Abstract
The enzymatic activities of three proteins encoded by the thienamycin gene cluster of Streptomyces cattleya (ThnR, ThnH, and ThnT) have been shown to incrementally cleave CoA to afford the active side-chain component of the β-lactam antibiotic thienamycin. These results supersede proposals based on earlier radiochemical incorporation experiments. For 20 years it has been thought that cysteine was directly incorporated into the antibiotic. Specific, stepwise truncation of CoA to 4-phosphopantetheine, pantetheine, and finally cysteamine was observed with ThnR, ThnH, and ThnT, respectively, in a series of coupled enzymatic assays. Pantetheinylated carbapenams were synthesized to address possible thienamycin biosynthetic intermediates and were shown to be effective substrates for the pantetheine-cleaving enzyme ThnT. Finally, a fourth gene, thnF, was shown to encode a protein capable of N-acetylating a model compound containing cysteamine in the presence of acetyl-CoA, consistent with the production of the S. cattleya cometabolite, N-acetylthienamycin. Taken together, these four enzymes are proposed to siphon CoA from primary metabolism to create the side chains for the predominant S. cattleya carbapenems, thienamycin and N-acetylthienamycin, in a process likely to be general for the broader class of these antibiotics.
Keywords: β-lactam antibiotics, carbapenem, pyrophosphatase, acylase, phosphatase
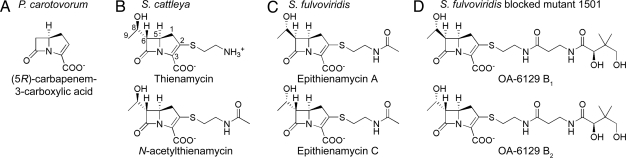
Thienamycin has been the hallmark of the carbapenem class of β-lactam antibiotics since its discovery in the 1970s from the soil actinomycete Streptomyces cattleya (1). Recognized for their broad antibacterial spectra and resistance to most classes of β-lactamases, the carbapenems are of growing importance in the treatment of infectious diseases in humans. Carbapenems are distinct from β-lactams such as pencillins and cephalosporins where carbon replaces sulfur at the C-1 position in the bicyclic nucleus (2). To date, >50 known naturally occurring carbapenems and carbapenams have been structurally characterized, yet most biosynthetic knowledge is limited to the simplest carbapenem, (5R)-carbapenem-3-carboxylic acid (2, 3) (Fig. 1A).
Fig. 1.
Selected carbapenems produced by P. carotovorum (A), S. cattleya (B), S. fulvoviridis A933 1719 (C), and S. fulvoviridis (D) blocked mutant 1501 in lieu of the epithienamycins. The antibiotic profile of the S. fulvoviridis mutant parallels that of Streptomyces sp. OA-6129.
The gene cluster for (5R)-carbapenem-3-carboxylic acid was first discovered in Pectobacterium carotovorum (formerly Erwinia carotovorum), but similar clusters have since been found in both Serratia marcescens and Photorhabdus luminescens (4, 5). Nine ORFs comprise the cluster, but only three genes (carA, carB, and carC) are absolutely required for antibiotic production (6). Although crystal structures for each are known, and detailed mechanistic insights have been determined for two of the three enzymes involved in creating the simple carbapenem, very little is known about the biosyntheses of the more highly substituted carbapenems such as thienamycin and the remainder of this antibiotic family (7, 8).
Early biochemical studies revealed that the primary metabolic sources of the thienamycin core paralleled those of the simple carbapenem, with glutamate giving rise to the pyrroline ring and acetate the β-lactam carbons (9). The C-6 hydroxyethyl side chain was shown to originate from methionine through two successive methylation events, presumably occurring by adenosylmethionine-utilizing enzymes (9, 10). This result was further supported by the dependence on vitamin B12 and Co2+ for thienamycin production (2, 11).
Initial insights into the origin of the thienamycin C-2 side chain were based on incorporation studies that suggested cysteine was directly incorporated into the carbapenem. [35S]- and [U-14C]cystine was shown to give >70% incorporation into thienamycin, whereas [35S]pantethine was very poorly incorporated into the antibiotic (9). These feeding studies were carried out in response to the discovery of the OA-6129 family of carbapenems, where an intact pantetheine side chain was observed in place of cysteamine (12). Interestingly, random mutagenesis of Streptomyces fulvoviridis A933 1719, a producer of the PS-5 series of carbapenems, resulted in a mutant that produced the OA-6129 series in lieu of PS-5 (Fig. 1 C and D) (13). In addition, A933 acylase from S. fulvoviridis was purified from the parent strain and shown to hydrolyze the pantetheine side chain of the OA-6129 series to produce the cysteamine side chain found in thienamycin (14).
Despite intense efforts made through the mid 1980s, further progress has been thwarted on thienamycin biosynthesis because of the poor transformation systems for the host strain as well as the instability and low titers of the antibiotic (15). However, in 2003, Nunez and coworkers (16) isolated and sequenced the thienamycin gene cluster in S. cattleya, reopening the field to further research. Here, we report the first biochemical analysis of enzymes encoded by the thienamycin gene cluster. Four enzymes are now shown to be involved in processing the C-2 side chain in thienamycin biosynthesis. Three of these have been demonstrated to incrementally truncate CoA to cysteamine, with the fourth able to cap the amine of a cysteamine-containing model substrate with acetate. Taken together, these enzymes are proposed to be responsible for the predominant C-2 side chains of thienamycin and N-acetylthienamycin produced by S. cattleya (14).
