Abstract
It is now almost 15 years since the publication of the first paper on text mining in the genomics domain, and decades since the first paper on text mining in the medical domain. Enormous progress has been made in the areas of information retrieval, evaluation methodologies and resource construction. Some problems, such as abbreviation-handling, can essentially be considered solved problems, and others, such as identification of gene mentions in text, seem likely to be solved soon. However, a number of problems at the frontiers of biomedical text mining continue to present interesting challenges and opportunities for great improvements and interesting research. In this article we review the current state of the art in biomedical text mining or ‘BioNLP’ in general, focusing primarily on papers published within the past year.
Keywords: text mining, natural language processing, information extraction, text summarization, image mining, question answering, literature-based discovery, evaluation, user orientation
INTRODUCTION
Biomedical text mining: context and objectives
One of the most common motivating claims for the necessity of biomedical text mining is the phenomenal growth of the biomedical literature, and the resulting need of biomedical scientists for assistance in assimilating the high rate of new publications. [In this article, we discuss the biological, rather than the medical/clinical literature, almost exclusively, due both to the subject matter of this journal, and to the difficulty of covering both topics in the allotted number of pages. We use the term biomedical nonetheless, since much of what we say about processing of biological text applies to medical text, as well (1)]. Hunter and Cohen [2] demonstrate that the growth in new PubMed/MEDLINE publications is exponential; at this rate of publication, it is difficult or impossible for biologists to keep up with the relevant publications in their own discipline, let alone publications in other, related disciplines. For bench scientists, published data is the best source for interpreting high-throughput experiments, but automated text processing methods are required to integrate them into the data analysis workflow [3]. For researchers in general, literature-based discovery has often been held out as a potential source of promising hypotheses. Model organism database curators are often implicitly, if not explicitly, the intended users of biomedical text mining systems, and their need for text mining technologies may be the greatest; recent work by Baumgartner et al. [4] suggests that at the current rate of annotation of genes and gene products, it will be years at best and decades at worst, before some of the manually curated genomic resources are complete without the development of automated curation aids such as could be supplied by text mining.
Methods and scope
This article surveys recent work in biomedical text mining over a period which ranges approximately from the end of 2005 (the date of publication of the most recent review of biomedical text mining in this journal [5]) to the beginning of 2007. We selected interesting publications by scanning the tables of contents of the following journals: Artificial Intelligence in Medicine, Bioinformatics, Biomedical Digital Libraries, BMC Bioinformatics, Genome Biology, Genome Research, Journal of the American Medical Informatics Association, Journal of Biomedical Informatics, Journal of Biomedical Science, Nature Reviews Genetics, Nucleic Acids Research, PLoS Computational Biology, PNAS and ACM Transactions on Information Systems. We did the same for conference or workshop proceedings from: PSB 2006, 2007, BioNLP 2006, NAACL 2006, COLING/ACL 2006, AMIA 2005, 2006 and ISMB 2006. We also issued bibliographic queries for: ‘text mining’ in Bioinformatics (MEDLINE), ‘biology’ or ‘medicine’ in ACM journals and PubMed ‘related articles,’ starting from the review papers [5–7].
We selected papers where text-based processing was involved. We included a few borderline papers where literature mining was based on manually assigned Medical Subject Headings (MeSH) keywords, or which relied only on information-retrieval methods. We focused on the biomedical domain, including a few borderline papers in the clinical domain. Because of the necessarily restricted focus of this survey, and of the extreme proficiency of the field, we could not do justice to the important work performed until 2005, nor to the totality of the activity which took place in 2006–07. We refer the interested reader to previous surveys [2, 5–15] (marked S in the bibliography) and to the above-mentioned journals and conferences.
Areas of research
Most biomedical text mining research relies, to varying degrees, on natural language processing methods and tools.
There are broader and stricter definitions of text mining (e.g. [16, 17]). On the strictest definition of the term, a text mining system must return knowledge that is not explicitly stated in text. On this definition, literature-based discovery (Section ‘Literature-based discovery’) and some summarization and question-answering systems would qualify as text mining. On a broader definition, any system that extracts information from text or performs functions that are necessary prerequisites for doing so, would be considered as text mining. This would include a range of application types, from named entity recognition to literature-based discovery, and many things in between.
