Abstract
Niemann–Pick C1-like protein (NPC1L1) mediates the absorption of dietary cholesterol in the proximal region of the intestine, a process that is blocked by cholesterol absorption inhibitors (CAIs), including ezetimibe (EZE). Using a proteomic approach, we demonstrate that NPC1L1 is the protein to which EZE and its analogs bind. Next, we determined the site of interaction of EZE analogs with NPC1L1 by exploiting the different binding affinities of mouse and dog NPC1L1 for the radioligand analog of EZE, [3H]AS. Chimeric and mutational studies indicate that high-affinity binding of [3H]AS to dog NPC1L1 depends on molecular determinants present in a 61-aa region of a large extracellular domain (loop C), where Phe-532 and Met-543 appear to be key contributors. These data suggest that the [3H]AS-binding site resides in the intestinal lumen and are consistent with preclinical data demonstrating in vivo efficacy of a minimally bioavailable CAI. Furthermore, these determinants of [3H]AS binding lie immediately adjacent to a hotspot of human NPC1L1 polymorphisms correlated with hypoabsorption of cholesterol. These observations, taken together with the recently described binding of cholesterol to the N terminus (loop A) of the close NPC1L1 homologue, NPC1, may provide a molecular basis for understanding EZE inhibition of NPC1L1-mediated cholesterol absorption. Specifically, EZE binding to an extracellular site distinct from where cholesterol binds prevents conformational changes in NPC1L1 that are necessary for the translocation of cholesterol across the membrane.
Whole-body cholesterol homeostasis is maintained through three major pathways: de novo synthesis, intestinal absorption, and biliary excretion. Absorption of dietary and biliary cholesterol occurs in the proximal jejunum of the small intestine (1), and this process is blocked by ezetimibe (EZE), a drug used for the treatment of hypercholesterolemia. EZE, a potent cholesterol and phytosterol uptake inhibitor, effectively lowers circulating plasma cholesterol in humans by 15–20% (2), and its coadministration with 3-hydroxy-3-methylglutaryl CoA (HMG CoA) reductase inhibitors (statins), inhibitors of cholesterol synthesis, leads to further reductions in cholesterol plasma levels (3).
By searching expressed sequence tag databases for the presence of a sterol-sensing domain (SSD), a plasma membrane secretion signal, and enriched expression in intestinal enterocytes, the Niemann–Pick C1-Like 1 (NPC1L1) protein was identified in 2004 as a potential candidate gene for the EZE-sensitive pathway of cholesterol absorption (4). Mice deficient in NPC1L1 were found to have ≈70% reduction in sterol absorption, with the residual component being insensitive to EZE (4), suggesting that NPC1L1 is a critical component of cholesterol uptake in enterocytes (4). The use of enterocyte brush border membranes (BBMs) from several species, including NPC1L1 KO mice (5), or membranes derived from cells expressing recombinant NPC1L1, has provided strong evidence for NPC1L1 being the target to which EZE binds (5).
More recently, in vitro studies have demonstrated EZE-sensitive cholesterol transport into McArdles RH7777 hepatoma (6), CaCo-2 (7), and MDCKII cells (8) overexpressing NPC1L1. However, although EZE-sensitive cholesterol transport correlates well with the amount of NPC1L1 expression (7, 8), it is not possible to determine whether NPC1L1 functions alone or as part of a multiprotein complex [SR-B1 (9–15), CD36 (14), CD13 (9), caveolin-1/annexin-2 (16)], facilitating the transfer of cholesterol from outside the cell to internal cholesterol pools (9, 15). Furthermore, it has been speculated that EZE may inhibit cholesterol transfer by binding to some of these other targets, in addition to its inhibition of NPC1L1 (9).
In the present study, we attempted to determine whether EZE binds to NPC1L1 directly by purifying an EZE-NPC1L1 complex and analyzing its constituents by mass spectrometry. Quantitative analysis unambiguously demonstrated that NPC1L1 is the only protein to account for EZE binding. Subsequently, using a chimera/mutagenesis approach that takes advantage of the large difference in affinities between dog and mouse NPC1L1 for EZE-like compounds, we identified two residues in a large extracellular loop of NPC1L1 that are mostly responsible for the large difference in affinity between the two species, and that reside adjacent to a hotspot of human polymorphisms associated with reduced cholesterol absorption (17). Based on the assumption that region(s) associated with cholesterol binding to NPC1L1 and its close homologue NPC1 are similar, we propose a model for the action of EZE on NPC1L1-mediated cholesterol absorption. In this model, cholesterol binds to an extracellular binding site on NPC1L1 and is transported across the membrane through an undisclosed mechanism. EZE binding to a distinct extracellular site in NPC1L1 prevents certain conformational changes in NPC1L1 that are necessary for cholesterol flux across the membrane. This model is consistent with the human genetic NPC1L1 polymorphisms and with preclinical data that indicate inhibitor absorption is not critical for in vivo efficacy of the drugs.
Results
Affinity Purification of Dog NPC1L1-Kv1.1:[3H]AS.
