Abstract
The aerobic catabolism of nicotinic acid (NA) is considered a model system for degradation of N-heterocyclic aromatic compounds, some of which are major environmental pollutants; however, the complete set of genes as well as the structural–functional relationships of most of the enzymes involved in this process are still unknown. We have characterized a gene cluster (nic genes) from Pseudomonas putida KT2440 responsible for the aerobic NA degradation in this bacterium and when expressed in heterologous hosts. The biochemistry of the NA degradation through the formation of 2,5-dihydroxypyridine and maleamic acid has been revisited, and some gene products become the prototype of new types of enzymes with unprecedented molecular architectures. Thus, the initial hydroxylation of NA is catalyzed by a two-component hydroxylase (NicAB) that constitutes the first member of the xanthine dehydrogenase family whose electron transport chain to molecular oxygen includes a cytochrome c domain. The Fe2+-dependent dioxygenase (NicX) converts 2,5-dihydroxypyridine into N-formylmaleamic acid, and it becomes the founding member of a new family of extradiol ring-cleavage dioxygenases. Further conversion of N-formylmaleamic acid to formic and maleamic acid is catalyzed by the NicD protein, the only deformylase described so far whose catalytic triad is similar to that of some members of the α/β-hydrolase fold superfamily. This work allows exploration of the existence of orthologous gene clusters in saprophytic bacteria and some pathogens, where they might stimulate studies on their role in virulence, and it provides a framework to develop new biotechnological processes for detoxification/biotransformation of N-heterocyclic aromatic compounds.
Keywords: ring-cleavage dioxygenase, nicotinic acid hydroxylase, heterocyclic compounds
Nicotinic acid (NA) is a carboxylic derivative of pyridine that is widely distributed in nature as part of pyridine cofactors (e.g., NAD and NADP) and alkaloids (e.g., nicotine and anabasine), and it is essential (vitamin B3) for those organisms that are not able to carry out its synthesis (1). NA is also a carbon and nitrogen source for different bacteria and some fungi (1, 2), and the biochemical pathways involved in the degradation of this N-heterocyclic aromatic compound have been used as a source of novel and unusual enzyme activities as well as metabolic intermediates, e.g., 6-hydroxynicotinic acid (6HNA) and 2,5-dihydroxypyridine (2,5DHP), that can funnel the degradation of toxic compounds of major environmental concern, such as nicotine and hydroxypyridines (1, 2), and are of pharmacological and agrochemical value (3). So far, the only fully elucidated NA degradation pathway is the anaerobic route from Eubacterium barkeri (4).
Although the aerobic NA degradation has been considered as a model system for the degradation of N-heterocyclic aromatic compounds and its biochemistry has been studied in different microorganisms for >50 years (1, 2, 5–7), the complete set of genes encoding this pathway as well as the structural–functional relationships of most of the enzymes involved in this process have not been reported and analyzed so far in any organism. In many bacteria, the aerobic degradation of NA produces 2,5DHP as a central intermediate, which is further degraded according to a scheme (maleamate pathway) that has been outlined (Fig. 1) (1, 2, 6, 7). In this work, we show that Pseudomonas putida KT2440, a well studied and paradigmatic bacterium renowned for its ability to metabolize a wide range of aromatic compounds (8, 9), is able to use NA aerobically as a sole carbon, nitrogen, and energy source. We have been able to identify and characterize in this bacterium a gene cluster (nic genes) responsible for the aerobic NA degradation. The biochemistry of aerobic NA degradation through the maleamate pathway has been revisited, and we have demonstrated that some gene products of this pathway become prototypes of new types of enzymes with unprecedented molecular architectures. Some ecological and evolutionary considerations regarding the nic genes are also discussed.
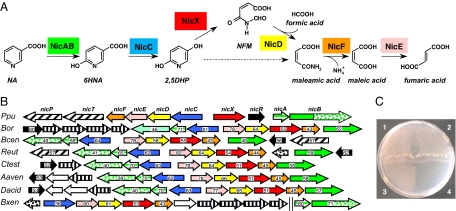
Fig. 1.
Aerobic NA degradation via the maleamate pathway. (A) Scheme of the aerobic NA degradation pathway in P. putida KT2440. NicAB, NA hydroxylase (green); NicC, 6HNA monooxygenase (blue); NicX, 2,5DHP dioxygenase (red); NicD, NFM deformylase (yellow); NicF, maleamate amidohydrolase (orange); NicE, maleate isomerase (pink). The hatched arrow shows the previously proposed 2,5DHP ring-cleavage reaction. (B) Genetic organization of the nic cluster in different bacteria. Genes are functionally annotated following the color code indicated in A. Regulatory genes encoding NicR-type and TetR-type regulators are indicated by black and dotted arrows, respectively. Solid, dotted, and striped green arrows indicate the nicB2 gene/domain, the nicB1 gene/domain, and the nicA gene, respectively. Porin and MFS transport genes are indicated by hatching arrows. Genes encoding subunits of putative ABC transporters are represented by vertically striped arrows. Numbers within the arrows indicate the percent amino acid sequence identity with the ortholog gene product from P. putida KT2440. The abbreviations and accession codes for the different gene clusters are detailed in SI Text. (C) Growth of P. putida KT2440 (portion 1), Pseudomonas sp. MT14 (pNIC) (portion 2), P. putida KT2440dnicB (portion 3), and Pseudomonas sp. MT14 (pBBR1MCS-5) (portion 4) in MC minimal medium containing 5 mM NA as the sole carbon source.