Most biomedical text mining systems include a module that recognizes biological entities or concepts in text (Section ‘Named entity recognition’) (sometimes normalized to unique identifiers in an ontology or other knowledge source). Relations between biological entities can then be detected (Section ‘Identifying relations between biomedical entities’). These are the two usual components of information extraction (Section ‘Extracting facts from texts’). Beyond information extraction (in Section ‘Beyond information extraction’), document summarization aims to identify and present succinctly the most important aspects of a document in order to save reading time (Section ‘Summarization’). The source documents are more and more often full-text articles, which generally include not only text, but also information-rich non-textual information such as tables and images (Section ‘Processing non-textual material’). The ‘Question answering’ section describes systems which strive to provide precise answers to naturally formulated questions. True text mining not only gives direct access to facts stated in texts, but also helps uncover indirect relationships between biological entities (Section ‘Literature-based discovery’), thereby directly addressing the problem of information overload.
The most important requirement of text mining (and arguably one of the most under-addressed to date) is to be oriented towards the user (section ‘Assessment and user-focused systems’). Evaluation of the quality of systems and results helps assess the confidence in the produced data (Section ‘Annotated text collections and large-scale evaluation’). And finally, actual studies of user needs should drive technical developments, rather than the opposite (Section ‘Understanding user needs’). The rest of this article is organized according to these areas.
EXTRACTING FACTS FROM TEXTS
Extracting explicitly stated facts from text was the goal of many of the earliest biologically oriented text mining applications (see [9, 12] for reviews of this early work). Systems with this goal are commonly known as information extraction or relation extraction applications. Such systems typically perform named entity recognition as an initial processing step.
Named entity recognition
Biological named entity recognition (NER) is a task that identifies the boundary of a substring and then maps the substring to a predefined category (e.g. Protein, Gene or Disease). The earliest NER systems typically applied rule-based approaches (e.g. [18]). As annotated corpora have become available, machine-learning approaches have become a mainstream of research. Although Conditional Random Fields (CRFs) have recently gained popularity for the NER task (e.g. [19])—Jin et al. [20] annotated over 1000 MEDLINE abstracts to recognize clinical descriptions of malignancy presented in text, trained on the annotated data with CRFs, and reported 0.84 F-measure—the choice of algorithm seems to matter less than the feature set [21]. High-performing systems have included a combination of data-driven features, such as character n-grams for tokens and word n-grams for context; linguistic solutions to the problem of boundary location for multi-word names, such as syntactic analysis and location of gene symbol definitions; and corpus-based methods such as Google searches for patterns like ‘X gene’.
Biological named entities are often ambiguous in their boundaries and categories. Olsson et al. [22] found that the differences in boundary criteria (e.g. ‘right match’ and ‘left match’) had an impact on NER performance, and proposed a variety of scoring criteria for different application needs. Dingare et al. [23] also examined the effect of variability in annotation consistency on system performance.
Many NER and information extraction systems make use of lists of terms of entities. Sandler et al. [24] constructed term lists using distributional clustering methods. The methods group words based on the contexts they appear in, including neighboring words and syntactic relations. Results suggested that automatically generated term lists significantly boost the performance of a CRF gene tagger. However, in most cases, unprocessed lists of gene names do not increase the performance of gene/protein NER systems, except in cases where their performance without external lists is unusually poor [21].
Tanabe et al. [25] constructed a semantic database called SemCat that consists of a large number of semantically categorized terms that come from biomedical knowledge resources (e.g. UMLS, GO and ChemID) and open-domain corpora (e.g. the Wall Street Journal corpus and Brown Corpus). SemCat data was used to train a priority model [26] which takes into consideration the position of words (a word to the right is more likely to determine the nature of the entity than a word to the left). The priority model out-performed two other baseline systems, achieving an F-measure of 0.96 for name classification.