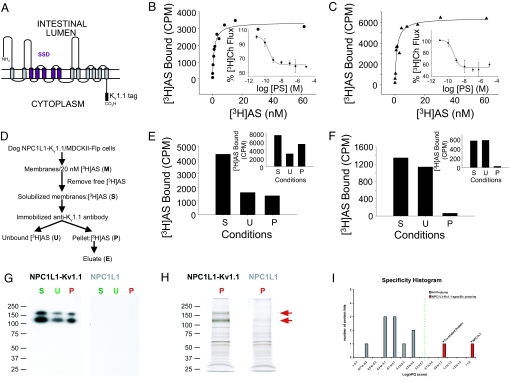
To determine whether EZE analogs bind directly to NPC1L1, we developed an affinity purification strategy for which an 18-aa sequence, previously used to effectively affinity purify native brain potassium channels (18), was engineered into the C terminus of dog NPC1L1 ((Fig. 1A), dog NPC1L1-Kv1.1). Under equilibrium-binding conditions, dog NPC1L1-Kv1.1 expressed in MDCKII-Flp cells (dog NPC1L1-Kv1.1/MDCKII-Flp cells) binds [3H]AS in a similar manner to dog NPC1L1 stably expressed in MDCKII-Flp cells (dog NPC1L1/MDCKII-Flp cells) [Kd of 1.12 ± 0.32 nM (n = 5) and 1.07 ± 0.18 nM (n = 3) for dog NPC1L1-Kv1.1 and dog NPC1L1, respectively; Fig. 1 B and C]. Furthermore, [3H]Ch flux into both MDCKII-Flp cell lines was blocked by PS (8) with similar characteristics [IC50 of 0.25 ± 0.03 nM (n = 3) and 0.34 ± 0.12 nM (n = 3) for dog NPC1L1-Kv1.1 and dog NPC1L1, respectively; Fig. 1 B and C Insets]. Together, these observations indicate that the introduction of the Kv1.1 tag does not significantly alter the functional properties of dog NPC1L1 expressed in MDCKII-Flp cells.
Fig. 1.
[3H]AS binds directly to dog NPC1L1-Kv1.1. (A) 2D model of dog NPC1L1-Kv1.1. The membrane topology of dog NPC1L1 was predicted with HMMTOP and TMHMM servers available through http://expasy.org/tools/#ptm and manually refined. The pentahelical SSD is highlighted in purple. (B and C) Dog NPC1L1-Kv1.1 is functional. Dog NPC1L1-Kv1.1/MDCKII-Flp (B) or dog NPC1L1/MDCKII-Flp (C) cells were seeded on 96-well plates and incubated with increasing concentrations of [3H]AS for 4 h at 37°C. Specific binding was fit to a single-site saturation model yielding Kd/Bmax values of 1.15 nM/3370 cpm for Dog NPC1L1-Kv1.1/MDCKII-Flp cells (●) and 1.28 nM/6436 cpm for dog NPC1L1/MDCKII-Flp cells (▲). (Inset) [3H]cholesterol flux into dog NPC1L1-Kv1.1/MDCKII-Flp cells (●) and dog NPC1L1/MDCKII-Flp cells (▲) was performed as described in the Experimental Procedures in the presence of increasing concentrations of PS. [3H]Ch flux was fit to a single-site inhibition model, yielding IC50 values of 0.21 (B) and 0.24 nM (C) for dog NPC1L1-Kv1.1/MDCKII-Flp cells (●) and dog NPC1L1/MDCKII-Flp cells (▲), respectively. (D) Strategy for affinity purification of dog NPC1L1-Kv1.1. (E and F) Immunoprecipitation of dog NPC1L1-Kv1.1 from membranes and cells. Membranes from dog NPC1L1-Kv1.1/MDCKII-Flp (E) or dog NPC1L1/MDCKII-Flp (F) cells were incubated with 20 nM [3H]AS overnight and solubilized with 1% digitonin/0.03% sodium taurocholate for 30 min at 4°C, as described in Experimental Procedures. Solubilization of 35% or 26% of membrane bound [3H]AS activity from either dog NPC1L1-Kv1.1 or dog NPC1L1 membranes, respectively, was obtained. Solubilized [3H]AS activity (S) was incubated with protein A Sepharose beads coated with an anti-Kv1.1 antibody for 3 h at 4°C. Unbound [3H]AS activity (U) was collected, and the beads were washed three times before determination of [3H]AS bound (P). [3H]AS in U and P has been corrected to account for the dissociation of bound [3H]AS (t1/2 ≈6 h) during the time of immunoprecipitation. [3H]AS recovered in P of dog NPC1L1/MDCKII-Flp is identical to that obtained in the absence of anti-Kv1.1 antibody. Dog NPC1L1-Kv1.1/MDCKII-Flp (E Inset) or dog NPC1L1/MDCKII-Flp (F Inset) cells were incubated with 20 nM [3H]AS overnight. Free [3H]AS was removed from cells by aspiration as described in Experimental Procedures, and the [3H]AS activity bound to cells was solubilized with 1% digitonin/0.03% sodium taurocholate for 30 min at 4°C. The solubilized [3H]AS activity (S) was immunoprecipitated as indicated in D. Data in E and F are representative of 10 and 8 independent experiments from membranes and cells, respectively. (G) Characterization of affinity-purified dog NPC1L1-Kv1.1. The solubilized (S), unbound (U), and purified (P) material of the immunoprecipitation from dog NPC1L1-Kv1.1/MDCKII-Flp and dog NPC1L1/MDCKII-Flp membranes was resolved by SDS/PAGE, transferred onto a PVDF membrane, and analyzed with an anti-Kv1.1 antibody. Two proteins of Mr 125 and 165 kDa are specifically recognized by the anti-Kv1.1 antibody in the three NPC1L1-Kv1.1 but not NPC1L1 samples. (H) Purified material (P) from G was resolved by SDS/PAGE and visualized by silver staining. Two bands at 125 and 165 kDa (indicated by red arrows) are present in the material purified from dog NPC1L1 Kv1.1 but not dog NPC1L1. (I) Quantification histogram displaying the relative specificities scored by relative peptide queries (rPQ) of affinity-purified proteins identified by LC-MS/MS sequencing of the gel lanes in H. * indicates proteins for which MS/MS spectra are assigned from dog NPC1L1 Kv1.1 but not dog NPC1L1-material. Values were 12 queries for NPC1L1 (gi 148223061) and 2 queries for Trx-related protein (gi 73963782).