Results and Discussion
The nic Genes Are Responsible for NA Catabolism in P. putida KT2440.
The in silico analysis of the P. putida KT2440 genome revealed a gene cluster, named nicTPFEDCXRAB (Fig. 1), some of whose gene products showed significant similarities to proteins involved in the catabolism of N-heterocyclic aromatic compounds [supporting information (SI) Table S1]. When P. putida KT2440 was tested with different N-heterocycles (pyridoxal, pyridoxamine, pyridine, 2- and 3-hydroxypyridine, NA, isonicotinic acid, picolinic acid, quinoline, and isoquinoline) as sole carbon and energy sources, we observed growth only in NA (Fig. 1C), thus implicating the nic cluster in NA catabolism. To confirm this assumption, several P. putida KT2440 knockout mutants were constructed (Table S2). Disruption of the nicA, nicB, nicC, nicD, and nicX genes did not allow the corresponding P. putida KT2440 mutant strains to grow in NA as the sole carbon source (Fig. 1C). The nic cluster also contained two genes, nicP and nicT, encoding a potential porin and permease transport system, respectively, and a regulatory gene (nicR) (Fig. 1 and Table S1; see also SI Text).
Growth in NA could be restored when the P. putida knockout mutants were complemented with a broad-host-range plasmid (pNIC) harboring a 14-kb DNA cassette containing the complete nic cluster (Table S2). Interestingly, the pNIC plasmid also conferred the ability to use NA as sole carbon source to other bacteria, such as Pseudomonas sp. MT14 (Table S2), that are unable to degrade this aromatic compound (Fig. 1C). All of these results demonstrate that the nic cluster identified in P. putida KT2440 constitutes the first complete set of genes reported so far in any organism that is responsible for aerobic NA degradation.
The nicAB Genes Encode the NA Hydroxylase.
The observation that the disruption of the nicA and nicB genes abolished growth of the corresponding P. putida KT2440 mutants in NA but did not affect their growth in 6HNA (data not shown), as well as the fact that the nicA and nicB gene products show significant sequence similarity with the subunits of some members of the xanthine dehydrogenase family (Table S1), suggested that these two genes might encode the NA hydroxylase from P. putida KT2440 (Fig. 1). To confirm the role of the nicA and nicB genes, they were cloned and expressed in plasmid pNicAB (Table S2). When plasmid pNicAB was transferred to Pseudomonas strains that are not able to oxidize NA, such as Pseudomonas fluorescens R2f and Pseudomonas sp. DSM6412 (Table S2), the recombinant strains acquired the ability to efficiently convert NA into a stoichiometric amount of 6HNA (Fig. S1). NA hydroxylase assays using crude extracts from P. fluorescens R2f (pNicAB) revealed that the activity increased significantly when an external electron acceptor like phenazine methosulfate (PMS) was added and decreased below detection levels under anaerobic conditions (Fig. 2A). The hydroxylase activity was maximal at 30°C and pH 7.5, and it was highly specific for NA (Table S3). These data indicate that genes nicAB encode the two-component NA hydroxylase that converts NA into 6HNA, constituting the first aerobic NA hydroxylase whose primary structure is known.
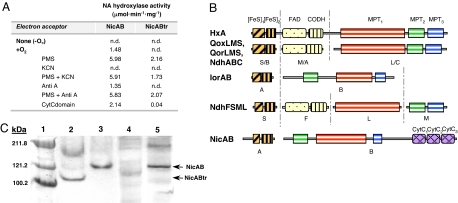
Fig. 2.
The NA hydroxylase from P. putida. (A) NA hydroxylase activity of the NicAB and NicABtr enzymes. The enzyme assays were carried out with crude extracts of P. fluorescens R2f harboring plasmids pNicAB (NicAB) or pNicABtr (NicABtr) under anaerobic (−O2) or aerobic (+O2) conditions and in the presence of 0.4 mM PMS (PMS); 0.4 mM PMS plus 50 μM KCN (PMS+KCN); 50 μM antimycin A (AntiA); 0.4 mM PMS plus 50 μM antimycin A (PMS+AntiA); and cytochrome c domain of NicB (CytCdomain). n.d., not detected. Results of one experiment are shown, and values were reproducible in three separate experiments with SD values of <10%. (B) Molecular architecture of different molybdohydroxylases acting on N-heterocyclic aromatic compounds. The descriptions, accession codes, and conserved domains for the different enzymes are detailed in SI Text. (C) Nondenaturing PAGE of NicAB and NicABtr proteins. Lane 1, Coomassie-prestained protein markers; lanes 2 and 4, 100 μg of total protein from a crude extract of P. fluorescens R2f (pNicABtr) expressing the NicABtr protein; lanes 3 and 5, 100 μg of total protein from a crude extract of P. fluorescens R2f (pNicAB) expressing the NicAB protein. Lanes 2 and 3, NA hydroxylase activity staining; lanes 4 and 5, heme staining.