Results and Discussion
The thienamycin gene cluster consists of 22 ORFs, only two of which have apparent homologs in the nine-gene cluster for the simple carbapenem (5R)-carbapenem-3-carboxylic acid. ThnE and ThnM show 31.5% and 23.7% sequence identity to the first two biosynthetic proteins in the carbapenem pathway, CarB and CarA, respectively (www.ebi.ac.uk/emboss/align/). In contrast, a homolog of the last essential protein, CarC, is not apparent within the thienamycin cluster. The lack of a CarC homolog suggests the existence of an alternative pathway for control of the bridgehead stereochemistry as well as carbapenam desaturation. It is of note that two probable α-ketoglutarate-dependent, nonheme iron oxygenases, ThnG and ThnQ, are encoded by the thienamycin cluster and are of the same superfamily as CarC. Despite the low sequence conservation for all enzymes involved in the formation of the carbapenem core, the metabolic precursors appear to be the same for both antibiotics. The major differences between the two clusters lie in the several genes needed to attach and tailor the thienamycin side chains.
To discern what genes might be responsible for generating the cysteamine side chain in thienamycin, a translated protein capable of amide hydrolysis to possibly possess the pantetheine-hydrolyzing activity of A933 acylase described earlier from S. fulfoviridis was sought. ThnT (GenBank accession no. CAD18988), encoded by the thienamycin gene cluster, has sequence similarity to the DmpA/OAT superfamily represented by amidohydrolases DmpA, BapA, and NylC (17). Although originally thought to be a part of the N-terminal nucleophile (n-tn) hydrolase superfamily, research now suggests the DmpA/OAT superfamily is related to n-tn hydrolases by convergent evolution (18). Both superfamilies contain heterodimeric proteins that cleave autocatalytically into the two subunits from a single polypeptide chain. The N-terminal residue of the β-chain (serine, threonine, or cysteine) is thought to be responsible for peptide cleavage and to be a catalytic residue in amide hydrolysis. It is noteworthy that DmpA and BapA have the unique ability to cleave amide bonds containing β-amino acids, which are typically resistant to enzymatic hydrolysis (19). Paralleling this ability, cleavage of pantetheine in the OA-6129 series of carbapenems by A933 acylase resulted in hydrolysis of the cysteamine–β-alanine amide bond. Without another amidohydrolase apparent in the thienamycin cluster, ThnT was thought to be the A933 acylase homolog reported to exist in S. cattleya (14).
Initial attempts to heterologously express thnT in Escherichia coli failed, presumably because of discrepancies in codon usage between G-C rich actinomyces coding regions and the more G-C neutral DNA of E. coli. Thus, the first 15 codons of thnT were optimized through the use of a unique DraIII site to ligate a synthetic double-stranded DNA linker of the optimized sequence into the beginning of the gene. Expression was finally achieved through the use of an N-terminal 6-histidine (6-His) tag, the codon-optimized thnT sequence, and the expression host E. coli Rosetta2(DE3), in which six rare E. coli tRNA synthetases are supplemented.
ThnT purified as a combination of the intact full-length proprotein and the predicted self-cleavage products characteristic of the DmpA/OAT superfamily [supporting information (SI) Fig. S1]. The cleavage occurred between residues N281 and T282 to produce 29,229.4-Da and 12,109.7-Da fragments, which were confirmed by MALDI-TOF mass spectrometric analysis. The heterodimeric subunits appear to remain associated after cleavage because both fragments copurified on nickel-chelate chromatography, and a single band was present on native PAGE (data not shown).
The ability of ThnT to hydrolyze pantetheine to cysteamine and pantothenic acid was detected through derivatization of the thiol-containing products with 2-chloro-1-methylquinolinium tetrafluoroborate (CMQT), followed by HPLC and ESI mass spectrometric analysis. CMQT reacts within minutes at room temperature with thiols in aqueous solutions at neutral pH to produce derivatized species with a shifted UV-absorption maximum at 348 nm with respect to the reagent maximum of 328 nm (20). Pantetheine and cysteamine derivatizations coincided with synthetic standards in retention time and in exact mass with the compounds detected in the ThnT reaction. Interestingly, ThnT was unable to hydrolyze 4-phosphopantetheine or CoA after incubation at 37°C for 24 h (data not shown). Because 4-phosphopantetheine and CoA levels are considerably higher than pantetheine in certain bacteria (21), enzymes capable of metabolizing CoA to pantetheine were sought within the thienamycin cluster.
ThnR (GenBank accession no. CAD18986) bears sequence similarity to the Nudix hydrolase family, showing 38% sequence similarity to a CoA pyrophosphatase from Deinococcus radiodurans (22). This family of enzymes hydrolyzes diphosphates of various nucleotide-containing molecules. Nudix hydrolases are characterized by a conserved Nudix box sequence and, in the case of Nudix enzymes, by using CoA as a substrate, a conserved NuCoA motif present directly upstream of the Nudix box (22). ThnR possesses slight variations of both motifs characteristic of a CoA pyrophosphatase and was thought to be the enzyme responsible for processing CoA or a CoA-containing precursor of thienamycin (Fig. S2).