To identify and optimize appropriate solubilization conditions, membranes were preincubated with [3H]AS, exposed to detergent, and, after centrifugation, the relative amounts of NPC1L1 bound [3H]AS in the supernatant and pellet fractions were determined (Fig. 1D). Incubation of the solubilized [3H]AS complex with an anti-Kv1.1 antibody immobilized to beads caused immunoprecipitation of almost 50% of the [3H]AS-binding sites of dog NPC1L1-Kv1.1 (Fig. 1E). In contrast, <4% of the [3H]AS-binding activity solubilized from dog NPC1L1 membranes could be immunoprecipitated, a value identical to that obtained using beads without anti-Kv1.1 antibody conjugated (data not shown) (Fig. 1F). In addition, experiments carried out with material solubilized from either dog NPC1L1-Kv1.1/MDCKII-Flp or dog NPC1L1/MDCKII-Flp cells indicated that >65% and <4% of the solubilized [3H]AS-binding sites could be immunoprecipitated by immobilized anti-Kv1.1 antibody, respectively (Fig. 1 E and F Insets).
Immunoprecipitation was monitored at the protein level by immunoblotting with an anti-Kv1.1 antibody and silver staining combined with LC-MS/MS sequencing. Immunoblotting analysis revealed that two bands, present in the immunoprecipitated material with apparent molecular weights of 125 and 165 kDa, are identical to those detected in solubilized and unbound material (Fig. 1G). The relative intensities of the bands indicate a relative purification efficiency (U/P) of ≈50%, in line with the values determined for [3H]AS-binding sites (Fig. 1E). No cross-reactive material was identified from identical dog NPC1L1 samples lacking the Kv1.1 tag (Fig. 1G). Silver staining of the immunoprecipitated material indicated the specific purification of two major proteins with apparent Mr of 125 and 165 kDa (Fig. 1H, arrows) from dog NPC1L1-Kv1.1. Consistent with these results, mass spectral analysis identified dog NPC1L1 with the highest significance and specificity in the dog NPC1L1-Kv1.1-tagged sample [12 matched MS/MS spectra vs. no peptide signal in the untagged dog NPC1L1 material; Fig. 1I, supporting information (SI) Table S1]. One additional protein was specifically identified in the dog NPC1L1-Kv1.1 material with much lower abundance (Trx-related protein, possibly a protein disulphide isomerase, with two matched MS/MS spectra, Fig. 1I, Table S1). In summary, NPC1L1 alone has been unambiguously identified as the protein underlying [3H]AS binding to solubilized dog NPC1L1-Kv1.1 expressed in MDCKII-Flp cells.
Binding of [3H]AS to Dog or Mouse NPC1L1 Expressed in TsA-201 Cells.
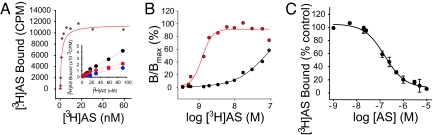
Given that [3H]AS binds directly to dog NPC1L1, we developed a strategy to identify residues involved in [3H]AS binding by comparing the binding properties of [3H]AS to high-affinity dog NPC1L1 and low-affinity mouse NPC1L1. Under equilibrium-binding conditions, [3H]AS binds to dog NPC1L1 transiently expressed in TsA-201 cells with a Kd of 1.04 ± 0.37 nM (n = 13) (Fig. 2 A and B, Table 1). TsA-201 cells transiently transfected with mouse NPC1L1 bind [3H]AS, but Kd values are difficult to estimate because of the relatively small component of specific to nonspecific [3H]AS binding present at the high radioligand concentrations necessary to saturate the low affinity mouse site (Fig. 2 A Inset and B). However, at radioligand concentrations (50–150 nM), where a reliable specific binding component can be measured, it is possible to characterize the pharmacological properties of the binding reaction. Under these conditions, the ability of β-lactams to compete with [3H]AS is consistent with binding of the radioligand to mouse NPC1L1 [PS (Ki = 50.2 nM) > EZE-Gluc (Ki = 1.21 μM) > EZE (Ki = 2.41 μM) > EZE and EZE-gluc enantiomers (Ki ND) [Fig. 2C, supporting information (SI) Fig. S1].
Fig. 2.
Binding of [3H]AS to TsA-201 cells expressing dog or mouse NPC1L1. (A) Saturation studies. TsA-201 cells were transiently transfected with dog NPC1L1 and incubated with increasing concentrations of [3H]AS for 4 h at 37°C as indicated in Experimental Procedures. Nonspecific binding is determined in the presence of 100 μM EZE-gluc. Specific binding, defined as the difference between total and nonspecific binding, is presented. Specific binding for dog NPC1L1 (red circle) is a saturable function of [3H]AS concentration and displays a Kd of 1.37 nM (Dog NPC1L1 Bmax = 11455 cpm). (Inset) TsA-201 cells were transiently transfected with mouse NPC1L1 and [3H]AS binding was determined as described in A. Total (black circle), specific (red circle), and nonspecific (blue circle) binding are presented. (B) Specific binding data from A are presented relative to the maximum receptor occupancy. (C) Pharmacology of [3H]AS binding to mouse NPC1L1. Cells were incubated with 150 nM [3H]AS in the presence or absence of increasing concentrations of PS (●) for 4 h at 37°C. Inhibition of binding was assessed relative to an untreated control. Specific binding was fit to a single-site inhibition model, yielding a Ki value of (●) 51 nM (PS).