Hydroxylases of N-heterocyclic aromatic compounds incorporate oxygen derived from a water molecule into the product, and they typically contain a redox center, formed by a molybdenum ion coordinated to an organic cofactor (molybdopterin cytosine dinucleotide, MCD), two [2Fe-2S] clusters, and usually FAD (Fig. 2B), which transports electrons from the reducing substrate (N-heterocyclic compound) to the oxidizing substrate (the electron acceptor) (5, 10). The NA hydroxylase small subunit (NicA, 23.8 kDa) harbors the 41-C-X4-C-G-X-C-Xn-C-59 and 100-C-G-X-C-X31-C-X-C-137 conserved motifs likely involved in binding of the two [2Fe-2S] clusters that are located in the small subunit of other multicomponent xanthine dehydrogenases (5, 10) (Fig. 2B). However, analysis of the amino acid sequence of the NA hydroxylase large subunit (NicB, 128 kDa) revealed some unique features not reported yet in other hydroxylases of N-heterocyclic aromatic compounds. Thus, the N-terminal region of NicB shows a molecular architecture similar to that of the IorB subunit of isoquinoline oxidoreductase, containing the active site and the MCD binding sites in an arrangement (MPT2-MPT1-MPT3) that differs from that observed (MPT1-MPT2-MPT3) in most members of the xanthine dehydrogenase family (5, 10) (Fig. 2B). The existence of a MCD cofactor in NicB and the fact that Escherichia coli cannot synthesize this particular cofactor (11) explain the lack of NA hydroxylase activity in E. coli (pNicAB) cells (data not shown). On the other hand, the C-terminal region of NicB (residues 750-1187) contains three conserved cytochrome c (CytC) heme-binding motifs (817-CAVCH-823, 963-CTACH-969, and 1087-CLGCH-1093) (12) (Fig. 2B) that have not been described previously in any member of the xanthine dehydrogenase/oxidase family. The involvement of a CytC mediating the electron transfer from the intramolecular redox center to the final acceptor (oxygen) would explain the absence in NicB of the FAD-binding site usually present in enzymes of the xanthine-dehydrogenase family (5, 10) (Fig. 2B) and the observation that the basal NA hydroxylase activity could not be increased by adding external electron acceptors such as the organic cofactors NADP, NAD, FMN, and FAD or the inorganic ions Fe3+ and NO3− (data not shown).
When a truncated NicB subunit lacking the CytC domain was constructed and expressed from plasmid pNicABtr, the NicABtr truncated enzyme did not show NA hydroxylase activity (Fig. 2A). However, NicABtr acquired significant activity when PMS was added as a terminal electron acceptor (Fig. 2A). The NA hydroxylase assay could be also performed when the NicAB and NicABtr enzymes were loaded in a nondenaturing PAGE, and two bands of the expected molecular size for each heterodimer were observed (Fig. 2C). Nevertheless, whereas the band corresponding to the wild-type NicAB enzyme could be also detected by a heme-staining assay, the corresponding band of the NicABtr enzyme was not detected (Fig. 2C), which agrees with the predicted absence of heme groups in the truncated protein. The NA hydroxylase activity was partially restored (NicABtr) and increased (NicAB) when these enzymes were complemented in trans with the CytC domain of NicB cloned and expressed in plasmid pVLTCytC (Fig. 2A), which strongly suggests that this region behaves indeed as a functional CytC domain. On the other hand, it was observed that the NA hydroxylase activity was inhibited in the presence of KCN, but the addition of antimycin A had no effect (Fig. 2A). As expected, neither KCN nor antimycin A inhibited the NA hydroxylase activity when PMS was used as terminal electron acceptor (Fig. 2A). Because it is well known that the CytC oxidase is inhibited by KCN but not by antimycin A (12), it is likely that the CytC domain of NA hydroxylase may transfer the electrons from the redox center to a CytC oxidase that uses molecular oxygen as final electron acceptor.
In summary, all of these data show that the two-component NicAB protein constitutes the prototype of a previously uncharacterized class of hydroxylases within the xanthine dehydrogenase family whose modular architecture includes a CytC domain as component of the electron transport chain to molecular oxygen. Because the NA hydroxylase is used in industry for the formation of 6HNA, the key precursor of several insecticides (e.g., imidachloprid) and herbicides (e.g., imazapyr) (3), its cloning and successful heterologous expression reported here are of biotechnological interest, revealing also that P. fluorescens R2f and Pseudomonas sp. DSM6412 can be suitable hosts for the functional expression of other enzymes that require the unusual MCD cofactor for their activity.
The nicC Gene Encodes the 6HNA Monooxygenase.