While NER categorizes biological entity occurrences in text, other methods can be used to assign categories to biological entities based on the set of texts in which they occur. For instance, Maguitman et al. [27] correctly assigned over 75% of 3663 proteins to one of 618 Pfam families, relying on the set of MEDLINE abstracts associated by SWISSPROT to each protein. Proteins with similar sets of abstracts were assumed to have the same Pfam family; the best results in this experiment were achieved by representing abstracts by the words they contain.
Identifying relations between biomedical entities
The basic facts that text mining systems generally aim to extract from the literature typically take the form of relations between two biological elements identified by NER. (As we discuss below in Section ‘Outlook for the future: what are the “new frontiers” for biomedical text mining?’, this is an area where improvement is called for, and wherein there has been progress in the recent past.) Work reviewed in this section shows an evolution in the distribution of extraction methods from co-occurrence and patterns to fuller parsing. Advances are made in assessing the quality of extracted facts. Finally, multiple types of relations are addressed in the literature, among which ‘contrasts’ between proteins.
The simplest way to detect relations between biomedical entities is to collect texts or sentences in which they co-occur. Co-occurrence statistics can provide high recall (if most co-occurrences are returned) but may have poor precision, and are now used more as a simple baseline method against which other methods are compared [28–30]. Pattern-based methods enforce more precise linguistic conditions for relation detection. Although they can theoretically be applied directly to raw text, sentence segmentation and part-of-speech (POS) tagging are performed in virtually all cases. Phrase chunkers are used in some instances to detect basic phrases (noun phrase, prepositional phrase, etc.). Patterns detect individual hypothetical instances of relations, which can be aggregated over a corpus. Bunescu et al. [28] learn the weights of patterns, based on word and POS features, which extract (unlabeled) confidence-rated gene/protein relations from individual sentences. Confidence of a relation for the whole corpus is computed as the maximum of its confidence values over all sentences. This method is combined with statistical co-occurrence extraction using pointwise mutual information, and the combined model performs better than any individual method.
An important advance in the recent past has been an increase in the attention paid to syntax. Fuller parsing methods produce more elaborate syntactic information. Syntactic structures are represented as constituent parse trees or dependency trees, and encode grammatical relations (subject, direct object, noun modifier, etc.) between phrases or words. Curran and Moens [29] have shown that for some information extraction tasks, using simpler methods on large corpora may be more effective than syntactically more elaborate but computationally more expensive methods on smaller corpora. However, when corpus size is bounded, as is for instance that of MEDLINE, and when the whole corpus can be parsed in a reasonable amount of time, complete syntactic analysis of sentence structure is expected to provide better results. The conjunction of the well-known increase in computing power and of sustained research into fast parsing algorithms now makes it feasible to apply complete syntactic analysis to very large corpora, and a resurgence of work in this area is a notable development in current work in biomedical text mining.
Fundel et al. [30] apply the Stanford Lexicalized Parser to produce dependency trees from MEDLINE abstracts. This information is complemented with gene and protein names obtained by the ProMiner [31] NER system, after chunking with fnTBL (http://nlp.cs.jhu.edu/~rflorian/fntbl/). The system applies three relation extraction rules to the obtained structure, also checking for negation and passive inversion, to detect gene/protein interactions. Recall/precision/F-measure figures of 85/79/82 were achieved on the LLL challenge data set (80-sentence test set [32]) and 78/79/78 on a 50-abstract subset of the Human Protein Reference Database (HPRD). Again, the system achieved significantly better precision and F-measure, at the expense of recall, than simple co-occurrence. More importantly, it also significantly outperformed all the approaches previously applied to the LLL-challenge. Fundel et al. also performed a large-scale feasibility test of complete syntactic analysis on 1 million MEDLINE abstracts, demonstrating that it was achieveable in only 1 week of processing time, given a 40-Xeon cluster. There has also been interesting work on an alternative form of syntactic representation known as dependency parsing. Rinaldi et al. [33] demonstrate that dependency parsing can be used to build an effective relation extraction application. The system is known as the Pro3Gres dependency parser. Processing begins with POS tagging, lemmatization, NP and VP chunking (LTCHUNK) and terminology detection. Pro3Gres combines a hand-written grammar with a statistical language model. Patterns with access to lexical, syntactic and semantic type information are applied to dependency trees. ‘Semantic’ patterns group several variant syntactic patterns, to take into account, e.g. the passive transformation. Evaluated on three relations (activate, bind and block) extracted from the GENIA corpus [34], a range of precision and recall values was achieved on various measures, ranging from precision of 52% (strict)–90% (correct relation with approximate boundaries) and recall of 40% (estimated lower bound)–60% (actually measured on a subset of the corpus).