Table 1.
[3H]AS binding to NPC1L1 chimeras and mutants
| NPC1L1 chimeras | Kd, nM | NPC1L1 mutants | Kd, nM |
|---|---|---|---|
| Dog NPC1L1 | 1.04 ± 0.37 (n = 13) | Dog NPC1L1 | 1.04 ± 0.37 (n = 13) |
| Mouse NPC1L1* | 51.2 ± 4.99 (n = 3) | Mouse NPC1L1* | 51.2 ± 4.99 (n = 3) |
| Chimera 1 | 49.1 ± 20 (n = 2) | ||
| Chimera 2 | 1.79 ± 0.54 (n = 4) | Y532F/Mouse | 13.25 ± 3.22 (n = 4) |
| Chimera 3 | 3.28 ± 1.4 (n = 3) | I543M/Mouse | 9.3 ± 1.86 (n = 4) |
| Chimera 4 | 1.73 ± 0.37 (n = 4) | Y532F/I543M/Mouse | 2.61 ± 1.2 (n = 3) |
| Chimera 5 | 1.2 ± 0.28 (n = 3) | ||
| Chimera 6 | 1.04 ± 0.29 (n = 4) | F532Y/Dog | 5.33 ± 1.86 (n = 3) |
| Chimera 7 | 2.89 ± 0.98 (n = 4) | M543I/Dog | 2.16 ± 0.4 (n = 3) |
| Chimera 8 | 79.9 ± 46.4 (n = 3) | F532Y/M543I/Dog | 18.35 ± 11.39 (n = 3) |
| Chimera 9 | 56.9 ± 0.9 (n = 2) | ||
| Chimera 10 | 51.45 ± 19.2 (n = 2) | ||
| Chimera 11 | 1.59 ± 0.13 (n = 4) |
[3H]AS binding to NPC1L1 constructs was determined as described in Experimental Procedures. Kdvalues represent the average ± standard error of indicated number of experiments.
*Ki values are reported for mouse NPC1L1.
Molecular Determinants of High-Affinity [3H]AS Binding Lie in Loop C.
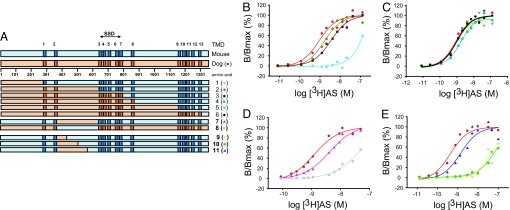
Because of the significant difference in [3H]AS-binding affinity between dog and mouse NPC1L1, a series of full-length dog/mouse NPC1L1 chimeras possessing progressively larger domains of dog NPC1L1 were generated and characterized to determine the region of NPC1L1 responsible for high-affinity ligand binding (Fig. 3A, Table S2). Results from these experiments are illustrated in Fig. 3B. Replacing the large N terminus of mouse NPC1L1 with dog NPC1L1 ((D1–264/M265–1333, chimera 1) did not significantly change the chimera's affinity for [3H]AS binding [Kd = 49.1 ± 20 nM (n = 2)]. In contrast, chimera 2, in which the N terminus, two transmembrane domains (TMDs) and loop C of mouse NPC1L1 have been replaced with the corresponding regions of dog NPC1L1 (D1–629/M630–1333, chimera 2) binds [3H]AS with similar characteristics to those of full length dog NPC1L1, Kds of 1.79 ± 0.54 nM (n = 4) and 1.04 ± 0.37 nM (n = 13) for chimera 2 and dog NPC1L1, respectively. As expected, chimeras 3 (D1–806/M807–1333), 4 (D1–873/M874–1333), 5 (D1–1102/M1104–1333), and 6 (D1–1268/M1270–1333), which progressively contain more dog NPC1L1 in the mouse NPC1L1 background, display similar [3H]AS-binding features to those of dog NPC1L1 [(Figs. 3 B and C), Kds of 3.28 ± 1.4 nM (n = 3), 1.73 ± 0.37 nM (n = 4), 1.2 ± 0.28 nM (n = 3), and 1.04 ± 0.29 nM (n = 4), respectively].
Fig. 3.
Binding of [3H]AS to dog/mouse NPC1L1 chimeras. (A) Schematic of dog/mouse chimeras. Dog (orange) and mouse (blue) NPC1L1 sequences are shown, with their predicted TMDs indicated in dark color. SSD, between amino acids 629 and 806 is indicated by an arrow. (B) [3H]AS binding to dog/mouse chimeras 1–3. Saturation studies. TsA-201 cells transfected with dog NPC1L1 (●) and chimeras 1 (▼), 2 (♦), or 3 (■) were incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding data were fit to a single class of [3H]AS-binding sites and are presented relative to the maximum receptor occupancy; dog NPC1L1 [(●) Kd 0.91 nM, Bmax 9,873 cpm], 1 [(▼) Kd 102 nM, Bmax 464 cpm], 2 [(♦) Kd 1.66 nM, Bmax 15250 cpm] and chimera 3 [(■) Kd 3.54 nM, Bmax 8,277 cpm]. (C) [3H]AS binding to dog/mouse chimeras 4–6. Saturation studies. TsA-201 cells transfected with dog NPC1L1 (●) and chimeras 4 (■), 5 (▼) or 6 (●) were incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding was fit to a single class of [3H]AS-binding sites and is presented relative to the maximum receptor occupancy; dog NPC1L1 [(●) Kd 0.91 nM, Bmax 9,873 cpm], 4 [(■) Kd 1.91 nM, Bmax 9,802 cpm], 5 [(▼) Kd 0.81 nM, Bmax 13,014 cpm] and 6 [(●) Kd 1.01 nM, Bmax 15,443 cpm]. (D) [3H]AS binding to dog/mouse loop C exchange chimeras 7–8. Saturation studies. TsA-201 cells were transfected with dog NPC1L1 (●) and loop C exchange chimeras 7 (▲) or 8 (■) and incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding data were fit to a single class of [3H]AS-binding sites and are presented relative to the maximum receptor occupancy; dog NPC1L1 [(●) Kd 1.23 nM, Bmax 8,597 cpm], 7 [(▲) Kd 3.38 nM, Bmax 43,683 cpm] and 8 [(■) Kd 63 nM, Bmax 2081 cpm]. (E) [3H]AS binding to dog/mouse loop C exchange chimeras 9–11. Saturation studies. TsA-201 cells were transfected with dog NPC1L1 (●) and chimeras 9 (▼), 10 (■), or 11 (▲) and incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding data were fit to a single class of [3H]AS-binding sites and are presented relative to the maximum receptor occupancy; dog NPC1L1 [(●) Kd 0.43 nM, Bmax 6,976 cpm], 9 [(▼) Kd 46.8 nM, Bmax 2,321 cpm], 10 [(■) Kd 71 nM, Bmax 5,759 cpm] and 11 [(▲) Kd 1.38 nM, Bmax 11,297 cpm].