The second enzymatic step in the maleamate pathway for the aerobic NA degradation is the oxidative decarboxylation of 6HNA to render 2,5DHP (6) (Fig. 1). This reaction is catalyzed by a 6HNA-3-monooxygenase (13), which shows 60% amino acid sequence identity with the product of the nicC gene (Table S1). The role of the nicC gene product as the 6HNA monooxygenase was in agreement with the observation that P. putida KT2440dnicC resting cells showed a complete conversion of NA to 6HNA (Fig. S1). On the other hand, when the nicC gene was cloned and expressed in plasmid pIZNicC (Tables S2 and S4), P. fluorescens R2f (pIZNicC) cell extracts revealed a 42-kDa protein that catalyzed the conversion of 6HNA to 2,5DHP when NADH and FAD were added to the assay (data not shown). These data indicate, therefore, that NicC is the 6HNA monooxygenase of the NA degradation pathway in P. putida KT2440.
NicX Is the First Member of a New Family of Extradiol Dioxygenases.
The third step in the aerobic NA degradation pathway is the one catalyzed by a 2,5DHP dioxygenase (Fig. 1) (6). Although the 2,5DHP dioxygenase activity had been characterized many years ago and 18O2 incorporation experiments confirmed that the pyridine ring was opened by insertion of both atoms of 18O2 (14, 15), the gene encoding the corresponding enzyme and the primary structure of the protein remained unknown. Although amino acid sequence analysis of the nic gene products did not reveal the presence of any putative ring-cleavage dioxygenase (Table S1), the P. putida KT2440dnicX mutant strain was shown to lack 2,5DHP dioxygenase activity (data not shown), pointing out that the nicX gene might encode such activity. To test this hypothesis, the nicX gene was overexpressed in E. coli BL21(DE3) cells containing plasmid pETNicX (Table S2). Cell extracts showed 2,5DHP dioxygenase activity (6 μmol·min−1·mg−1), revealing that the nicX gene product was the ring-cleavage enzyme in the NA degradation pathway from P. putida KT2440.
The NicX protein was purified and analyzed by SDS/PAGE, showing a band whose apparent molecular mass (39 kDa) (Fig. 3D) corresponded with that predicted for the nicX gene product. Analysis by inductively coupled plasma emission spectroscopy (ICP) of the metal content of purified NicX indicated the absence of any metal; however, after Fe2+-dependent reactivation of the enzyme (25 μM), 19.8 μM iron was detected, indicating that the ratio of moles of iron to moles of a monomer of NicX was near 1.0. Some biochemical properties of NicX (see SI Text), e.g., 1 mol of oxygen consumption per mole of 2,5DHP, and a strict substrate specificity for 2,5DHP with Km and Vmax values of 70 μM and 2.3 μM·min−1·mg−1, respectively, resemble those reported for the 2,5DHP dioxygenase from P. putida N9 (14).
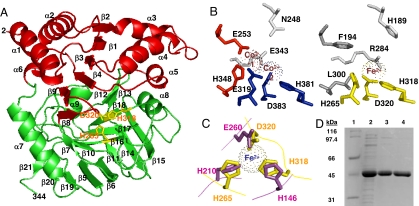
Fig. 3.
The NicX dioxygenase. (A) Three-dimensional structural model of NicX. The N-terminal domain (red) and the C-terminal domain (green) are drawn as a ribbon. Putative catalytic residues (H265, H318, and D320) are indicated in yellow. (B) Comparison between the active site of the aminopeptidase AmpS, harboring the Co1 (red) and Co2 (blue) binding sites, and the predicted Fe2+-binding site of NicX (yellow). (C) Superposition of the Fe2+-binding site of 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) from B. xenovorans LB400 (violet) and that predicted for NicX (yellow). (D) Analysis by SDS/PAGE of the purification of NicX from E. coli BL21(DE3) (pETNicX) cells. Lane 1, protein markers; lane 2, soluble crude extract; lanes 3 and 4, protein fraction after DEAE-cellulose and hydroxyapathite chromatography, respectively.
The nature of the product generated by the 2,5DHP dioxygenase is controversial. Initial studies revealed that 2,5DHP ring cleavage was performed between C5 and C6, suggesting that N-formylmaleamic acid (NFM) was the actual ring-cleavage product, although it could not be detected (6). Further work suggested that the hydrolytic cleavage of NFM to form maleamate was also catalyzed by the 2,5DHP dioxygenase enzyme (15, 16). Therefore, to determine unequivocally the chemical structure of the product generated by the 2,5DHP dioxygenase, a combination of NMR spectroscopy and mass spectrometry was used to follow the course of the reaction catalyzed by purified NicX. The reaction of 2,5DHP with the dioxygenase enzyme revealed the complete transformation of this heterocyclic compound into a single opening product whose 1H NMR and 13C NMR spectra are consistent with NFM as the ring-cleavage product (Fig. S2). On the other hand, negative mass spectrometry (electrospray) experiments revealed that the [M-H]− peak at a m/z value of 110 due to 2,5DHP completely disappeared upon treatment with NicX, and a new major peak at a m/z value of 142 was observed (Fig. S2). The molecular ion at the m/z value of 142 corresponds to the ring-cleavage product of 2,5DHP by addition of two oxygen atoms generating the corresponding N-formyl derivative, i.e., NFM. Therefore, all of these data show that NicX catalyzes the dioxygenolytic ring cleavage of 2,5DHP between carbons 5 and 6 generating NFM as final product. In light of this finding, the previously suggested dioxygenase-mediated decarboxylation of NFM to form maleamate and formate (15, 16) can be now explained as a result of a deformylase activity (see below) that was contaminating the purified 2,5DHP dioxygenase enzyme.
