Abstract
Hydrogenases, which catalyze H2 to H+ conversion as part of the bioenergetic metabolism of many microorganisms, are among the metalloenzymes for which a gas-substrate tunnel has been described by using crystallography and molecular dynamics. However, the correlation between protein structure and gas-diffusion kinetics is unexplored. Here, we introduce two quantitative methods for probing the rates of diffusion within hydrogenases. One uses protein film voltammetry to resolve the kinetics of binding and release of the competitive inhibitor CO; the other is based on interpreting the yield in the isotope exchange assay. We study structurally characterized mutants of a NiFe hydrogenase, and we show that two mutations, which significantly narrow the tunnel near the entrance of the catalytic center, decrease the rates of diffusion of CO and H2 toward and from the active site by up to 2 orders of magnitude. This proves the existence of a functional channel, which matches the hydrophobic cavity found in the crystal. However, the changes in diffusion rates do not fully correlate with the obstruction induced by the mutation and deduced from the x-ray structures. Our results demonstrate the necessity of measuring diffusion rates and emphasize the role of side-chain dynamics in determining these.
Keywords: crystallography, structure/function relationships, substrate tunnel, protein film voltammetry, isotope exchange
Enzyme channels for substrates are elongated cavities, or “tunnels,” which either connect the active site to the solvent or guide intermediates from one active site to another in multifunctional enzymes (1). As far as redox catalysis is concerned, the best-documented example is certainly the 70-Å-long channel that connects the two active sites involved in CO production and utilization in acetyl-CoA synthase/CO dehydrogenase (ACS-CODH) (2, 3). In addition, channels dedicated to the transport of O2, N2, or H2 are supposed to exist in heme-copper oxidase (4–6), nitrogenase (7), lipoxygenase (8), photosystem II (9), and both NiFe and FeFe hydrogenases (10), to cite but a few. These channels were often discovered by searching for hydrophobic cavities in x-ray structures hence their qualification as “static.” Their affinity for gas molecules was sometimes confirmed by showing that they can bind Xenon in the crystal (10, 11). Alternatively, molecular dynamics (MD) calculations can evaluate the dynamics of gas permeation through the protein matrix (11–14). This approach unveils the role of conformational flexibility and questions the requirement for a static channel to transport small diatomic molecules. In the case of the FeFe hydrogenase mentioned above, such simulations revealed a “dynamic” channel that had not been detected as a cavity in the frozen crystal (12). Whether the channel is static or dynamically formed, it may consist of a single or multiple routes for gas transport.
There are examples of enzymes for which site-directed mutagenesis studies suggest there is no static channel for gas transport. For example, that no single mutation was found to impact O2 access to the active site of copper-containing amine oxidase has been taken as evidence of the existence of multiple dynamic pathways (13). In a few cases, however, mutations of some residues located in a putative gas channel did affect the enzyme's kinetic properties. For example, certain mutations in enzymes that use molecular oxygen as substrate increase the Km for O2 (4, 5, 8, 15). Brzezinski and coworkers (6) demonstrated that a glycine-to-valine mutation almost completely obstructs the oxygen channel of cytochrome c oxidase; in this study, the delayed access of substrate O2 and inhibitor CO to the active-site heme was probed by using time-resolved UV-vis spectroscopy. From these observations, they inferred that the protein is rigid in the region of this residue, because otherwise fluctuations would counter the blockage introduced by the mutation. Several alanine mutations were also found to block the long channel in ACS-CODH (2, 3). However, the structure of the mutants has been determined in only one of the above cases (15). This structural information is important, because mutations may have unexpected structural consequences; for example, a glycine to phenylalanine mutation in the channel of carbamoyl-phosphate synthetase causes a large conformational change that creates an escape route for ammonia directly to the bulk solvent (16).
The existence of a H2 channel in NiFe hydrogenase has been proposed on the basis of crystallographic studies and MD simulations (10, 11). The latter suggested preferred routes for H2 access to the active sites, which match the hydrophobic cavities found in the x-ray structures. However, these simulations cannot yield diffusion rates. Learning about gas diffusion in hydrogenase is not only of academic interest; kinetic data are needed, because it is believed that the tunnel is also used by the inhibitor oxygen, and oxygen sensitivity is the major obstacle for using these enzymes in biotechnological devices, either for H2 oxidation in biofuel cells or for H2 production by oxygenic photosynthetic microorganisms. According to some reports, the structure of the gas channel is a determinant of oxygen sensitivity in hydrogenases (17–19), but whether the rate of inhibition is limited by O2 access to the active site or by its reaction at the active site is not known.
Here, we introduce and use two methods for determining the rates of gas diffusion in hydrogenases, and we demonstrate that two double mutations in a NiFe enzyme affect both the width of the gas channel and the kinetics of intramolecular diffusion.