These observations implicating loop C in high-affinity [3H]AS binding are consistent with the physiochemical properties of [3H]AS and its low levels of nonspecific binding, which suggest a hydrophilic- rather than a membrane-based binding site (8). Given that the SSD of dog and mouse NPC1L1 resides in a pentahelical membrane region (amino acids 629–806), and that the homologous region to the cholesterol-binding region of NPC1 (19, 20), loop A, resides between amino acids 1–240, it appears that the molecular determinants for [3H]AS binding (between 265–629) are distinct from those involved in cholesterol binding.
To further pursue the molecular determinants for [3H]AS binding, other chimeras were generated. Replacing loop C of mouse NPC1L1 with dog NPC1L1 (M1–386/D387–629/M630–1333, chimera 7) led to a significant increase in [3H]AS-binding affinity [(Fig. 3D), Kds of 1.04 ± 0.37 nM (n = 13) and 2.89 ± 0.98 nM (n = 4) for dog NPC1L1 and chimera 7, respectively], and consistent with this finding, when the extracellular loop of dog NPC1L1 was replaced with the equivalent region of mouse NPC1L1 (D1–386/M387–629/D630–1325, chimera 8), a loss of [3H]AS binding occurred [(Fig. 3D), Kds of 1.04 ± 0.37 nM (n = 13) and 79.9 ± 46.4 nM (n = 3) for dog NPC1L1 and chimera 8, respectively]. These data, taken together, suggest that the major molecular determinants of high-affinity [3H]AS binding are present in loop C of dog NPC1L1.
To determine whether loop C alone is capable of binding [3H]AS, we attempted to express amino acids 374–634 of dog NPC1L1 in bacteria as a DsbC or GST fusion protein or in mammalian cells as a CD8-loop C or maxiK β-loop C fusion protein (data not shown). However, specific [3H]AS binding could not be detected with any of these constructs after transient transfection in TsA201 cells. Although our arbitrary choice of fusion protein junctions near the predicted TMD2 and 3 helix:loop interfaces may have affected the correct folding and/or stability of the loop C fusion proteins, it is also possible that loop C requires interaction(s) with other region/domain(s) of NPC1L1 for correct folding, stability and/or [3H]AS binding.
Loop C is predicted to be >250 aa in length, and therefore, in an effort to narrow the molecular determinants of high-affinity [3H]AS binding, a new set of chimeras were designed [chimeras 9 (M1–386/D387–434/M435–1333), 10 (M1–386/D387–509/M510–1333), and 11 (M1–386/D387–571/M572–1333)], in which progressively larger sections of mouse NPC1L1 loop C were replaced with those from dog NPC1L1 loop C (Fig. 3E). Results from these experiments clearly indicate that the region of loop C containing amino acids 510–571 is critical for high-affinity [3H]AS binding [(Fig. 3E), Kds of 56.9 ± 0.9 nM (n = 2), 51.45 ± 19.2 (n = 2), and 1.59 ± 0.13 nM (n = 4), respectively].
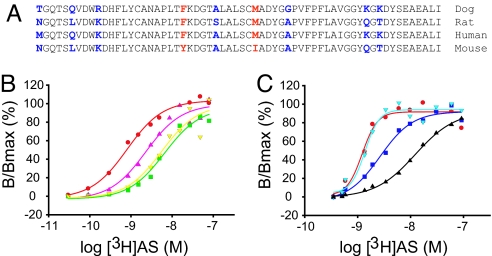
Gain and Loss of High-Affinity [3H]AS-Binding Mutations in NPC1L1.