As mentioned above, the amino acid sequence of NicX does not show any significant similarity to that of other ring-cleavage dioxygenases described so far. Instead, NicX shows amino acid sequence similarity to members of the MQ family of proteins such as the aminopeptidases AmpS and AmpT from Staphylococcus aureus (17) and Thermus thermophilus (18), respectively. Since the three-dimensional structures of AmpS and AmpT have been recently determined (17, 18), homology modeling for NicX was accomplished. According to this model, the protein is composed of two globular domains (Fig. 3A). The N-terminal domain (residues 1–123 and 177–191) is predicted to have a Rossmann fold with a central six-stranded mixed β-sheet (β2-β1-β3-β4-β9-β8) flanked by α-helices (α1, α2, α3, α4, α5, and α6) on both sides. The C-terminal domain is organized into two antiparallel-β-sheets (β21-β20-β19-β5-β6-β11-β14-β15 and β7-β10-β16-β17-β18-β12-β13) that form a pseudo β-barrel capped by the α7 and α8 helices (Fig. 3A). The active site of AmpS contains two high-affinity Co(II) binding sites, i.e., Co1 constituted by residues E253 and H348 and Co2 formed by residues E319, H381, and D383 (17). According to the structural model of NicX, residues corresponding to the first Co(II) binding site are F194 and L300, indicating that the Co1 site is lost in this protein. However, residues H265, H318, and D320 located on the surface of one of the β-sheets at the C-terminal domain may constitute the iron coordination sphere in NicX replacing the equivalent Co2 site of AmpS (Fig. 3B). Despite the phylogenetic diversity of extradiol ring-cleavage dioxygenases, the iron (II) binding motif constituted by two histidines and one carboxylate group is a common feature of this family of enzymes (19). In this sense, the modeled active site of NicX fits perfectly the geometry of the canonical 2-His-1-carboxylate metal binding motif found in classical extradiol ring-cleavage dioxygenases (19). Crucial side-chain atoms coordinating the metal are in the same spatial position despite coming from different local architectures (Fig. 3C). It is worth noting that although 2,5DHP is not a canonical peptidic substrate like those recognized by the aminopeptidases AmpS and AmpT, it may acquire a peptidic-like structure with an amide bond (5-hydroxy-2-pyridone) as a result of its keto-enolic tautomerization in the active site of the ring-cleavage enzyme. This fact might explain the similarity found between the active site of NicX and that of such aminopeptidases. In summary, NicX is proposed as the founding member of a new family of extradiol ring-cleavage dioxygenases that evolved from the MQ family of metalloproteases with partial replacement of active site residues to obtain new facial catalytic triad.
The nicD Gene Encodes a N-Formylmaleamate Deformylase.
We have shown above that the 2,5DHP dioxygenase generates NFM as final product. To check whether nicD was responsible for the required deformylase activity, this gene was overexpressed from plasmid pETNicD (Table S2). SDS/PAGE analysis of crude extracts from E. coli BL21(DE3) cells harboring plasmid pETNicD showed the presence of an intense band corresponding to a protein of 29 kDa (data not shown), in good agreement with that predicted for the product of the nicD gene (29.1 kDa). Interestingly, these extracts were able to transform NFM to maleamic and formic acids as shown by HPLC and 1H NMR analyses (Fig. S3). These data reveal that deformylation of NFM to generate formic and maleamic acid is catalyzed by the NicD enzyme in a reaction that was not outlined in the proposed NA degradation pathway (Fig. 1).
The primary structure of NicD shows significant similarity to that of members of the α/β-hydrolase-fold superfamily of enzymes (Table S1 and Fig. S4). A homology model was proposed for the structure of NicD based on the known three-dimensional structure of the lactamase from Aureobacterium sp. (20) (Fig. S5). The α/β-hydrolases share a conserved catalytic triad formed by a nucleophile residue (usually Ser), an acidic residue, and a histidine (21). The sequence 98-G-H-S-M-G-104 in NicD perfectly matches the consensus motif for the nucleophile residue (Fig. S4). The conserved histidine in α/β-hydrolases is located in a loop after the β8 strand, and it corresponds to H245 in NicD (Figs. S4 and S5). The acidic residue in α/β-hydrolases is generally placed in a loop that follows the β7 strand; however, a multiple sequence alignment revealed that the corresponding position is occupied by residue G217 in NicD (Fig. S4), being the two closest acidic residues, E221 and D224, in an α-helix located outside the predicted active site pocket (Fig. S5). However, a detailed analysis of the predicted 3D structure for NicD revealed that another acid residue, D125, located after the β6 chain faces the putative active site (Fig. S5). Interestingly, this position is equivalent to that found for the acidic residue in some α/β-hydrolases (21) (Fig. S4).