ThnR expression was achieved in Rosetta2 either as an N-terminal 6-His tag fusion or as a C-terminal 6-His tag construct with the first 12-aa codon-optimized. Hydrolysis of CoA within the diphosphate moiety yielding 4-phosphopantetheine and adenosine 3′,5′-diphosphate was determined through CMQT derivatization. Importantly, ThnR was unable to yield pantetheine from CoA for subsequent ThnT cleavage, hinting that yet another enzyme in the cluster was responsible for connecting the activities of the two enzymes.
One ORF in the thienamycin cluster, thnH, encoded a protein with weak homology to the haloacid dehalogenase (HAD) superfamily of hydrolases, which was hypothesized to hydrolyze the phosphate from 4-phosphopantetheine (23). As with other genes in the cluster, thnH would not express in any of the E. coli hosts tested. The codons of thnH were suboptimal, with rare E. coli codons spread throughout the 675-bp gene. Therefore, the whole of thnH was codon-optimized and harmonized for E. coli expression through the annealing of overlapping 40-bp primers by using the polymerase cycling assembly (PCA) method (24). Success was achieved by separating the gene into two synthons of ≈375 bp, followed by overlap extension of the two partial sequences to create the full-length, codon-optimized synthetic sequence. Expression was attained in E. coli Bl21(DE3) pLysS, because the construct seemed to be toxic and inhibited growth of Rosetta2 before induction.
As hoped, ThnH (GenBank accession no. CAD18976) hydrolysis of 4-phosphopantetheine to yield pantetheine and phosphate was initially detected by using a phosphate-molybdate assay (data not shown) and further supported by HPLC and ESI mass spectrometric analysis (25). Thus, three enzymes are required from the thienamycin cluster to fully process CoA to cysteamine, the active side-chain component of thienamycin.
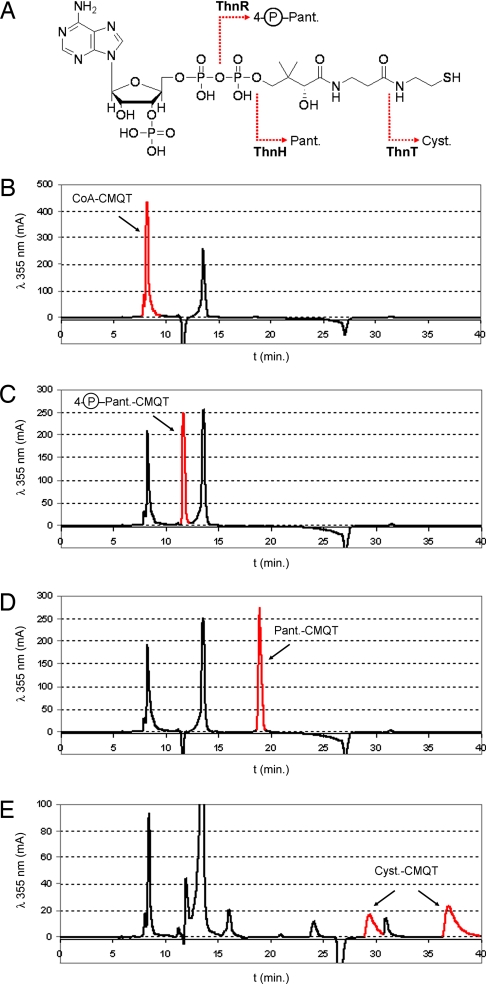
The ability for ThnR, ThnH, and ThnT to work in concert to completely and specifically process CoA to cysteamine was tested in a series of coupled enzyme assays. In addition to the three-enzyme coupled assay, reactions containing two of the three enzymes were incubated with CoA and MgCl2 for 3 hours at 37°C. As shown in Fig. 2 A–E, all enzymes acted specifically to cleave only the molecules previously identified as substrates. The buildup of appropriate intermediates in CoA processing was present without the detection of subsequently hydrolyzed products. Interestingly, CoA prederivatized with CMQT was not a substrate for ThnR, suggesting a potential specificity toward CoA bearing only its free thiol. ThnH had a markedly reduced cleavage rate for prederivatized 4-phosphopantetheine, whereas ThnT easily cleaved all prederivatized pantetheine under the conditions tested (Fig. S3). From these results, it was still uncertain at which stage of CoA processing the side chain is attached to the carbapenam or carbapenem core during thienamycin biosynthesis. Based on the apparent structural homology to the S. fulvoviridis A933 1719 mutant 1501 OA-6129 series of carbapenems, it is reasonable to suggest attachment of the C-2 thienamycin side chain occurs before pantetheine cleavage.
Fig. 2.