Results
Biochemical, Spectroscopic, and Structural Characterization of the Mutants.
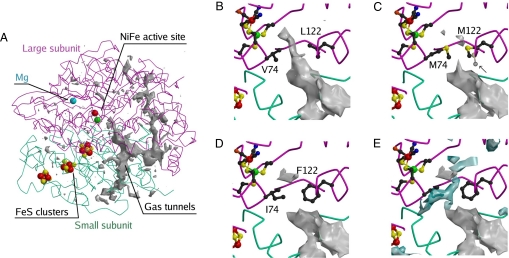
Fig. 1A shows the hydrophobic cavity calculated in WT NiFe hydrogenase using Protein Data Bank (PDB) coordinates 1YQW and a probe radius of 0.8 Å (20). As described earlier (17), the entrance to the active site is flanked by two conserved residues, V74 and L122 (Fig. 1B). We designed the L122M-V74M (MM) mutant in an attempt to change significantly the shape of the tunnel at this position and the L122F-V74I (FI) mutant to mimic the tunnel of oxygen-tolerant NiFe regulatory hydrogenases (17–19). The mutations did not affect the purification yield (≈0.7 mg of purified protein per liter of culture).
Fig. 1.
Structural models of the three enzymes. A gives an overview of the tunnel network; B is a closeup of the tunnel near the active site in the WT. C, D, and E are closeups of the MM and FI mutants, as indicated. In C, an arrow points to the second conformation of M122. A conserved hydrophilic cavity is shown in blue in E.
The enzymes were assayed for H2 oxidation with 50 mM oxidized methyl viologen (saturating conditions) and under 1 atm of H2 (Table 1). The apparent first-order rate constants (kcatapp) measured under 1 atm of H2 can be converted to apparent specific activities using 1 s−1 = 0.66 μmol of H2 oxidized per min/mg of protein (the molecular weight of hydrogenase is 90 kDa). The Km values for H2 were measured by using protein film voltammetry (21) and showed no significant temperature dependence over the 5–40°C range. The values of kcat, extrapolated to infinite concentration of H2 from the data in Table 1 using kcat = kcatapp (1+Km/[H2]), are similar for the three enzymes. Therefore, the lower value of the apparent first-order rate constant under 1 atm of H2 (kcatapp) in the case of the MM mutant merely results from the Km for H2 being large.
Table 1.
Kinetic properties of the WT and mutant enzyme
| Unit | kcatapp under 1 atm of H2*, s−1 | Km†, m atm (H2) | kcat‡, s−1 | kinCO§, s−1 atm(CO)−1 | koutCO§, s−1 | kout/k¶ |
|---|---|---|---|---|---|---|
| WT | 750 ± 90 | 10 | 760 ± 95 | 1 × 104 to 2 × 104 | 2 to 10 | 1.25 |
| FI | 800 ± 30 | 50 | 840 ± 50 | 2 × 103 | 4 × 10−1 | 10−1 |
| MM | 590 ± 30 | 200 | 675 ± 90 | 65 | 4 × 10−3 | 2 × 10−2 |
*Apparent first-order rates constants for H2 oxidation, determined in solution assays in the presence of 1 atm H2 and 50 mM oxidized methyl viologen, at T = 30°C, pH = 8. A turnover frequency of 1 s−1 is equivalent to 0.66 μmol of H2 oxidized per min and per mg of enzyme.
†Michaelis constant relative to H2 (21). The accuracy is of the order of 50%.
‡Values of kcat extrapolated using kcat = kcatapp (1+Km/[H2]).
§Rate constants relative to CO binding (kinCO) and release (koutCO) determined from fitting data such as those in Fig. 3 (27). For the WT, the values have been extrapolated to 30°C from the data in Fig. 4. ″One atm of CO″ refers to the concentration of CO dissolved in a solution that is equilibrated under 1 atm of CO at 25°C.
¶Rate of dihydrogen release from the active site divided by the rate of H+/D+ exchange at the active site, determined from interpreting isotope exchange assays such as those in Fig. 5. We used the model and the method that corrects for the slow gas consumption by the mass spectrometer, both described in SI Text.