To assess the proposed catalytic triad of NicD, the residues S101, D125, H245, and E221 were substituted through site-directed mutagenesis by Ala residues. The NicD mutant proteins were overexpressed as the wild-type protein, and their specific activities were compared. Whereas substitutions S101A, D125A, and H245A lead to a complete loss of the deformylase activity, the NicD mutant protein harboring the substitution E221A retained ≈70% of the wild-type activity (Fig. S5). Thus, these data suggest that residues S101, D125, and H245 are essential for the enzyme activity, constituting the catalytic triad of the NicD deformylase.
Although some members of the α/β-hydrolase superfamily are able to break a C–N linkage in a peptidic bond, e.g., serine-carboxypeptidase and proline iminopeptidase, no deformylase activity had been reported so far within this superfamily (21). The NicD protein, therefore, expands further the wide functional diversity associated to the α/β-hydrolase common fold.
The nicF and nicE Gene Products Catalyze the Last Two Steps of the Maleamate Pathway.
The primary structure of the nicF gene product is similar to that of members of the isochorismatase superfamily such as the N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. and the pyrazinamidase (nicotinamidase) from Pyrococcus horikoshii (22) (Table S1), two enzymes that catalyze the substitution of the amine group in an amide bond by a hydroxyl group. The catalytic mechanism of these proteins involves a cysteine residue as nucleophile, an aspartic acid, and a lysine (22). Because these three residues were conserved in the primary structure of NicF (D31, K121, and C154), it was tempting to speculate that this enzyme is the maleamate amidase that transforms maleamic acid into maleic acid plus ammonia in the NA degradation pathway (Fig. 1), thus explaining the growth of P. putida KT2440 when cultivated in NA as sole nitrogen source. To confirm the predicted role for NicF, the nicF gene was overexpressed from plasmid pETNicF (Table S2). Crude extracts of E. coli BL21 (DE3) (pETNicF) cells showed maleamate amidohydrolase activity (19.3 μmol·min−1·mg−1), indicating that the nicF gene encodes the maleamate amidohydrolase of the NA degradation pathway in P. putida KT2440. Although another amidohydrolase has been described in the anaerobic NA degradation pathway (4), this enamidase recognizes a different substrate (1,4,5,6-tetrahydro-6-oxonicotinate), and it does not show any amino acid sequence similarity to NicF, indicating that these two enzymes belong to different protein families.
The maleamate pathway ends with the isomerization of maleic acid to fumaric acid, a Krebs cycle intermediate (6) (Fig. 1). Because the nicE gene product shows a significant amino acid sequence similarity with a previously characterized maleate cis/trans-isomerase (Table S1) and contains the two conserved Cys residues (C104 and C222 in NicE) involved in the catalytic activity (23), the nicE gene was assigned as that encoding the last step of the aerobic NA degradation pathway in P. putida KT2440.
Distribution of the nic Genes in Other Bacteria and Evolutionary Considerations.
Within the Pseudomonas genus the nic genes are present only in strains of P. putida, e.g., KT2440, F1, GB-1, and W619, where they show high sequence identity (>95%) and the same organization. Although a few P. fluorescens strains (7, 24) have been reported to use NA, it cannot be ruled out that these strains are not properly classified and/or that they harbor a mobile genetic element, such as the NIC plasmid from a P. putida strain (25), that confers the ability to degrade NA. Outside the Pseudomonas genus, orthologs of the nic genes were found only in the genomes of β-proteobacteria from the Burkholderiales family (Fig. 1B). In most β-Proteobacteria the nicB2 and nicB1 genes encode proteins that correspond to N-terminal (first 740 residues) and C-terminal (last 480 residues) regions of the nicB gene product from Pseudomonas, respectively (Fig. 1B). This observation suggests that the large subunit (NicB) of the NA hydroxylase from Pseudomonas has a modular architecture of two domains that have evolved as two different proteins in β-Proteobacteria: (i) the N-terminal domain (NicB2 protein) would be involved in binding to the substrate and MCD cofactor, and (ii) the C-terminal CytC domain (NicB1 protein) would participate in the electron transfer to the CytC oxidase. Nevertheless, the nicAB genes from Burkholderia xenovorans constitute an exception because they retain the same molecular architecture as the corresponding nicAB genes from P. putida and they are located not within the nic cluster (Fig. 1B) but in an additional cluster predicted to be involved in an alternative NA degradation pathway via 1,4,5,6,-tetrahydro-6-oxonicotinate that is also found in some α-Proteobacteria (4). Most nic clusters harbor a gene (nicR) that encodes a MarR-type transcriptional regulator and a putative NA transport system of the MFS superfamily (nicT) or an ABC transporter (nicT1-T4) (Fig. 1B).