Full parsing for relation extraction is applied to the whole of MEDLINE by the GENIA group [35]. Fast techniques for probabilistic HPSG parsing [36] are used to parse the 1.4 billion word MEDLINE corpus in 9 days on a double cluster of 340 Xeon CPUs. The system is evaluated by directly querying the resulting predicate-based representation and comparing the results with traditional, IR-style keyword search through MEDLINE sentences. An improvement in precision of over 80% is reported for most queries, with a recall of 30–50% relative to keyword search. Such a method enables users to quickly identify precise biological information in MEDLINE (the system can be accessed at http://www-tsujii.is.s.u-tokyo.ac.jp/info-pubmed/ [37]).
Syntactic analysis can be complemented by semantic role labeling, a step which assigns roles (e.g. location, time, etc.) to sentence elements and can help further improve relation extraction. Tsai et al. [38] train a role labeling system on a specifically prepared extract of the GENIA corpus [39], which they call BioProp, where the predicate-argument structures of 30 frequently used biomedical verbs predicates are annotated. Their system is much more effective at extracting arguments in biomedical text than a general-purpose (newswire-oriented) semantic role labeling system, especially for adjunct arguments such as location and manner (e.g. how to conduct an experiment), and obtains a global F-measure of 87%.
An important issue in text mining is the quality of extracted facts. Would it be possible to automatically determine that quality? Two strategies are suggested. Masseroli et al. [40] study a criterion which helps to identify less reliable extraction contexts, boosting precision at the expense of recall. They show that a shorter distance between predicate and argument increases the precision of predications extracted by their SemGen system (when a predicate is a verb or a preposition). Precision of gene–gene relations increases from 42 to 71% if distance is constrained to be minimal, and precision of gene–disease relations also increases from 74 to 88%. Incidentally, a shorter distance also helps in filtering results from co-occurrence-based extraction, although to a much lesser extent.
Rodriguez-Esteban et al. [41] go one step further to automatically mimic human evaluation of molecular interaction statements extracted by their GeneWays system. To prepare training data, evaluators annotated approximately 45 000 unique statements as correct or not, and if incorrect, specified the type of error. An automatic classifier was then trained on this data; the features used consisted of system output such as dictionary-based information, word metrics, punctuation, terms, and POS tags, as well as human-assigned evaluation annotations from the training set. The best results were obtained with a maximum entropy classifier, with an area under the ROC curve close to 0.95. The important point here is that this ‘artificial intelligence curator’ performs slightly better than any of the four human evaluators that prepared the data.
The rich body of work on relation extraction addresses various kinds of relations, including genes/proteins, protein point mutations [42], protein binding sites [43], gene-disease [44], phenotypic context [45, 46] and mutations [47]. Kim et al. [48] investigate a quite different kind of relation: contrasts between proteins, e.g. NAT1 binds eIF4A but not eIF4E. Starting from MEDLINE abstracts which contain the word ‘not’, they apply their own POS-tagger and NP chunker, then detect contrastive negation patterns such as ‘A but not B’, where A and B must be parallel (i.e. include similar words and phrases). Contrastive information is then extracted from the nonparallel parts of A and B. Identified proteins are grounded with respect to Swiss-Prot entries. Applied to 2.5 million MEDLINE abstracts, they produced 41 471 protein-protein contrasts (they can be examined at http://biocontrasts.bio-pathway.org/), with a precision of 97% estimated on a 100 pairs random sample. Incidentally, their POS tagger, when compared to the reference MedPost, trades −5 points of precision for a 10× factor in speed, so that the system processes an abstract in 0.038 s (on a Sun Fire V440).