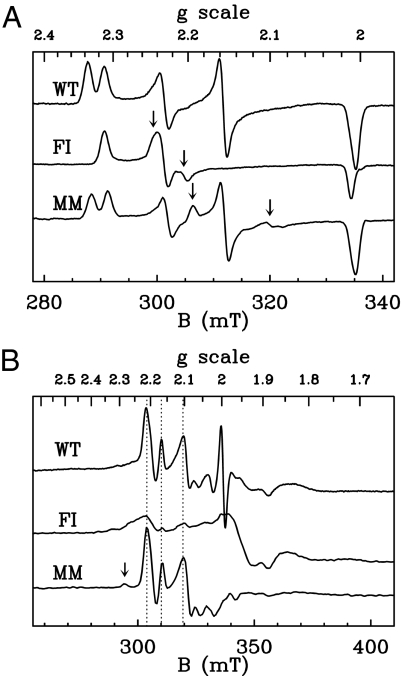
We used EPR spectroscopy to assess the integrity of the metal centers. In the oxidized as-prepared state, both mutants exhibit spectra similar to those reported for the WT enzyme (22). The oxidized active site normally gives a superposition of signals called Ni-A (g = 2.31, 2.23, 2.01) and Ni-B (g = 2.34, 2.16, 2.01). Fig. 2A and supporting information (SI) Table S1 show that the mutations have very little effect on the g values of these species, although we could detect only traces of Ni-B (besides Ni-A) in the FI mutant samples. The mutants showed additional spectral contributions (pointed to by arrows in Fig. 2A), which disappeared upon reduction, accounting for ≈0.03 and 0.18 spin per molecule in the MM and FI enzymes, respectively. In reductive potentiometric titrations of the mutants, the usual Ni-C signature at g = 2.19, 2.14, 2.01 was observed in the same potential range as for the WT enzyme (not shown). This signal accounts for up to 0.3 and 0.25 spin per molecule for the WT and MM variant, and at low temperature, it shows the same spin–spin interaction pattern with the proximal [4Fe4S] cluster (Fig. 2B) (23). In contrast, the Ni-C signal was much smaller in the case of the FI variant (<2%), which may result from the double-mutation slightly affecting the redox properties of the active site in this mutant. However, we observed no correlation between the maximal intensity of the Ni-C signal and the kinetic properties of the three enzymes. The mutations did not affect the signals of the FeS centers (not shown). In summary, this EPR study shows that the FI enzyme only differs from the WT in that a new signature of the oxidized active site accounts for a minor fraction of the Ni (although the normal Ni-A state dominates), and we could detect only small amounts of a normal Ni-C state in reduced samples. The MM mutant, which has the most interesting phenotype from the point of view of diffusion kinetics (see below), behaves essentially like the WT enzyme.
Fig. 2.
EPR characterization of the three enzymes in the oxidized (A) and reduced (B) states. Microwave power 10 mW, modulation amplitude 1 mT, T = 100 K (A) or 6 K (B). Arrows in A indicate the positions of the spectral lines at g = 2.245 and 2.205 (FI mutant) and g = 2.195 and 2.1 (MM mutant), which are absent in the WT enzyme. In B, vertical dashed lines mark the positions of the most intense lines of the low-temperature Ni-C signal, and the arrow points to a small contribution of the so-called Ni-L2 species. The signal at g = 2 arises from redox mediators.
The structures of the two mutants were solved to determine the impact of the mutations on the shape of the gas channel (Fig. 1). The changes induced by the mutations are highly localized; no significant modifications are detected elsewhere in the structure. The two mutated residues are well ordered in the FI structure. However, in the MM structure, both M74 and M122 side chains show some disorder, as significant negative peaks appeared in the difference Fourier (Fobs − Fcalc) electron-density map when their side chains were included in the model with full occupancy. For M122, two partially occupied conformations are observed (≈50% each), whereas for M74, only a major conformation with 80–90% occupancy is detected. In the WT enzyme, the channel is open between residues 74 and 122 when the probe radius (Rp) used for visualizing the cavity is 1.1 Å or smaller, whereas for MM and FI, the channel is blocked at that position unless Rp is smaller than 0.6 and 0.45 Å, respectively. Clearly, these tunnels would be too narrow to let gases through if the structure were static; the Van der Waals radius of hydrogen is reported as 1.0 ± 0.1 Å (24). Dynamic residue rearrangements could be facilitated by movements of nearby structurally conserved water molecules in a hydrophilic cavity that is located near residue 74 (Fig. 1E).
The Kinetics of CO Binding and Release Obtained from Electrochemical Experiments.