Because plants are the principal source of NA in the environment, it was not surprising to find nic genes in soil bacteria that are able to colonize the plant rhizosphere, such as P. putida, Burkholderia, or Ralstonia strains. On the contrary, it was unexpected to find conserved nic clusters in pathogens such as Bordetella spp. that are able to colonize mammalian respiratory tract. The presence of the nic genes in the genome of several Bordetella spp. provides some clues to the ability of these strains to metabolize NA and to control the NA levels that regulate their infectiveness (26).
Concluding Remarks.
The complete set of genes involved in aerobic NA degradation via the maleamate pathway has been studied at the molecular level and successfully expressed in heterologous hosts. Some unique proteins, such as the two-component NA hydroxylase and the 2,5DHP dioxygenase, constitute the founding members of previously undescribed classes of enzymes. These results provide also a framework for developing new biotechnological processes that use N-heterocyclic aromatic compounds, such as 6HNA and 2,5DHP, as building blocks, and for detoxification of nicotine in tobacco wastes (3, 27). The nic genes from P. putida are also useful to explore the existence of homologous gene clusters in saprophytic bacteria and some animal pathogens where they might stimulate studies on their role in virulence.
Materials and Methods
Bacterial Strains, Plasmids, Growth Conditions, and Molecular Biology.
The bacterial strains and plasmids used in this study are listed in Table S2. The construction of recombinant plasmids and P. putida KT2440 mutants harboring disrupted nic genes is detailed in SI Text. Bacteria were grown in LB medium (28) at 37°C (E. coli) or 30°C (Pseudomonas). Pseudomonas strains were also grown at 30°C in M63 or MC minimal media (29) containing citrate (0.2%), NA (5 mM), or 6HNA (5 mM). Standard molecular biology techniques were performed as previously described (28).
Enzymatic Assays.
NA hydroxylase and 6HNA 3-monooxygenase were assayed in resting cells and cell extracts as detailed in SI Text. The 2,5DHP dioxygenase (NicX) was assayed spectrophotometrically by measuring the disappearance of 200 μM 2,5DHP at 320 nm (ε = 5,200 M−1·cm−1) as previously described (14). Because purified NicX is inactive, the protein (5 μg) was reactivated by incubation with 10 mM DTT and 0.5 mM FeSO4 for 15 min at 25°C. Kinetic parameters and the substrate specificity were calculated by measuring the substrate-dependent oxygen consumption rate of the reaction mixture at 25°C using a DW1 Hansa-Tech Oxygen Electrode. NFM deformylase activity (NicD) was measured by coupling deformylase activity to a commercial formate dehydrogenase enzyme from Candida boidinii (Sigma) and monitoring NADH formation as indicated by the manufacturer. The two products of the NicD-catalyzed reaction, i.e., formate and maleate, were identified by HPLC and RMN analyses. Maleamate amidohydrolase (NicF) was assayed by coupling NH4+ formation to a commercial glutamate dehydrogenase (Sigma) and by measuring NADPH oxidation at 340 nm (ε = 6,220 M−1·cm−1), in the presence of α-ketoglutarate.
Other Techniques.
The overproduction and purification of NicX were carried out as indicated in SI Text. Proteins were analyzed by SDS/PAGE and Coomassie-stained as described previously (28). The protein concentration in cell extracts was determined by the method of Bradford by using BSA as the standard (29). NA hydroxylase activity staining and heme staining in nondenaturing PAGE were performed with nitroblue tetrazolium and dimethoxybenzidine dihydrocholoride, respectively, as previously described (7, 30). Fe2+ was determined by ICP using a ICP-OES Optima 2000DV equipment (PerkinElmer). The different techniques used for metabolites detection are detailed in SI Text. Three-dimensional modeling of NicX and NicD was performed as indicated in SI Text, and figures were generated with PyMOL (DeLano Scientific).
Supplementary Material
Acknowledgments.
We thank M. Morales, A. Valencia, and M. Zazo for technical assistance; E. Rial for the oxygen-consumption experiments; and D. Stjepandic and J. D. Hoheisel for the gift of the P. putida cosmid library. We are indebted to V. de Lorenzo for the critical reading of the manuscript. This work was supported by grants QLK3-CT2000-00170, GEN2001-4698-C05-02 and GEN2006-27750-C5-3-E. K.G. acknowledges an EMBO Installation Grant. J.I.J. was the recipient of a I3P predoctoral fellowship from the Consejo Superior de Investigaciones Científicas.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802273105/DCSupplemental.