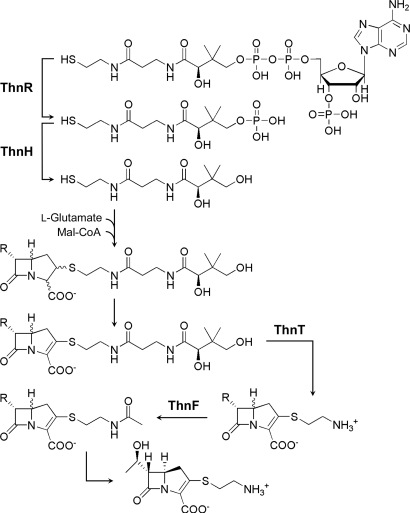
The stepwise truncation of CoA. (A) Representation of the incremental truncation of CoA by ThnR, ThnH, and ThnT. ThnR cleaves CoA to 4-phosphopantetheine, whereas ThnH subsequently cleaves the phosphate to produce pantetheine, and ThnT hydrolyzes pantetheine to yield cysteamine. (B–E) HPLC analyses of coupled enzyme assays to cleave CoA before CMQT derivatization: ThnH and ThnT (B), ThnR and ThnT, ThnR and ThnH (D), and ThnR, ThnH, and ThnT (E).The 4-phosphopantetheine is labeled as 4-P-Pant.; pantetheine is labeled as Pant.; cysteamine is labeled as Cyst. Peaks corresponding to enzymatic products are highlighted in red.
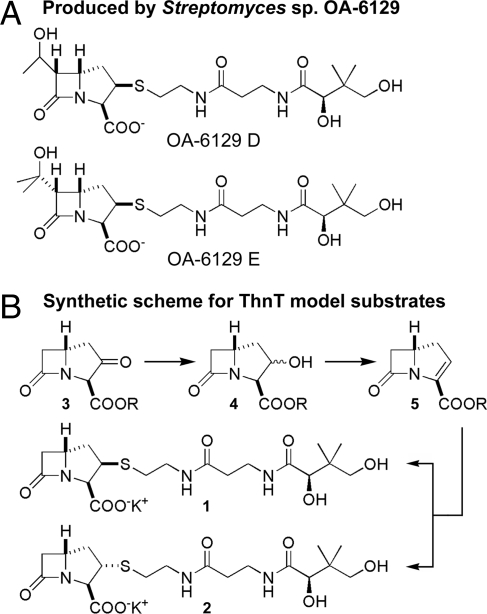
To link the truncation of CoA or, more specifically, pantetheine, to potential thienamycin biosynthetic precursors, 2-pantetheinyl carbapenams were selected to test the ability of ThnT to hydrolyze the pantetheinyl side chains after linkage to the β-lactam antibiotic core. Pantetheinylated carbapenams have been isolated in trace amounts from Streptomyces sp. OA-6129 (Fig. 3A) (26). Given that clear homologs of CarB and CarA, which are responsible for the biosynthesis of the carbapenam nucleus, correspond to ThnE and ThnM from the thienamycin gene cluster, we deduced that simple carbapenam formation was likely an early step. Although we now know the precursors of both the C-6 and C-2 side chains, the timing of their attachments, bicyclic ring inversion, and desaturation to the carbapenem are not known. To examine test substrates for the ThnT reaction that presented minimum steric demand but contained the probable carbapenam core, two potential substrates, 2-pantetheinyl carbapenams 1 and 2, were prepared (Fig. 3B). The synthesis proceeded from l-aspartic acid to 2-oxo-carbapenam 3 (R = p-methoxybenzyl) by established procedures (27, 28). Reduction of 3 gave a diastereomeric mixture of alcohols 4 (10:1 ratio), which underwent elimination in the presence of methanesulfonyl chloride and base to the carbapenem 5 (R = p-methoxybenzyl) (29). Heteroconjugate addition of pantetheine acetonide (30) to the carbapenem afforded a 2:1 mixture of (3R)-thioether products (31), which could be separated by HPLC and deprotected with trifluoroacetic acid and anisole (32) to give the (2R,3R,5R)- and (2S,3R,5R)-2-pantetheinyl carbapenams 1 and 2, respectively. Both the desired cysteaminyl carbapenams and the β-lactam-hydrolyzed products were detected by dansyl chloride derivatization, followed by HPLC-ESI mass spectrometry (Fig. S4). A preference for the absolute configuration of the pantetheinyl side chains at C-2 was not seen under the conditions tested. The acceptance of either thioether configuration does not allow discrimination between a pantetheinyl carbapenam or carbapenem as the true substrate of ThnT. Thus, ThnT seems to parallel the activity of hydrolyzing pantetheine-containing carbapenem substrates previously observed for the A933 acylase from S. fulvoviridis.
Fig. 3.
Natural and synthetic pantetheinylated carbapenams. (A) Carbapenams naturally produced by Streptomyces sp. OA-6129. (B) Scheme for the synthesis of 2-pantetheinyl carbapenams to mimic possible biosynthetic intermediates susceptible to ThnT.
In addition to the hydrolyzing activity of the A933 acylase, the enzyme was also shown to acylate the amine of select carbapenems with a variety of acyl-CoAs (14). This acylase was presumed to create acetylated carbapenems such as epithienamycin A and C produced in S. fulvoviridis (Fig. 1C). These two enzymatic functions were reported as originating from a single enzyme despite the fact that a mutant strain of S. fulvoviridis producing the OA-6129 series of pantetheinylated carbapenems lost the hydrolyzing activity of the A933 acylase and yet retained acylating activity. This result suggests either the A933 acylase was a multidomainal protein or that another protein copurified with the A933 acylase.