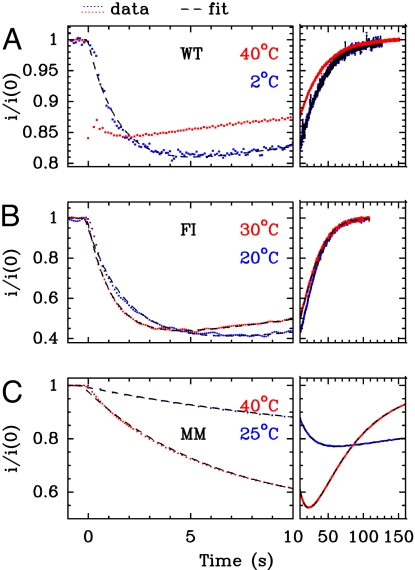
Carbon monoxide substitutes for H2 at the active site of NiFe hydrogenase in a competitive manner (21). The kinetics of CO inhibition can be probed by using protein film voltammetry (PFV), a technique where the enzyme is adsorbed onto an electrode that acts as a sink or a source of electrons. Electron transfer is direct (no mediators are used), and H2-oxidation activity is measured as a current (25, 26). This is particularly convenient for detecting rapid changes in turnover rates that follow changes in inhibitor concentration. We have developed a method to characterize quantitatively the kinetics of inhibition of hydrogenase, which consists in injecting in the electrochemical cell aliquots of a solution saturated with CO, whereas the cell is continuously degassed by bubbling H2. The transient change in activity is continuously monitored as the concentration of inhibitor decreases; that this decay is exactly exponential makes data modeling straightforward (21, 26, 27). Fig. 3A shows that, in the case of the WT at T = 40°C, the decrease in activity after CO injection is faster than the mixing time of ≈0.1 s; the inhibition kinetics is too fast to be resolved. Also, the recovery of activity follows exactly the decrease in CO concentration as the latter is flushed away from the cell (21). The binding and release of CO slow down as the temperature is lowered, and at T ≤ 15°C, the decrease in activity is clearly delayed from the time of injection (blue trace in Fig. 3A). This effect is much more pronounced in the case of the double mutants, even at 20°C (Fig. 3 B and C), and the MM variant exhibits the greatest inertia.
Fig. 3.
Comparison of the kinetics of CO inhibition of H2 oxidation in PFV experiments (26). The current i has been normalized by its value i(0), measured before CO was added. Left shows the short-term change in current, whereas the end of the relaxation is shown on Right. The dimensionless volumic fractions of solutions saturated under 1 atm of CO at 25°C and injected at time 0 (see SI Text) were x = 7 × 10−3 (A, WT), 12 × 10−3 (B, FI), 2.5 × 10−3 (C, MM). Electrode rotation rate 2 krpm, pH 7, T as indicated. The fits of the data to Eq. 1 in SI Text are shown as dashed black lines.
Our quantitative analysis (27) is based on the assumption that CO binding is a bimolecular process, whereas CO release follows first-order kinetics:
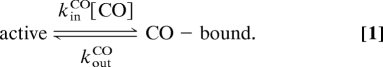 |
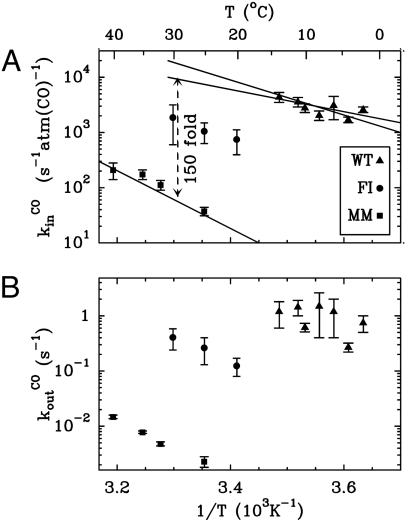
From this scheme, with the exponential decay of [CO] taken into account, [CO] = [CO]0exp(−t/τ), the change in turnover rate against time can be predicted as a function of the pseudo first-order rate constant kinCO[CO]0, the first-order rate constant koutCO, and τ (Eq. 3 in SI Text). When data like those in Fig. 3 were fitted to this equation, the relation between kinCO[CO]0 and the amount of CO injected was found to be linear for all three enzymes (data not shown), confirming our hypothesis of bimolecular inhibition kinetics. We corrected the values of kinCO to take into account that, because the inhibition by CO is competitive, H2 protects the active site and lowers the rate of CO binding by a factor of (1+[H2]/Km) (equation 3 in ref. 27). We repeated the experiments over a range of temperatures, so that the activated nature of the processes could be demonstrated (Fig. 4), and all rates could be measured at, or extrapolated to, 30°C (Table 1).
Fig. 4.
Temperature dependence of the rates of CO binding (A) and release (B) determined from data such as those in Fig. 3.
Fig. 4 shows that CO binding and release rates are 2 orders of magnitude slower in the MM mutant than in the WT enzyme, and that the FI enzyme has an intermediate phenotype. As observed by crystallography, EPR, and from the solution assays of H2 oxidation, the two mutants have intact and functional active sites. Therefore, we conclude that CO binding and release at the active site are not affected by the mutations, and that the retardations observed in Fig. 3 result from hindrance of intramolecular transport. The rate of CO intake exhibits a slightly smaller temperature dependence for the WT than for the MM mutant (Fig. 4A), suggesting that in the WT, either CO diffusion has a lower activation energy (in addition to being faster), or the step that determines the inhibition rate is different. For instance, the rate of inhibition could be limited by the binding at the active site in the WT and by intraprotein mass transport in the mutant.