References
- 1.Kaiser JP, Feng Y, Bollag JM. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev. 1996;60:483–498. doi: 10.1128/mr.60.3.483-498.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fetzner S. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl Microbiol Biotechnol. 1998;49:237–250. [Google Scholar]
- 3.Yoshida T, Nagasawa T. Enzymatic functionalization of aromatic N-heterocycles: Hydroxylation and carboxylation. J Biosci Bioeng. 2000;89:111–118. doi: 10.1016/s1389-1723(00)88723-x. [DOI] [PubMed] [Google Scholar]
- 4.Alhapel A, et al. Molecular and functional analysis of nicotinate catabolism in Eubacterium barkeri. Proc Natl Acad Sci USA. 2006;103:12341–12346. doi: 10.1073/pnas.0601635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fetzner S. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: Molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften. 2000;87:59–69. doi: 10.1007/s001140050011. [DOI] [PubMed] [Google Scholar]
- 6.Behrman EJ, Stanier RY. The bacterial oxidation of nicotinic acid. J Biol Chem. 1957;228:923–945. [PubMed] [Google Scholar]
- 7.Hurh B, Yamane T, Nagasawa T. Purification and characterization of nicotinic acid dehydrogenase from Pseudomonas fluorescens TN5. J Ferment Bioeng. 1994;78:19–26. [Google Scholar]
- 8.Nelson KE, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 9.Jiménez JI, Miñambres B, García JL, Díaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol. 2002;4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- 10.Hille R. Molybdenum-containing hydroxylases. Arch Biochem Biophys. 2005;433:107–116. doi: 10.1016/j.abb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Frerichs-Deeken U, et al. Functional expression of the quinoline 2-oxidoreductase genes (qorMSL) in Pseudomonas putida KT2440 pUF1 and in P. putida 86–1 Δqor pUF1 and analysis of the Qor proteins. Eur J Biochem. 2003;270:1567–1577. doi: 10.1046/j.1432-1033.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 12.Cramer WA, Knaff DB. In: Energy Transduction in Biological Membranes. Cantor CR, editor. New York: Springer; 1990. [Google Scholar]
- 13.Nakano H, et al. Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. Eur J Biochem. 1999;260:120–126. doi: 10.1046/j.1432-1327.1999.00124.x. [DOI] [PubMed] [Google Scholar]
- 14.Gauthier JJ, Rittenberg SC. The metabolism of nicotinic acid. I. Purification and properties of 2,5-dihydroxypyridine oxygenase from Pseudomonas putida N-9. J Biol Chem. 1971;246:3737–3742. [PubMed] [Google Scholar]
- 15.Gauthier JJ, Rittenberg SC. The metabolism of nicotinic acid. II. 2,5-dihydroxypyridine oxidation, product formation, and oxygen 18 incorporation. J Biol Chem. 1971;246:3743–3748. [PubMed] [Google Scholar]
- 16.Behrman EJ. The bacterial oxidation of nicotinic acid. N-formylmaleamic and N-formylfumaramic acids. Arch Microbiol. 1976;110:87–90. doi: 10.1007/BF00416972. [DOI] [PubMed] [Google Scholar]
- 17.Odintsov SG, et al. Staphylococcus aureus aminopeptidase S is a founding member of a new peptidase clan. J Biol Chem. 2005;280:27792–27799. doi: 10.1074/jbc.M502023200. [DOI] [PubMed] [Google Scholar]
- 18.Odintsov SG, et al. Substrate access to the active sites in aminopeptidase T, a representative of a new metallopeptidase clan. J Mol Biol. 2005;354:403–412. doi: 10.1016/j.jmb.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 19.Vaillancourt FH, Bolin JT, Eltis LD. The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 20.Line K, Isupov MN, Littlechild JA. The crystal structure of a (−) γ-lactamase from an Aureobacterium species reveals a tetrahedral intermediate in the active site. J Mol Biol. 2004;338:519–532. doi: 10.1016/j.jmb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Holmquist M. Alpha/beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 22.Du X, et al. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry. 2001;40:14166–14172. doi: 10.1021/bi0115479. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama K, et al. Analysis of oxidation sensitivity of maleate cis-trans isomerase from Serratia marcescens. Biosci Biotechnol Biochem. 2000;64:1477–1485. doi: 10.1271/bbb.64.1477. [DOI] [PubMed] [Google Scholar]
- 24.Stanier RY, Palleroni NJ, Doudoroff M. The aerobic Pseudomonads: A taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 25.Thacker R, Rorvig O, Kahlon P, Gunsalus IC. NIC, a conjugative nicotine-nicotinate degradative plasmid in Pseudomonas convexa. J Bacteriol. 1978;135:289–290. doi: 10.1128/jb.135.1.289-290.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melton AR, Weiss AA. Characterization of environmental regulators of Bordetella pertussis. Infect Immun. 1993;61:807–815. doi: 10.1128/iai.61.3.807-815.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SN, et al. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol Lett. 2004;26:1493–1496. doi: 10.1023/B:BILE.0000044450.16235.65. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell DV. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 29.Nogales J, Canales A, Jiménez-Barbero J, García JL, Díaz E. Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J Biol Chem. 2005;280:35382–35390. doi: 10.1074/jbc.M502585200. [DOI] [PubMed] [Google Scholar]
- 30.Francis RT, Jr, Becker RR. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal Biochem. 1984;136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.