BEYOND INFORMATION EXTRACTION
This section describes systems that go beyond information extraction into areas that meet the strictest definition of text mining, as well as systems that deal with additional data types other than text, per se. While the input to information extraction systems are typically single sentences, the inputs to these systems are typically a full document—usually at least an abstract, sometimes a full journal article, and in rare cases, a collection of documents (as in multi-document summarization, discussed below). Another contrast with information extraction systems is that the outputs of these systems are not restricted to simple statements about relations between entities.
Summarization
The goal of automatic text summarization is to identify the most important aspects of one or more documents and present these aspects succinctly and coherently. In recent evaluation paradigms, these aspects are perceived as important ‘nuggets of information’ if they satisfy the need for information expressed in the form of complex questions on a topic of interest. The interest in topic-specific (also known as targetted summarization) summarization in the open domain (i.e. when applied to non-domain-specific general English text, typically from newswire stories) is exemplified by the Document Understanding Conference evaluations [49], and in the clinical domain by experiments in summarizing the best treatments for a given disease [50]. [Traditional ‘generic’ summaries make no assumptions about the intended use of the summary, other than a distinction between indicative summaries (whose only goal is to help the reader make a decision about whether or not they would be interested in reading the summarized document) and informative summaries (whose goal is to actually deliver information from the summarized document to the reader. Targeted/focused summaries, on the other hand, aim to satisfy a unique information need, often expressed as a query].
In targeted summarization of biological literature, Ling et al. [51] developed a method for generating structured summaries characterizing six aspects of a gene: (i) Gene products, (ii) Expression location, (iii) Sequence information, (iv) Wild-type function and phenotypic information, (v) Mutant phenotype and (vi) Genetic interaction. The summary frames are populated by retrieving relevant MEDLINE abstracts and extracting sentences containing information about a given aspect of the target gene. Similarly, to combining evidence in determining most informative sentences about the outcomes of treatments [50], Ling et al. [51] score sentences combining their marks for category relevance, document relevance and location of the sentence in the abstract. This extraction method achieved 50–70% precision in identifying the above six aspects for a test set of 10 randomly selected genes.
The task of succinctly describing a gene function using MEDLINE abstracts is carried out manually when providing Gene References Into Function (GeneRIF for genes described in Entrez Gene database). The TREC 2003 Genomics Track [52] included a task on the prediction of GeneRIFs. Lu et al. [53] suggest performing this task using summarization techniques combined with GO annotations associated with the existing Entrez Gene entries. The authors then further develop their method into an innovative application of summarization to a real-life task: a summary revision approach to detect low-quality and obsolete GeneRIFs, achieving 89% precision and 79% recall in this task, and producing qualitatively more useful GeneRIFs than other methods.
More recently, Baumgartner et al. [4] have applied a summarization approach to the BioCreative 2006 sentence selection subtask of the protein–protein interaction task. Their extractive summarization approach to finding the best sentence describing a protein–protein interaction achieved a 19% correct rate, the best achieved in this challenge; the second-place system scored 6%.
In addition to development of summarization techniques, there is ongoing research on providing better access to facts extracted from text and linking the facts and associated knowledge in databases. EBIMed [54] and GeneLibrarian [55] are new additions to such services as iHOP [56], MedMiner [57], Chilibot [58] and others (http://www.oxfordjournals.org/nar/webserver/cap/).
Related to summarization is the task of describing the main topics of a text using MeSH terms, as performed by human indexers for the MEDLINE database. Névéol et al. [59] strive to facilitate this manual process by improving the automatic generation of suggested MeSH terms; the NLM indexers use them in the indexing process. This work focuses on the novel task of assigning combinations of MeSH descriptors and qualifiers, rather than just assigning single MeSH descriptors, to a citation.
Categorization of a document into one of a set of predefined classes (e.g. GO codes) is another application related to summarization (see, e.g. [11] for more detail). A successful assignment of GO codes to genes was achieved by Stoica and Hearst [60], who assigns GO terms by searching biomedical text for GO codes assigned to orthologues of the target gene. Fyshe and Szafron [61] categorize document abstracts with respect to the sub-cellular localization of proteins, employing GO as an additional source of information. Categorization of document abstracts is also one of the components of Höglund et al.'s [62] method for predicting sub-cellular localization.