Kinetics of Substrate Diffusion from Isotope-Exchange Measurements.
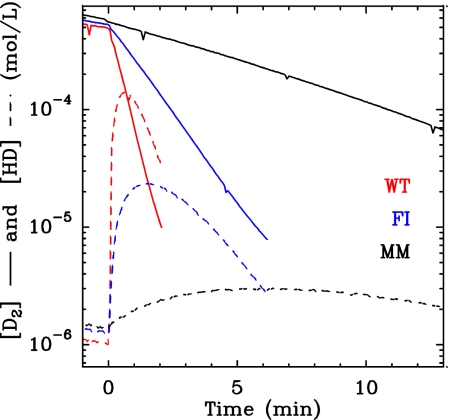
Hydrogenase activity can be assayed by using mass spectrometry to measure the rates of D2, HD, and H2 exchange in the absence of a redox partner (28). Fig. 5 shows typical results for D2 and HD concentration changes (plain and dashed lines, respectively) when the assay is initiated by adding the enzyme to a solution of D2. HD is present in the solution only transiently, and eventually bulk D2 is quantitatively replaced with H2. This is in agreement with a reaction mechanism according to which HD is an intermediate between D2 and H2: (i) D2 reversibly binds to the active site where it is heterolytically cleaved; (ii) H+ from the solvent substitutes for D+; and (iii) the D+ of the resulting HD species is eventually replaced with H+, generating H2. The latter substitution can occur either right away or after HD has been transiently released into the solvent, where it can be detected by mass spectrometry. Slowing the transport of H2, D2, and HD in the channel should decrease the probability that HD exits the enzyme before it further reacts to give H2. Therefore, HD production should be lower in the mutants where the diffusion is slowed, because HD is trapped at the active site. This is indeed observed in Fig. 5, which shows that the mutants, particularly the MM variant, produce much less HD than the WT enzyme. Consistently, Fig. 5 also shows that D2 consumption is slower for the mutants, despite the fact that the enzyme concentration was greater.
Fig. 5.
Comparison of the kinetics of isotope exchange. The concentrations of [D2] and [HD] are plotted as plain and dashed lines, respectively. The experiments were started by adding the enzyme at t = 0. pH 7, T = 30°C, enzyme concentration 2.7 μg/ml (WT), 5.6 μg/ml (FI), and 7.1 μg/ml (MM).
We quantitatively analyzed the results in Fig. 5 by using the analytical solution of the simple kinetic model described in the SI Text. According to this model, the maximal concentration of HD depends only on the ratio k′ = k/kout, where kout is the first-order rate of release to the solvent of dihydrogen as either D2, HD, or H2, and k is the first-order rate of H+/D+ exchange at the active site.
We denote [D2]0 the initial concentration of D2. As anticipated above, the maximal value of [HD] decreases upon increasing k/kout (e.g., by slowing the diffusion process while keeping k constant). The working curve in Fig. S2 can be used to determine this ratio from the value of [HD]max. The model also supports the experimental observation that the concentration of D2 (Fig. 5) and the isotopic content T = [D2]+[HD]/2 (not shown) decay exponentially over time. If kD and kT are the corresponding first-order rate constants, the value of k′ can alternatively be deduced from the following expression:
It is remarkable that the above combination of rate constants simply gives k/kout, whereas each of the measurable rate constants (kD and kT) is a complex function of all parameters in the model (see Eq. 7 in SI Text) and thus cannot be directly interpreted. We used the methods defined by Eqs. 2 and 3 to determine k′, and they gave consistent results; the latter is particularly useful when the isotope exchange activity is small, and the experiment is stopped before [HD] reaches its maximum value.
That the mutations do not significantly decrease the maximal rate of H2 oxidation suggests they have little or no effect on the value of k. Therefore, the variation of kout/k should reflect mainly the change in diffusion kinetics. Indeed, Table 1 shows a strong correlation between the values of kout/k, as determined from the isotope exchange measurement, and koutCO (for CO release) as determined by PFV. This agreement between the two independent measurements is remarkable, considering how simple our models are.
Discussion
Twenty years have passed since the first enzyme tunnel was identified by x-ray crystallography (29), and recent advances in computer modeling have made it possible to postulate gas migration pathways in several proteins. However, these theoretical results have not been confronted with experimental observations, because methods for determining the rates of diffusion inside proteins are lacking. It is not always established whether small diatomic molecules transit through static tunnels (6) or dynamic pathways (12), and to the best of our knowledge, there is only one reported example of a structurally characterized mutant of an enzyme showing impeded intramolecular diffusion (15).