By analogy to S. fulvoviridis, in S. cattleya, the final addition to the thienamycin side chain known to occur is acetylation to yield N-acetylthienamycin, a carbapenem coproduced with thienamycin in wild-type cultures (Fig. 1B). Because ThnT, the presumed A933 acylase homolog, was unable to acetylate cysteamine-containing substrates with acetyl-CoA (data not shown), an enzyme capable of capping the reactive primary amine of thienamycin (or more likely a thienamycin precursor) with acetyl-CoA was sought within the cluster. ThnF (GenBank accession no. CAD18974) was found to have weak homology to the GCN5-related N-acetyltransferase (GNAT) superfamily, which utilizes acetyl-CoA to transfer acetyl groups to various primary amine-containing compounds. The GNAT superfamily contains four conserved sequence domains not readily identified in ThnF (Fig. S5). Despite the lack of apparent sequence conservation, thnF was cloned and expressed as an N-terminal 6-His fusion in Rosetta2 to test for possible acetyltransferase activity.
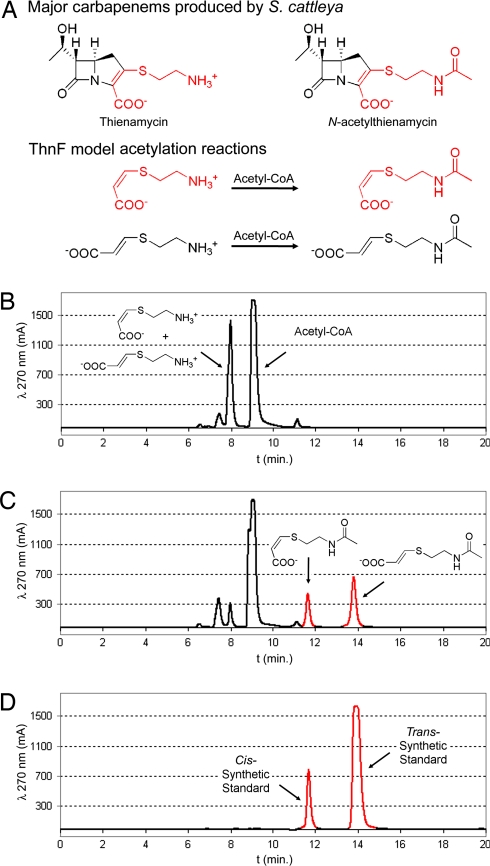
Initial efforts to detect acetyltransferase activity with cysteamine as the substrate were unsuccessful. Under the conditions tested, free thiol-containing potential substrates readily underwent transthioesterification with the acetyl group of acetyl-CoA. Cysteamine was especially problematic, however, because the molecule not only underwent transthioesterification, but also intramolecular rearrangement presumably occurred to produce N-acetylcysteamine in the presence of acetyl-CoA alone. Therefore, model substrates cis- and trans-carboxyethylenecysteamine [cis- and trans-3-(2-aminoethylthio)acrylic acid] were synthesized and tested to eliminate the unwanted reactivity of the substrate itself and to better serve as carbapenem precursors (Fig. 4A).
Fig. 4.
ThnF model reactions and HPLC analysis. (A) ThnF model reactions with cis- and trans-carboxyethylenecysteamine based on the two predominant carbapenems produced by S. cattleya. (B–D) HPLC analyses of ThnF reactions with the model substrates and acetyl-CoA. (B) Reaction control without enzyme. (C) ThnF reaction. (D) HPLC trace of synthetic cis- and trans-carboxyethylene-N-acetylcysteamine product standards.
ThnF was able to efficiently convert both the cis- and trans-carboxyethylenecysteamine to the corresponding acetylated products in the presence of acetyl-CoA (Fig. 4 B-D). Acetylation of the trans-model substrate was not expected. This result can be partially explained through the observation that spontaneous isomerization of the cis- to the trans-carboxyethylenecysteamine occurs in acidic aqueous conditions and, therefore, could mask the specificity of ThnF for the two substrates. Nonetheless, a preference for coplanarity of the carboxylic acid with the cysteaminyl side chain seems to be governing ThnF activity because coupled reactions of ThnT and ThnF with the 2-pantetheinyl carbapenams did not yield any acetylated products (data not shown). These results could be interpreted to suggest acetylation occurs after desaturation of the carbapenam ring in thienamycin biosynthesis, although further investigation will be needed to address this question.
The incremental truncation of CoA to cysteamine by ThnR, ThnH, and ThnT marks an advance in understanding the enzymes encoded by a substituted carbapenem gene cluster. CoA has been shown to be the source of the thienamycin cysteaminyl side chain instead of the previously proposed direct incorporation of cysteine. In addition, ThnF has been shown most likely responsible for the formation of N-acetylthienamycin through acetylation of the reactive primary amine in thienamycin. However, ThnF is not likely to be the terminal step in thienamycin biosynthesis because an enzyme capable of deacetylating N-acetylthienamycin to thienamycin was found and purified from wild-type cultures of S. cattleya (33). The discovery of an S. cattleya mutant strain that solely produced N-acetylthienamycin further supports this hypothesis (34). Thus, a tentative biogenetic pathway to the cysteaminyl side chain from CoA can be proposed for thienamycin biosynthesis (Fig. 5). The vinyl thioether side chain confers both increased chemical stability and potency required for the clinical usefulness of carbapenems. The efficient truncation of the common cellular component, CoA, by this cassette of proteins paves the way to apply metabolic engineering methods for either partial synthesis or modification of substituted carbapenem antibiotics.
