of MDC1 abolished the MDC1–NBS1 interaction and the retention of NBS1 to DNA DSBs. Smaller deletions only partially decreased the MDC1–NBS1 interaction and NBS1 foci formation, suggesting the six clusters work in a redundant manner. Second, inhibition of the CK2 activity decreased the MDC1–NBS1 interaction and abolished the recruitment of NBS1 to sites of DNA damage. Finally, a phosphopeptide mimicking CK2 phosphorylation (pSDpTDXD/E) could specifically interact with GST-fusion proteins containing the FHA and BRCT domains of NBS1. Phosphorylation of these SDTDXD/E clusters by CK2 is unlikely to be induced after DNA damage because the MDC1–NBS1 interaction is constitutive (16, 17), and CK2 is not known to be activated after DNA damage. Whether the CK2-dependent phosphorylation of MDC1 and the subsequent MDC1–NBS1 interaction are subjected to regulation by other mechanisms requires further investigation.
We and others have shown that MDC1 directly binds γ-H2AX through its BRCT domains and helps to amplify ATM signaling (15, 24). Several reports also suggest that NBS1 foci formation requires H2AX and MDC1 (15–19, 34). These studies and our current study strongly suggest that MDC1 serves as a mediator between NBS1 and γ-H2AX. Moreover, recent studies demonstrated that the E3 ubiquitin ligase RNF8 interacts with MDC1 in a phosphorylation-dependent manner at a region (residues 698–768) containing three TQXF clusters (29–32). This region is different from the region (residues 200–420) required for NBS1 binding. It seems that phosphorylation of MDC1 at different segments regulates the assembly of distinct subsets of cell cycle checkpoint and repair proteins after DNA damage and therefore ensuring the proper execution of multiple cellular responses to DNA damage (Fig. 5D).
NBS1 contains a FHA and tandem BRCT domains in juxtaposition. Consistent with previous reports, our data suggested that both FHA and the BRCT1 domain of NBS1 are required for its foci formation after DNA damage and its interaction with MDC1. Intriguingly, we found that the BRCT2 domain of NBS1 is also required for its binding to MDC1 and foci formation after DNA damage. These results suggest that each of these domains (FHA, BRCT1, and BRCT2) is necessary but not individually sufficient for mediating its interaction to MDC1 and foci formation after DNA damage. It is not clear how the FHA and tandem BRCT domains of NBS1 bind to the pSDpTDXD/E motif. In contrast to BRCT domains that recognize both pSer- and pThr-containing sequences, FHA domains appear to recognize only pThr-containing motifs. It is tempting to speculate that the FHA domain and the tandem BRCT domain of NBS1 form a compact structure and recognize the double phosphorylated pSDpTDXD/E motifs of MDC1. Structural studies will provide molecular details of the MDC1–NBS1 interaction in the future.
In summary, our data reveal a critical role of MDC1 in promoting the recruitment of NBS1 to DSB sites. The interaction between MDC1 and NBS1 is mediated through CK2 phosphorylation of six SDTDXD/E clusters of MDC1 and the FHA-BRCT domains of NBS1. Functionally, the MDC1–NBS1 interaction is required for the intra-S-phase cell cycle checkpoint control after DNA damage.
Materials and Methods
Plasmids and Reagents.
Human MDC1 cDNA was cloned into a pcDNA3 derivative vector containing HA tag. Human NBS1 cDNA was cloned into a pIRES2 vector with an N-terminal FLAG tag. All deletion mutants were generated by using the QuickChange site-directed mutagenesis kit (Stratagene) and verified by sequencing. Puromycin and CK2 inhibitor DMAT were purchased from Sigma.
Cell Lines.
HeLa cells were purchased from the American Type Culture Collections and maintained in RPMI medium 1640 supplemented with 10% FBS. MDC1-deficient MEF cells were maintained in DMEM with 10% FBS and 1 mM pyruvate. NBS1-deficient cell line NBST (John Petrini, Sloan–Kettering Institute, New York) was maintained in DMEM with 10% FBS. All MDC1 and NBS1 stable reconstitution cell lines were generated by puromycin selection. Cells were irradiated by using a JL Shepherd 137Cs source at the indicated doses.
Antibodies and Transfections.
Monoclonal anti-FLAG and anti-HA antibodies were purchased from Sigma and Covance, respectively. Antibody to pThr was purchased from Cell Signaling. Antibodies against γ-H2AX, BRCA1, MDC1, and 53BP1 were described in refs. 15 and 29. Andre Nussenzweig (National Institutes of Health, Bethesda, MD) provided mouse NBS1 and MRE11 antibodies. Transfections were performed by using Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions.
Coimmunoprecipitation Assay.