We have proposed two experimental methods for probing intramolecular gas-transport kinetics in hydrogenases, which we applied to two double mutants where the end of the gas tunnel is significantly narrowed (Fig. 1). The mutations strongly increase the Michaelis constant for H2, with no appreciable effect on the value of kcat (extrapolated to infinite concentration of H2 in Table 1). This phenotype is expected when a mutation slows substrate access to the active site (4, 5, 8, 15).
The first method may be useful in other studies assessing the dynamics of gas diffusion in redox enzymes that are inhibited by O2, NO, CO, or CO2. It takes advantage of the high temporal resolution of PFV-monitored H2 oxidation to measure the rates of binding and release of the competitive inhibitor CO. In the WT enzyme and at room temperature, binding of CO is fast; it is just below the diffusion limit of 108 s−1·M−1 [≈105 s−1·atm(CO)−1; the solubility of CO is 0.96 mM·atm−1]. The two double mutations induce spectacular delays in both binding and release of CO (Fig. 3 and Table 1). The rate constants for both forward and backward CO transport decrease by more than 2 orders of magnitude when the tunnel-surrounding residues V74 and L122 are changed to methionine, whereas the double FI mutant has an intermediate phenotype. Noteworthy, the mutations affect the rates of CO binding and release in approximately the same manner, so they have a smaller effect on the binding affinity, which is the ratio of the two. This is as expected for a mutation that affects the channel for CO access, but not the free energy of binding at the active site (4, 5, 14).
A second indication that intramolecular diffusion in NiFe hydrogenase is slowed by the mutations comes from examining the yield in the isotope-exchange reaction. HD is an intermediate along the reaction pathway from D2 to H2, and because HD escape from the active site competes with the formation of H2, the slower the transport, the less HD dissociates from the enzyme and can be detected. Indeed, the two double mutants produce less HD than the WT enzyme (Fig. 5). Our kinetic analysis relates the maximum concentration of [HD] to the rate of diffusion to the bulk, and the results agree well with the independent measurements of the kinetics of CO release. Hence, the two mutations affect the rates of transport of H2 and CO in the same manner, although the former diffuses much more quickly; for example, in the MM variant at 30°C, the pseudofirst-order rate of CO binding under 1 atm of CO (65 s−1 in Table 1) is 1 order of magnitude slower than the apparent first-order rate constant (kcatapp = 600 s−1 under 1 atm of H2), which incorporates H2 binding.
A major outcome of our study is that it demonstrates the existence of a specific path for gas access to the active site of NiFe hydrogenase that coincides with the end of the tunnel found in the x-ray structure (17). Furthermore, the methods we propose are invaluable, because they allow for the calculation of diffusion rates in both directions, making it possible to establish how these rates are affected by mutations. The diameter of the tunnel's bottleneck correlates only partially with the observed rates of diffusion; the tunnel is widest in the WT enzyme, which shows the highest rates, but gas diffusion is slowest in the MM enzyme, despite the fact that the tunnel is narrower in the FI mutant. This clearly demonstrates the importance of experimentally determining diffusion rates. In addition to the main hydrophobic tunnel, a nearby “wet” hydrophilic cavity (Fig. 1E) may generate a deviation for gas diffusion, but only if the bound internal water molecules are displaced. Movements of the water molecules in the wet cavity could also allow the mutated residues to rearrange. In any case, it is clear that protein dynamics plays a major role. This is particularly evident in the mutants, because without side-chain flexibility, the access to the active site should be fully blocked. We conclude that, although the cavity search in the static crystallographic model successfully identified the main gas pathway, thermal fluctuations define the kinetics of gas diffusion.
Recently, Cohen and Schulten (14) have computed O2 diffusion maps in a number of globins, to determine which structural features promote the formation of transient gas tunnels (14). They concluded that rather large and/or flexible hydrophobic residues such as Phe and Ile have the highest propensity to form gas pathways, because they promote the formation of transient cavities. Our results do not advocate this mechanism in the case of NiFe hydrogenase, because we found that both CO and HD diffusions are slowed upon introduction of these amino acids. This effect is consistent with the earlier observation that substituting these bulky residues for smaller amino acids in oxygen-tolerant hydrogenases increases their sensitivity to O2, suggesting that they obstruct the tunnel (18, 19). We are now using site-directed mutagenesis and the methods introduced above to systematically study how the detailed structure of the gas tunnels affects both gas diffusion and inhibition by oxygen in NiFe (21) and FeFe (30) hydrogenases.