Fig. 5.
Proposed pathway for thienamycin biosynthesis based on the apparent substrate preferences of enzymes ThnR, ThnH, ThnT, and ThnF. R denotes a C-6 thienamycin side chain of hydrogen, methyl, ethyl, or hydroxyethyl. Malonyl-CoA is labeled as Mal-CoA.
Materials and Methods
Cloning.
Genes thnR, thnH, thnT, and thnF were amplified from genomic S. cattleya DNA (NRRL 8057) by using PCR primers (Sigma–Genosys) listed in Table S1. Genes thnR, thnT, and thnF were cloned into pET28b(+) (Novagen) to create N-terminal 6-His tag fusions, whereas thnH was ligated into pET29a(+) to create a C-terminal 6-His tag construct. Positive clones were transformed into E. coli Rosetta2(DE3)-competent cells (Novagen).
thnT Codon Optimization.
The first 15 codons of thnT were optimized for expression in E. coli by using a DraIII cut site within the thnT sequence. Primers thnT-codon-F and thnT-codon-R were annealed to create a DNA linker with sticky ends for NdeI and DraIII cut sites. The DNA linker and appropriately cut pET29b plasmid were then ligated at approximately a 5:1 molar ratio and subsequently transformed into DH5α-competent cells for sequence analysis.
thnH Codon Optimization.
The entire thnH sequence was synthetically codon-optimized by using the PCA method (24, 35). The web site http://software.kosan.com/GeMS was used to codon optimize thnH as well as split the gene into 40-bp strands with 20-bp overlaps. Gene thnH was prepared in two synthons of ≈375 bp to maximize success of the PCA reaction. The 50-μl PCA mixtures contained 20 mM Tris·HCl (pH 8.8), 10 mM KCl, 6 mM (NH4)2SO4, 0.1% (vol/vol) Triton X-100, 0.1 mg/ml bovine albumin, 0.2 mM each dNTP, 1.25 units Pfu turbo DNA polymerase (Stratagene), 2.0 ng/μl concentrations of each primer, and 5% (vol/vol) DMSO and was run at 96°C for 9 min (hot start after 2 min), with 25 cycles at 96°C for 30 s, gradient annealings of 45–54°C for 30 s, and 72°C for 1 min. Subsequent PCRs were run with the outermost primers for an additional 35 cycles with 2 μl from each PCA reaction to further amplify each synthon. To create full-length thnH, 1 μl of each synthon PCR was amplified in another PCR with only the outside primers of the gene.
Protein Expression and Purification.
ThnR, ThnT, and ThnF were expressed in E. coli Rosetta2(DE3) cells with 1 mM IPTG induction at 18°C for 16 h. ThnH was expressed in E. coli Bl21(DE3) pLysS cells. Nickel-NTA resin (Qiagen) was used to purify each protein according to the manufacturer's specifications.
Coupled Enzyme and ThnF Activity Assays.
All 500-μl reactions were performed in 50 mM Tris (pH 7.5) and incubated for 3 h at 37°C before derivatization. ThnR (10 μM), 2.0 μM ThnH, and 2.0 μM ThnT were incubated in various combinations with 2.5 mM CoA and 10.0 mM MgCl2. ThnF activity assays contained 10 μM enzyme, 2.0 mM concentrations each of cis- and trans-carboxyethylenecysteamine, 2.5 mM acetyl-CoA, and 10.0 mM MgCl2 in 100 mM Tris (pH 7.5).
Thiol Derivatization and HPLC Analysis.
Thiol-containing compounds were derivatized with CMQT, which was synthesized as described (20). Briefly, a typical derivatization reaction contained 200 μl of 2.5 mM thiol-containing compound and 100 μl of 25 mM CMQT in 100 mM Tris (pH 7.5). The reaction was mixed and acidified after 5–10 min with 40 μl of 72% (wt/vol) trichloroacetic acid. Standards (10 mM) of CoA, 4-phosphopantetheine, pantetheine, and cysteamine were derivatized and independently injected onto an Agilent 1100 Series HPLC in 20-μl aliquots onto a Phenomenex Luna 5-μm CN 100 Å (250 × 10.0 mm) column. A flow rate of 3.0 ml/min was used with a mobile phase consisting of acetonitrile (solvent A) and 10 mM NH4C2H3O2 pH 5.0 (solvent B) adjusted with NaOH. A method of 95–85% solvent B from 0 to 20 min, 30% solvent B at 40 min, and 95% solvent B from 45 to 50 min was used for separation of all analytes. Monitoring at 355 nm, the retention times of each derivatized standard were: CoA-CMQT, 8.2 min; 4-phosphopantetheine-CMQT, 11.7 min; pantetheine-CMQT, 18.9 min; and cysteamine-CMQT at 29.3 and 36.9 min. Masses for the derivatized samples were: CoA-CMQT (C31H44N8O16P3S+) theoretical (m/z 909.18), synthetic standard (m/z 909.13), observed (m/z 909.04); 4-phosphopantetheine-CMQT (C21H31N3O7PS+) theoretical (m/z 500.16), synthetic standard (m/z 500.00), observed (m/z 499.98); pantetheine-CMQT (C21H30N3O4S+) theoretical (m/z 420.20), synthetic standard (m/z 420.23), observed (m/z 420.24); cysteamine-CMQT (C12H15N2S+) theoretical (m/z 219.10), synthetic standard (m/z 218.46), observed (m/z 218.11).