Alignment of several NPC1L1 species with different [3H]AS-binding affinities across the 510- to 571-aa region identifies nine positions of sequence variation (Fig. 4A). Although dog, rat, and human NPC1L1 bind [3H]AS with affinities of ≈1, 5, and 10 nM, respectively, mouse NPC1L1 binds [3H]AS with significantly lower affinity. Therefore, we searched for amino acid positions identical in dog, rat, and human NPC1L1 and different in mouse NPC1L1, reasoning that these substitutions may be responsible for the changes in [3H]AS-binding affinity. This led to the identification of two positions, amino acids 532 and 543, as candidates for high-affinity [3H]AS binding in dog NPC1L1. Therefore, these two dog residues were then replaced individually or together into the full-length mouse NPC1L1. Individual substitutions at either position significantly increased [3H]AS-binding affinity to mouse NPC1L1 [(Fig. 4B), Kds of 1.04 ± 0.37 nM (n = 13), 13.25 ± 3.22 nM (n = 4), and 9.3 ± 1.86 nM (n = 4) for dog NPC1L1, Tyr-532→Phe and Ile-543→Met, respectively]. Furthermore, in the double mouse mutant Tyr-532→Phe/Ile-543→Met, [3H]AS-binding affinity is only 3-fold lower than that of dog NPC1L1 [(Fig. 4B), Kd 2.61 ± 1.2 nM (n = 3)]. Interestingly, in the reciprocal experiment, substitution of dog NPC1L1 residues at either position 532 or 543 with the corresponding mouse residues has only a minor effect on [3H]AS binding [(Fig. 4C), Kds of 1.04 ± 0.37 nM (n = 13), 5.33 ± 1.86 nM (n = 3) and 2.16 ± 0.4 nM (n = 3) for dog NPC1L1, Phe-532→Tyr and Met-543→Ile, respectively]. However, in the double dog NPC1L1 mutant, Phe-532→Tyr/Met-543→Ile, [3H]AS-binding affinity appears to be significantly altered [(Fig. 4C), Kd 18.35 ± 11.39 nM (n = 3)].
Fig. 4.
Two residues in loop C are critical for high-affinity [3H]AS binding to NPC1L1. (A) Sequence alignment of a 61-aa subdivision across multiple species of NPC1L1. The amino acid sequence of NPC1L1 (residues 510–571) corresponding to high-affinity (dog, rat, and human) or low-affinity (mouse) [3H]AS binding is shown. Nonconserved positions for which there is a correlation between ligand binding affinity and amino acid identity are highlighted in red, whereas those nonconserved positions where there is no correlation between ligand-binding affinity and amino acid identity are highlighted in blue. (B) Gain of [3H]AS-binding NPC1L1 mutants. TsA-201 cells were transfected with dog NPC1L1 (red circle) or mouse NPC1L1 with the mutations Tyr-532→Phe (green square), Ile-543→Met (yellow inverted triangle), or Tyr-532→Phe/Ile-543→Met (magenta triangle) and incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding data were fit to a single class of [3H]AS-binding sites and are presented relative to the maximum receptor occupancy; dog NPC1L1 [(red circle) Kd 0.81 nM, Bmax 3,878 cpm], Tyr-532→Phe [(green square) Kd 6.6 nM, Bmax 7,873 cpm], Ile-543→Met [(yellow inverted triangle) Kd 5.6 nM, Bmax 11,313 cpm], and Tyr-532→Phe/Ile-543→Met [(magenta triangle) Kd 2.29 nM, Bmax 12,701 cpm]. (C) Loss of [3H]AS-binding NPC1L1 mutants. TsA-201 cells were seeded and transfected with dog NPC1L1 (red circle) or dog NPC1L1 with the mutations Phe-532→Tyr (dark blue square), Met-543→Ile (cyan inverted triangle), or Phe-532→Tyr/Met-543→Ile (black triangle) and incubated with increasing concentrations of [3H]AS, as indicated in Experimental Procedures. Specific binding data were fit to a single class of [3H]AS-binding sites and are presented relative to the maximum receptor occupancy; dog NPC1L1 [(red circle) Kd 1.23 nM, Bmax 11,445 cpm], Phe-532→Tyr [(dark blue square) Kd 2.69 nM, Bmax 8,101 cpm], Met-543→Ile [(cyan inverted triangle) Kd 1.38 nM, Bmax 8,258 cpm] and Phe-532→Tyr/Met-543→Ile [(black triangle) Kd 12.4 nM, Bmax 7,940 cpm].
Although there are subtle differences in the extent of change in [3H]AS binding between reciprocal dog→mouse and mouse→dog NPC1L1 chimeras/mutations, these variations are relatively minor in energetic terms. However, given that substituting residues and/or regions of dog NPC1L1 loop C into mouse NPC1L1 (e.g., chimeras 2, 7, 11, and the double mouse mutant Tyr-532→Phe/Ile-543→Met) did not result in high-affinity [3H]AS binding indistinguishable from wild-type dog NPC1L1, we cannot completely exclude the possibility that other regions of NPC1L1 may also contribute to [3H]AS binding.
Discussion
The current study used biochemical and proteomic approaches to determine the nature of the interaction between NPC1L1 and EZE analogs. The results showing that the noncovalent [3H]AS-NPC1L1 complex can be affinity purified from MDCKII cells, a pharmacologically validated system for EZE-sensitive cholesterol flux (8), provide compelling evidence that EZE analogs bind to NPC1L1 directly.
Immunoblot analysis showed good quantitative correlation of NPC1L1 with [3H]AS binding during NPC1L1 affinity purification. In line with the specific band pattern obtained from silver staining of the affinity-purified material, comprehensive high-sensitivity nano-LC-MS/MS sequencing identified dog NPC1L1 as the dominant specifically purified protein. In addition, Trx-related protein, a homolog of ER protein disulphide isomerases, has been identified as a putative low-abundance interaction partner. Although the physiological relevance of this protein is unclear, it may provide an efficient means of folding the Cys-rich luminal loops of NPC1L1. Notably, our findings provided no evidence that support physical interaction between NPC1L1 and SR-B1, CD36, CD13, caveolin-1/annexin-2 (9), or the existence of other yet-unknown NPC1L1 interaction partners in MDCKII cells.