Materials and Methods
More detailed materials and methods may be found in SI Text.
Protein Production and Purification.
The WT and variants of the NiFe hydrogenase from Desulfovibrio fructosovorans were homologously produced. The D. fructosovorans and Escherichia coli strains, plasmids, growth conditions, and site-directed mutagenesis methods used in this study are described in ref. 22. The Strep tag II sequence (IBA) was introduced at the 5′ terminus of the large subunit gene (hynB). The two-step purification procedure (affinity, then anion-exchange chromatography) is described in detail in SI Text.
Assays and Biophysical Methods.
The assays of H2 oxidation were carried out after the enzymes were activated (22). The isotope-exchange assay is described in ref. 28. The electrochemical and spectroscopic characterizations were carried out as in refs. 21, 22, 26, 27.
Crystal Structure Determination.
Crystals of the double mutants of D. fructosovorans NiFe-hydrogenase were obtained and stored in liquid N2 as described for the S499A mutant (31). Diffraction data were collected at the European Synchrotron Radiation Facility at 100 K on a square Area Detector Systems Corporation Q315R detector, using beamline ID23–1 for the MM mutant and beamline ID14-4 for the FI mutant. Diffraction spots were integrated, scaled and subjected to zero-dose correction (32) with the XDS package (33). Intensity data statistics are given in Table S2. Both crystal structures were refined with REFMAC (34), as described in ref. 31. Refinement statistics are shown in Table S3. The tunnels were calculated with the program CAVENV (20).
Supplementary Material
Acknowledgments.
We thank Pierre Ceccaldi and Nicolas Martinez for taking part in this study and Athel Cornish Bowden (Centre National de la Recherche Scientifique, Marseilles, France) for fruitful discussions. This work was funded by the Centre National de la Recherche Scientifique, Commissariat à l'Energie Atomique, Agence Nationale de la Recherche, Action Concertée Incitative ECD110, the University of Provence, and the City of Marseilles and supported by the Pôle de Compétitivité Capénergies. The Groupe de Recherche 2977 (“Bio-hydrogène”) defrayed the publication fees of this article.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CUR and 3CUS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803689105/DCSupplemental.
References
- 1.Weeks A, Lund L, Raushel FM. Tunneling of intermediates in enzyme-catalyzed reactions. Curr Opin Chem Biol. 2006;10:465–472. doi: 10.1016/j.cbpa.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Tan X, Loke H-K, Fitch S, Lindahl PA. The tunnel of acetyl-coenzyme A synthase/CO dehydrogenase regulates delivery of CO to the active site. J Am Chem Soc. 2005;127:5833–5839. doi: 10.1021/ja043701v. [DOI] [PubMed] [Google Scholar]
- 3.Tan X, Volbeda A, Fontecilla-Camps JC, Lindahl PA. Function of the tunnel in acetylcoenzyme A synthase/CO dehydrogenase. J Biol Inorg Chem. 2006;11:371–378. doi: 10.1007/s00775-006-0086-9. [DOI] [PubMed] [Google Scholar]
- 4.Riistama S, et al. Channelling of dioxygen into the respiratory enzyme. Biochim Biophys Acta. 1996;1275:1–4. doi: 10.1016/0005-2728(96)00040-0. [DOI] [PubMed] [Google Scholar]
- 5.Riistama S, et al. Binding of O2 and its reduction are both retarded by replacement of valine 279 by isoleucine in cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39:6365–6372. doi: 10.1021/bi000123w. [DOI] [PubMed] [Google Scholar]
- 6.Salomonsson L, Lee A, Gennis RB, Brzezinski P. A single-amino-acid lid renders a gas-tight compartment within a membrane-bound transporter. Proc Natl Acad Sci USA. 2004;101:11617–11621. doi: 10.1073/pnas.0402242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igarashi RY, Seefeldt LC. Nitrogen fixation: The mechanism of the Mo-dependent nitrogenase. Crit Rev Biochem. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 8.Saam J, et al. Molecular dioxygen enters the active site of 12/15-lipoxygenase via dynamic oxygen access channels. Proc Natl Acad Sci USA. 2007;104:13319–13324. doi: 10.1073/pnas.0702401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray JW, Barber J. Structural characteristics of channels and pathways in photosystem II including the identification of an oxygen channel. J Struct Biol. 2007;159:228–237. doi: 10.1016/j.jsb.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Structure/function relationships of NiFe- and FeFe-hydrogenases. Chem Rev. 2007;107:4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 11.Montet Y, et al. Gas access to the active site of NiFe hydrogenases probed by X-ray crystallography and molecular dynamics. Nat Struct Mol Biol. 1997;4:523–526. doi: 10.1038/nsb0797-523. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J, et al. Finding gas diffusion pathways in proteins: Application to O2 and H2 transport in CpI FeFe-hydrogenase and the role of packing defects. Structure (London) 2005;13:1321–1329. doi: 10.1016/j.str.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Johnson BJ, et al. Exploring molecular oxygen pathways in Hansenula polymorpha copper-containing amine oxidase. J Biol Chem. 2007;282:17767–17776. doi: 10.1074/jbc.M701308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen J, Schulten K. O2 migration pathways are not conserved across proteins of a similar fold. Biophys J. 2007;93:3591–3600. doi: 10.1529/biophysj.107.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, et al. The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase. Biochemistry. 2008;47:5368–5377. doi: 10.1021/bi800228w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thoden JB, Huang X, Raushel FM, Holden HM. Carbamoyl-phosphate synthetase: Creation of an escape route for ammonia. J Biol Chem. 2002;277:39722–39727. doi: 10.1074/jbc.M206915200. [DOI] [PubMed] [Google Scholar]
- 17.Volbeda A, et al. High-resolution crystallographic analysis of Desulfovibrio fructosovorans NiFe hydrogenase. Int J Hyd Energ. 2002;27:1449–1461. [Google Scholar]
- 18.Buhrke T, Lenz O, Krauss N, Friedrich B. Oxygen tolerance of the H2-sensing NiFe hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J Biol Chem. 2005;280:23791–23796. doi: 10.1074/jbc.M503260200. [DOI] [PubMed] [Google Scholar]
- 19.Duché O, Elsen S, Cournac L, Colbeau A. Enlarging the gas access channel to the active site renders the regulatory hydrogenase HupUV of Rhodobacter capsulatus O2 sensitive without affecting its transductory activity. FEBS J. 2005;272:3899–3908. doi: 10.1111/j.1742-4658.2005.04806.x. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Project. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Léger C, et al. Inhibition and aerobic inactivation kinetics of Desulfovibrio fructosovorans NiFe hydrogenase studied by protein film voltammetry. J Am Chem Soc. 2004;126:12162–12172. doi: 10.1021/ja046548d. [DOI] [PubMed] [Google Scholar]
- 22.Dementin S, et al. A glutamate is the essential proton transfer gate during the catalytic cycle of the NiFe hydrogenase. J Biol Chem. 2004;279:10508–10513. doi: 10.1074/jbc.M312716200. [DOI] [PubMed] [Google Scholar]
- 23.Guigliarelli B, et al. Structural organization of the Ni and [4Fe4S] centers in the active form of Desulfovibrio gigas hydrogenase: Analysis of the magnetic interactions by EPR spectroscopy. Biochemistry. 1995;34:4781–4790. doi: 10.1021/bi00014a036. [DOI] [PubMed] [Google Scholar]
- 24.Mandal PK, Arunan E. Hydrogen bond radii for the hydrogen halides and van der Waals radius of hydrogen. J Chem Phys. 2001;114:3880–3882. [Google Scholar]
- 25.Vincent KA, Parkin A, Armstrong FA. Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem Rev. 2007;107:4366–4413. doi: 10.1021/cr050191u. [DOI] [PubMed] [Google Scholar]
- 26.Léger C, Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem Rev. 2008;108:2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- 27.Almeida MG, et al. A needle in a haystack: The active site of the membrane-bound complex cytochrome c nitrite reductase. FEBS Lett. 2007;581:284–288. doi: 10.1016/j.febslet.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Cournac L, Guedeney L, Peltier G, Vignais PM. Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC 6803 deficient in the type I NADPH-dehydrogenase complex. J Bacteriol. 2004;186:1737–1746. doi: 10.1128/JB.186.6.1737-1746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyde CC, et al. Three dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 30.Baffert C, et al. Hydrogen-activating enzymes: Activity does not correlate with oxygen sensitivity. Angew Chem Int Ed. 2008;47:2052–2054. doi: 10.1002/anie.200704313. [DOI] [PubMed] [Google Scholar]
- 31.Volbeda A, et al. Structural differences between the ready and unready oxidized states of NiFe hydrogenases. J Biol Inorg Chem. 2005;10:239–249. doi: 10.1007/s00775-005-0632-x. [DOI] [PubMed] [Google Scholar]
- 32.Diederichs K, McSweeney S, Ravelli RB. Zero-dose extrapolation as part of macromolecular synchrotron data reduction. Acta Crystallogr D. 2003;59:903–909. doi: 10.1107/s0907444903006516. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch W. In: International Tables for Crystallography. Rossmann MG, Arnold E, editors. Vol F. The Netherlands: Kluwer, Dordrecht; 2001. pp. 218–225. [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.