Synthesis of Substrates.
The 4-phosphopantetheine was synthesized essentially as described by Mandel (36). Cis- and trans-carboxyethylenecysteamine, as well as cis- and trans-carboxyethylene-N-acetylcysteamine [cis- and trans-3-(2-acetamidoethylthio)acrylic acid] were prepared according to published procedures (37). Details of the syntheses of (2R,3R,5R)- and (2S,3R,5R)-2-pantetheinyl carbapenems 1 and 2, respectively, can be found in SI Text and Fig. S6.
ThnF HPLC Assay.
Aliquots (100 μl) were injected onto a Phenomenex Luna 5-μm C18 100 Å (250 × 10.0 mm) column. A flow rate of 1.5 ml/min was used with a mobile phase of acetonitrile (solvent A) and dH2O with 0.1% (vol/vol) TFA (solvent B). Monitoring at 270 nm, a method of 80% solvent B from 0 to 10 min, 50% solvent B at 30 min, 80% solvent B at 40–45 min was used for separation. The cis- and trans-model substrates coeluted at 8.0 min whereas the enzymatic product cis-carboxyethylene-N-acetylcysteamine eluted at 11.6 min and the trans-product eluted at 13.8 min.
Supplementary Material
Acknowledgments.
We thank Prof. Maurice J. Bessman and Dr. Wenlian Xu for help discussions and technical assistance with Nudix hydrolases. MALDI-TOF mass spectrometry was performed by the Proteomics Core Facility at The Johns Hopkins University. This work was supported by National Institutes of Health Research Grant AI014937.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804500105/DCSupplemental.
References
- 1.Kahan JS, et al. Thienamycin, a new beta-lactam antibiotic. I. Discovery, taxonomy, isolation and physical properties. J Antibiot (Tokyo) 1979;32:1–12. doi: 10.7164/antibiotics.32.1. [DOI] [PubMed] [Google Scholar]
- 2.Williamson JM. The biosynthesis of thienamycin and related carbapenems. Crit Rev Biotechnol. 1986;4:111–131. [Google Scholar]
- 3.Demain AL, Elander RP. The beta-lactam antibiotics: Past, present, and future. Antonie Van Leeuwenhoek. 1999;75:5–19. doi: 10.1023/a:1001738823146. [DOI] [PubMed] [Google Scholar]
- 4.Coulthurst SJ, Barnard AM, Salmond GP. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol. 2005;3:295–306. doi: 10.1038/nrmicro1128. [DOI] [PubMed] [Google Scholar]
- 5.McGowan SJ, et al. Analysis of bacterial carbapenem antibiotic production genes reveals a novel beta-lactam biosynthesis pathway. Mol Microbiol. 1996;22:415–426. [PubMed] [Google Scholar]
- 6.Li R, Stapon A, Blanchfield JT, Townsend CA. Three unusual reactions mediate carbapenem and carbapenam biosynthesis. J Am Chem Soc. 2000;122:9296–9297. [Google Scholar]
- 7.Clifton IJ, et al. Crystal structure of carbapenem synthase (CarC) J Biol Chem. 2003;278:20843–20850. doi: 10.1074/jbc.M213054200. [DOI] [PubMed] [Google Scholar]
- 8.Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. Crystal structure of carbapenam synthetase (CarA) J Biol Chem. 2003;278:40996–41002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]
- 9.Williamson JM, et al. Biosynthesis of the beta-lactam antibiotic, thienamycin, by Streptomyces cattleya. J Biol Chem. 1985;260:4637–4647. [PubMed] [Google Scholar]
- 10.Houck DR, Kobayashi K, Williamson JM, Floss HG. Stereochemistry of methylation in thienamycin biosynthesis: Example of a methyl transfer from methionine with retention of configuration. J Am Chem Soc. 1986;108:5365–5366. [Google Scholar]
- 11.Chen TS, Arison BH, Ruby CL, Dombrowski AW, Inamine ES. A cofactor for thienamycin biosynthesis produced by a blocked mutant of Streptomyces cattleya. J Ind Microbiol. 1993;12:66–67. doi: 10.1007/BF01570130. [DOI] [PubMed] [Google Scholar]
- 12.Okabe M, et al. Studies on the OA-6129 group of antibiotics, new carbapenem compounds. I. Taxonomy, isolation and physical properties. J Antibiot (Tokyo) 1982;35:1255–1263. doi: 10.7164/antibiotics.35.1255. [DOI] [PubMed] [Google Scholar]
- 13.Fukagawa Y, et al. Studies on the biosynthesis of carbapenem antibiotics. I. Biosynthetic significance of the OA-6129 group of carbapenem compounds as the direct precursors for PS-5, epithienamycins A and C and MM 17880. J Antibiot (Tokyo) 1984;37:1388–1393. doi: 10.7164/antibiotics.37.1388. [DOI] [PubMed] [Google Scholar]
- 14.Kubo K, Ishikura T, Fukagawa Y. Studies on the biosynthesis of carbapenem antibiotics. II. Isolation and functions of a specific acylase involved in the depantothenylation of the OA-6129 compounds. J Antibiot (Tokyo) 1984;37:1394–1402. doi: 10.7164/antibiotics.37.1394. [DOI] [PubMed] [Google Scholar]