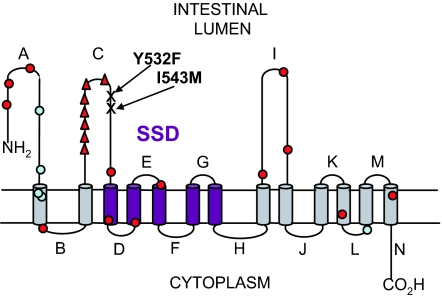
Alignment of NPC1L1 with mammalian and prokaryotic homologues, for which structural information is available, indicates that NPC1L1 is likely to consist of 13 TMD with an extracellular N terminus and cytoplasmic C terminus (21). Based on our demonstration that dog NPC1L1 binds [3H]AS directly, we exploited the differences in affinity between dog and mouse NPC1L1 for EZE analogs in a chimeric approach to identify a region of extracellular loop C between positions 510 and 571 that is critical for high-affinity [3H]AS binding (Fig. 5). In particular, two amino acids present in this region of dog NPC1L1 (Phe-532 and Met-543) and conserved in human NPC1L1 appear to represent key molecular determinants for the high-affinity interaction of [3H]AS and EZE analogs.
Fig. 5.
2D model of human NPC1L1. The membrane topology of human NPC1L1 was predicted with HMMTOP and TMHMM servers available through http://expasy.org/tools/#ptm and manually refined. Predicted loops are labeled throughout the protein sequence (A–N). The pentahelical SSD is highlighted in purple. Key residues for [3H]AS binding (Phe/Tyr-532 and Met/Ile-543) and sequence variants found exclusively in low (red circle) or high (light blue circle) absorbers of cholesterol as identified in the Dallas Heart study (17) are shown. The hotspot of hypoabsorption polymorphisms adjacent to key residues determining [3H]AS binding are highlighted (red triangle).
Physiologically, an extracellular binding site for transmembrane domains (CAIs) would explain the efficacy of a minimally absorbed CAI from microbia (<0.4% bioavailability) (22, 23) that is currently being developed for the treatment of hypercholesterolemia. Although the compound possesses minimal absorption, it has comparable preclinical efficacy to EZE, which is absorbed into the systemic circulation (22). These observations are consistent with the notion that CAIs bind to extracellular region(s) of NPC1L1 in the intestinal lumen and block the ability of NPC1L1 to transport cholesterol into the enterocyte cytoplasm. In addition, these data do not support the notion from other studies that inhibition of NPC1L1 by these agents occurs from the cytoplasm of the enterocyte (24).
Until recently, the only direct information on the regions and residues of NPC1L1 implicated in cholesterol absorption was implied from human genetics (17). Sequencing of NPC1L1 coding regions in individuals from the Dallas Heart Study who had the highest and lowest cholesterol absorption, determined by ratio of campesterol to lathosterol (Ca:L), identified rare nonsynonymous variants in human NPC1L1 associated with hypoabsorption of cholesterol (17). Although most of the hypoabsorption polymorphisms are distributed evenly throughout the primary sequence of human NPC1L1, a single hotspot exists in loop C, close to the two residues that we have shown to be critical for [3H]AS binding (Fig. 5). These observations are compatible with the notion that individual changes in the conformation of loop C at distinct and independent sites (polymorphisms and [3H]AS-binding determinants) translate into an effect on cholesterol transport without these sites communicating with each other. However, heterologous expression of human NPC1L1 loop C hypoabsorption polymorphisms in CHO-K1 cells have recently been associated with low levels of protein expression (25), and our preliminary data indicate that the affinity of [3H]AS for these loop C hypoabsorption polymorphisms is not significantly altered, whereas site density appears to be reduced (data not shown), providing an alternative explanation for the hypoabsorption phenotype of these polymorphisms. Although the functional consequences of the loop C polymorphisms and [3H]AS-binding mutations require further elucidation, these data clearly illustrate the importance of loop C in the overall mechanism of cholesterol absorption.
Interestingly, a recent study on NPC1, the closest homologue of NPC1L1 in the SSD-containing proteins, demonstrates that cholesterol binds to its N-terminal domain (19, 20). Briefly, [3H]Ch binds with the same affinity to full-length human NPC1 and to an N-terminal 240-aa region. Although the physiological significance of cholesterol binding to NPC1 function is unclear (19, 20), these unexpected observations challenge the dogma that cholesterol would bind only to the SSD of SSD-containing proteins (26). Although direct binding of cholesterol to the full-length and N-terminal domain of NPC1L1 will be necessary to evaluate their contributions to cholesterol binding and/or function, the results with NPC1 allow us to hypothesize that the N terminus, and not loop C, of NPC1L1 may be involved in cholesterol binding.
Thus, combining the cholesterol-binding data on human NPC1 (19, 20) with our biochemical and proteomic EZE analog binding, human genetics (17), and preclinical pharmacology (22), we propose a model for NPC1L1-mediated cholesterol flux. In this model, cholesterol binds to the N-terminal region of NPC1L1 and is transported across the membrane through an undisclosed mechanism. Many nonsynonymous polymorphisms in loop C or binding of EZE analogs to a distinct site in loop C would interfere with the conformation of the protein and prevent cholesterol transport. Although the model implies that NPC1L1 functions as a classical transporter, such a mechanism is equally compatible with a receptor-mediated process, and the actual process remains to be elucidated.
Experimental Procedures
Stable MDCKII-Flp cell lines expressing dog NPC1L1 or dog NPC1L1-Kv1.1 were generated as described (8). Cell-based [3H]AS binding or [3H]cholesterol flux was performed as described (8). Membranes were prepared by sucrose gradient and solubilization, immunoprecipitation, immunoblot, silver-stained gel, and mass spectral characterization were performed as described in SI Text. Generation of dog/mouse NPC1L1 chimeras and point mutations was performed as described in SI Text. Further experimental details can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Drs. Herb Bull, Raymond Evers, Margarita Garcia-Calvo, Euan MacIntyre, Carl Sparrow, and Sam Wright for continuous support during the course of this work.