For coimmunoprecipitation assays, cells were lysed with NETN buffer (20 mM Tris·HCl at pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF, and 1 μg/ml each of pepstatin A and aprotinin, on ice for 10 min. After removal of cell debris by centrifugation, the whole lysates were incubated with protein-A agarose, coupled with anti-MDC1 antibodies or S-protein agarose beads (Novagen) for 2 h at 4°C. The immunocomplexes were then washed with NETN buffer three times, boiled in 2× Laemmli buffer, and applied to SDS/PAGE. Immunoblotting followed standard procedures.
In Vitro GST Pull-Down and Peptide Pull-Down Assay.
Bacterially expressed GST-fusion proteins or GST alone (2 μg) was immobilized on glutathione-Sepharose 4B beads and incubated for 2 h at 4°C with lysates prepared from cells transiently transfected with plasmids encoding the indicated proteins. After washing with NETN buffer, the samples were separated by SDS/PAGE and analyzed by Western blotting. Biotinylated nonphospho- or phosphopeptides PGFID(p)SD(p)TDVEEE were synthesized by the Mayo Proteomics Core Facility. The peptides were coupled to streptavidin-Sepharose and incubated with GST-NBS1 fragments. Proteins retained on the Sepharose were eluted with biotin (2 mM) and subjected to SDS/PAGE and immunoblotting with anti-GST-antibodies.
Immunofluorescence Staining.
Cells grown on coverslips were fixed with 3% paraformaldehyde in PBS containing 50 mM sucrose at room temperature for 10 min. After permeabilization with 0.5% Triton X-100 buffer containing 20 mM Hepes pH 7.4, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose at room temperature for 5 min, cells were incubated with primary antibodies at 37°C for 20 min. After washing with PBS, cells were incubated with FITC- or rhodamine-conjugated secondary antibodies at 37°C for 20 min. Nuclei were counterstained with DAPI. After a final wash with PBS, coverslips were mounted with glycerol containing para-phenylenediamine.
Identification of MDC1 Phosphorylation Sites by Mass Spectrometry Analysis.
Cell lysates prepared from 293T cells stably expressing S/Flag-MDC1 were subjected to immunoprecipitation by S-protein agarose. The bound proteins were then washed with NETN buffer two times, boiled in Laemmli buffer, and subjected to SDS/PAGE. After Coomassie blue staining, the protein band was excised and subjected to mass spectrometry analysis for mapping phosphorylation sites on MDC1 (Taplin Biological Mass Spectrometry Facility, Harvard Medical School, Boston).
Radioresistant DNA Synthesis Assay.
Radioresistant DNA synthesis was assayed as described in ref. 33. In brief, cells were irradiated at the indicated dose (0, 10, 20, or 40 Gy) or left untreated. Twenty minutes later, cells were pulsed with [3H]thymidine and collected 40 min later by using a Cell Harvester (Molecular Devices). The 3H incorporation into DNA was measured in a liquid scintillation counter.
Acknowledgments.
We thank Dr. John Petrini for the NBST cells and Dr. Andre Nussenzweig for anti-mouse NBS1 and MRE11 antibodies. This work was supported in part by grants from the National Institutes of Health (to J.C.). Z.L was supported by a Susan G. Komen Breast Cancer Foundation Research Grant and the Mayo Cancer Center start-up fund. J.C. is the recipient of an Era of Hope Scholars award from the Department of Defense and a member of Mayo Clinic Breast Specialized Program of Research Excellence program.
Note Added in Proof.
While this work was under review, three related papers were published (35–37). These studies agree with our conclusion and support a role of MDC1/NBS1 interaction in DNA damage checkpoint control.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Varon R, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 2.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 4.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 5.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson CT, et al. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horejsi Z, et al. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–3127. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 11.Lim DS, et al. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 12.Yazdi PT, et al. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker E, Meyer V, Madaoui H, Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- 15.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 17.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 18.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Stern DF. NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. FASEB J. 2003;17:1842–1848. doi: 10.1096/fj.03-0310com. [DOI] [PubMed] [Google Scholar]
- 20.Tauchi H, et al. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J Biol Chem. 2001;276:12–15. doi: 10.1074/jbc.C000578200. [DOI] [PubMed] [Google Scholar]
- 21.Zhao S, Renthal W, Lee EY. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 2002;30:4815–4822. doi: 10.1093/nar/gkf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi J, et al. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 23.Lu R, Niida H, Nakanishi M. Human SAD1 kinase is involved in UV-induced DNA damage checkpoint function. J Biol Chem. 2004;279:31164–31170. doi: 10.1074/jbc.M404728200. [DOI] [PubMed] [Google Scholar]
- 24.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 26.Pagano MA, et al. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: A novel powerful and selective inhibitor of protein kinase CK2. Biochem Biophys Res Commun. 2004;321:1040–1044. doi: 10.1016/j.bbrc.2004.07.067. [DOI] [PubMed] [Google Scholar]
- 27.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 34.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melander F, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spycher C, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman JR, Jackson SP. Phospho-dependent interactions between NSB1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008 doi: 10.1038/embor.2008.103. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]