- 15.Buchan T, et al. Mutants of Streptomyces cattleya defective in the synthesis of a factor required for thienamycin production. J Antibiot (Tokyo) 1994;47:992–1000. doi: 10.7164/antibiotics.47.992. [DOI] [PubMed] [Google Scholar]
- 16.Nunez LE, Mendez C, Brana AF, Blanco G, Salas JA. The biosynthetic gene cluster for the beta-lactam carbapenem thienamycin in Streptomyces cattleya. Chem Biol. 2003;10:301–311. doi: 10.1016/s1074-5521(03)00069-3. [DOI] [PubMed] [Google Scholar]
- 17.Negoro S, Kakudo S, Urabe I, Okada H. A new nylon oligomer degradation gene (nylC) on plasmid pOAD2 from a Flavobacterium sp. J Bacteriol. 1992;174:7948–7953. doi: 10.1128/jb.174.24.7948-7953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng H, Grishin NV. DOM-fold: A structure with crossing loops found in DmpA, ornithine acetyltransferase, and molybdenum cofactor-binding domain. Protein Sci. 2005;14:1902–1910. doi: 10.1110/ps.051364905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heck T, et al. Enzymatic degradation of beta- and mixed alpha,beta-oligopeptides. Chem Biodivers. 2006;3:1325–1348. doi: 10.1002/cbdv.200690136. [DOI] [PubMed] [Google Scholar]
- 20.Bald E, Rafal G. 2-chloro-1-methylquinolinium tetrafluoroborate as an effective and thiol specific UV-tagging reagent for liquid chromatography. J Liq Chrom Rel Technol. 2001;24:1323–1339. [Google Scholar]
- 21.Jackowski S, Rock CO. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang LW, et al. Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: A member of the Nudix family. J Bacteriol. 2003;185:4110–4118. doi: 10.1128/JB.185.14.4110-4118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin EV, Tatusov RL. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol. 1994;244:125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 24.Kodumal SJ, et al. Total synthesis of long DNA sequences: Synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ames BN, Dubin DT. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- 26.Yoshioka T, et al. Structures of OA-6129D and E, new carbapenam antibiotics. J Antibiot (Tokyo) 1984;38:211–217. doi: 10.7164/antibiotics.37.211. [DOI] [PubMed] [Google Scholar]
- 27.Reider PJ, Grabowski EJ. Total synthesis of thienamycin—A new approach from aspartic-acid. Tetrahedron Lett. 1982;23:2293–2296. [Google Scholar]
- 28.Ueda Y, Roberge G, Vinet V. A simple method of preparing trimethylsilyl-enol and tert-butyldimethylsilyl-enol ethers of alpha-diazoacetoacetates and their use in the synthesis of a chiral precursor to thienamycin analogs. Can J Chem. 1984;62:2936–2940. [Google Scholar]
- 29.Shibuya M, Kubota S. Synthesis of 1,1-dimethylcarba-2-penem derivatives via a Dieckmann-type cyclization. Tetrahedron Lett. 1981;22:3611–3614. [Google Scholar]
- 30.Patil G. Patent Cooperation Treaty Appl WO95/11893. 1995. May 4, 1995. [Google Scholar]
- 31.Bateson JH, Hickling RI, Smale TC, Southgate R. Olivanic acid analogs. 6. Biomimetic synthesis of (+/−)-Ps-5, (+/−)-6-Epi-Ps-5, and (+/−)-Benzyl Mm22381. J Chem Soc Perk T. 1990;1:1793–1801. [Google Scholar]
- 32.Lee M, Hesek D, Mobashery S. A practical synthesis of nitrocefin. J Org Chem. 2005;70:367–369. doi: 10.1021/jo0487395. [DOI] [PubMed] [Google Scholar]
- 33.Uyeda M, Demain AL. Deacetylation of N-acetylthienamycin to thienamycin by a cell-free extract of Streptomyces cattleya, the thienamycin producer. J Ind Microbiol. 1987;1:341–347. [Google Scholar]
- 34.Rosi D, et al. Mutants of Streptomyces cattleya producing N-acetyl and deshydroxy carbapenems related to thienamycin. J Antibiot (Tokyo) 1981;34:341–343. doi: 10.7164/antibiotics.34.341. [DOI] [PubMed] [Google Scholar]
- 35.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 36.Mandel AL, La Clair JJ, Burkart MD. Modular synthesis of pantetheine and phosphopantetheine. Org Lett. 2004;6:4801–4803. doi: 10.1021/ol048853+. [DOI] [PubMed] [Google Scholar]
- 37.Funabiki K, Tamura K, Ishihara T, Yamanaka H. Tandem intermolecular-intramolecular Michael addition of bifunctional hetero nucleophiles to polyfluoro-2-alkynoic acids—Facile synthesis of polyfluoroalkylated azaheterocycles. B Chem Soc Jpn. 1994;67:3021–3029. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.