Footnotes
Conflict of interest statement: The authors declare they are employees of Merck and Co., Inc., and potentially own stock and/or hold stock options in the company.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800936105/DCSupplemental.
References
- 1.Grundy SM. Absorption and metabolism of dietary cholesterol. Annu Rev Nutr. 1983;3:71–96. doi: 10.1146/annurev.nu.03.070183.000443. [DOI] [PubMed] [Google Scholar]
- 2.Knopp RH, et al. Effects of ezetimibe, a new cholesterol absorption inhibitor, on plasma lipids in patients with primary hypercholesterolemia. Eur Heart J. 2003;24:729–741. doi: 10.1016/s0195-668x(02)00807-2. [DOI] [PubMed] [Google Scholar]
- 3.Ballantyne CM, et al. Effect of ezetimibe coadministered with atorvastatin in 628 patients with primary hypercholesterolemia: a prospective, randomized, double-blind trial. Circulation. 2003;107:2409–2415. doi: 10.1161/01.CIR.0000068312.21969.C8. [DOI] [PubMed] [Google Scholar]
- 4.Altmann SW, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Calvo M, et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci USA. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L, et al. Cholesterol-regulated translocation of NPC1L1 to the cell surface facilitates free cholesterol uptake. J Biol Chem. 2006;281:6616–6624. doi: 10.1074/jbc.M511123200. [DOI] [PubMed] [Google Scholar]
- 7.Yamanashi Y, et al. NPC1L1 over-expression facilitates ezetimibe-sensitive cholesterol and {beta}-sitosterol uptake in CaCo-2 cells. J Pharmacol Exp Ther. 2007;320:559–564. doi: 10.1124/jpet.106.114181. [DOI] [PubMed] [Google Scholar]
- 8.Weinglass AB, et al. MDCKII cells: A pharmacologically validated system for NPC1L1-mediated cholesterol uptake. Mol Pharmacol. 2008;73:1072–1084. doi: 10.1124/mol.107.043844. [DOI] [PubMed] [Google Scholar]
- 9.Labonte ED, et al. Class B type I scavenger receptor is responsible for the high affinity cholesterol binding activity of intestinal brush border membrane vesicles. Biochim Biophys Acta. 2007;1771:1132–1139. doi: 10.1016/j.bbalip.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knopfel M, et al. Multiple plasma membrane receptors but not NPC1L1 mediate high-affinity, ezetimibe-sensitive cholesterol uptake into the intestinal brush border membrane. Biochim Biophys Acta. 2007;1771:1140–1147. doi: 10.1016/j.bbalip.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Altmann SW, et al. The identification of intestinal scavenger receptor class B, type I (SR-BI) by expression cloning and its role in cholesterol absorption. Biochim Biophys Acta. 2002;1580:77–93. doi: 10.1016/s1388-1981(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 12.Bietrix F, et al. Accelerated lipid absorption in mice overexpressing intestinal SR-BI. J Biol Chem. 2006;281:7214–7219. doi: 10.1074/jbc.M508868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardones P, et al. Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. J Lipid Res. 2001;42:170–180. [PubMed] [Google Scholar]
- 14.van Bennekum A, et al. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry. 2005;44:4517–4525. doi: 10.1021/bi0484320. [DOI] [PubMed] [Google Scholar]
- 15.Hui DY, et al. Development and Physiological Regulation of Intestinal Lipid Absorption. III. Intestinal Transporters and Cholesterol Absorption. Am J Physiol. 2008;294:G839–G843. doi: 10.1152/ajpgi.00061.2008. [DOI] [PubMed] [Google Scholar]
- 16.Smart EJ, et al. Annexin 2-caveolin 1 complex is a target of ezetimibe and regulates intestinal cholesterol transport. Proc Natl Acad Sci USA. 2004;101:3450–3455. doi: 10.1073/pnas.0400441101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Cohen JC, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte U, et al. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvbeta1. Neuron. 49:697–706. doi: 10.1016/j.neuron.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Infante RE, et al. Purified NPC1 Protein: II. LOCALIZATION OF STEROL BINDING TO A 240-AMINO ACID SOLUBLE LUMINAL LOOP. J Biol Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 20.Infante RE, et al. Purified NPC1 Protein: I. BINDING OF CHOLESTEROL AND OXYSTEROLS TO A 1278-AMINO ACID MEMBRANE PROTEIN. J Biol Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 21.Davies JP, et al. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 22.Johnston JM, et al. Early clinical characterization of MD-0727, a novel cholesterol absorption inhibitor, in healthy subjects. Atherosclerosis (Suppl) 2006;7:558. [Google Scholar]
- 23.Bai X. New azetidinone cholesterol absorption inhibitors. Exp Opin Ther Pat. 2007;17:791–797. [Google Scholar]
- 24.Oswald S, et al. Intestinal expression of P-glycoprotein (ABCB1), multidrug resistance associated protein 2 (ABCC2), and uridine diphosphate-glucuronosyltransferase 1A1 predicts the disposition and modulates the effects of the cholesterol absorption inhibitor ezetimibe in humans. Clin Pharmacol Ther. 2006;79:206–217. doi: 10.1016/j.clpt.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Fahmi S, et al. Functional characterization of genetic variants in NPC1L1 supports the sequencing extremes strategy to identify complex trait genes. Hum Mol Genet. 2008;17:2101–2107. doi: 10.1093/hmg/ddn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radhakrishnan A, et al. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. Weinglass. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.