There seems to be steady ongoing research in biomedical text summarization. It would now be desirable to see more real-life applications of summarization, more research in task-driven summarization and research in coherent multi-document generative summarization.
Processing non-textual material
To date, most work on biomedical language processing systems has been applied to textual information only, and does not provide access to other important data, such as images (e.g. figures). Recent years have been marked by emerging research interests in applying image processing as well as natural language processing approaches to analyze figure images and their associated text [63–68] or to take into account specific forms of text such as chemical compounds [69].
The Subcellular Location Image Finder (SLIF) system [63, 64, 68] is the first system that targets images in biomedical literature. SLIF extracts and analyzes a specific type of image, i.e. the fluorescence microscope images from biomedical full-text articles. It utilizes geometric moments, textual measures and morphological image processing to extract all figure images from biomedical full-text journal articles, to identify those figures that depict fluorescence microscope images and then to identify numerical features (i.e. computing SLF6 features and then converting the outputs to a single numerical score) that capture sub-cellular location. The precision/recall of figure-caption extraction was 98/77%. Figures are decomposed into panels by recursively subdividing the figure by looking for horizontal and vertical white-space partitions. The decomposition achieved a precision of 73% and a recall of 60%. Fluorescence microscope images are identified using a k-nearest neighbor classifier with the gray-scale histogram as features; this achieved 97% precision with 92% recall. Multi-cell images are segmented into single cell images. The resulting binary images contain objects which correspond to the cells. The algorithm achieved a precision/recall of 62/32%. Subcellular location features (SLF) are produced to summarize the localization pattern of each cell. All methods demonstrated their robustness to variations introduced in experiment preparation, cell type and microscopy method, and image alternations introduced during publication. SLIF developed different methods to align image panels to their corresponding sub-captions [64, 68].
Rafkind et al. [67] defined five categories of images that appear in biomedical full-text articles (Figures 1–5), and applied the supervised machine learning algorithm Support Vector Machines (SVMs) to classify figure images automatically into these categories. Given a total of 554 annotated figure images, the classifiers achieved a 50.74% F-score when applying image features alone (intensity and edge-based features) and a 68.54% F-score when applying text features (bag-of-words and n-grams obtained from the captions). When fusing image features with text, the combined classifier achieved an F-score of 73.66%.
Figure 1.
Image category defined in [67]: Image-of-thing, from cover page of [70].
Figure 5.
Image category defined in [67]: Mix, from cover page of [74].
Shatkay et al. [65] developed a hierarchical image classification scheme for figure images. Figure images are classified into Graphical, Experimental and Other. Graphical figures are classified into Bar Chart, Line Chart and Other Diagrams. Experimental figures are classified into Gel Electrophoresis, Fluorescence Microscopy and Other Microscopy. With a total of 1600 annotated figure images, they applied SVM classifiers to achieve 95% accuracy for separating Graphical from Experimental figures, and 93% accuracy for separating the three types of Experimental figures. Forty-six image features (e.g. histograms and edge direction histogram) were used for the classification task. They found that the text categorization task can benefit from the integration of those image features.
Although images provide important biomedical experimental evidence [66], they are usually incomprehensible by humans without corresponding associated text. To this end, Yu [75] examined three types of associated text: image captions, associated sentences that appear in the abstract and associated sentences that appear in the full-text body, and concluded that sentences in the abstract can be used to summarize image content and that other associated text typically describes only experimental procedures and does not include the indications or conclusions of an experiment. Yu and Lee [66] randomly selected a total of 329 bioscience articles published in the journals Cell, EMBO, Journal of Biological Chemistry and Proceedings of the National Academy of Sciences (PNAS). For each article, they emailed the corresponding author and invited him/her to identify abstract sentence(s) which summarize image content within the same article. A total of 119 scientists (either the first or the corresponding author) from 19 countries participated voluntarily in the annotation and produced a total of 114 annotated articles, in which 87.9% figure images and 85.3% table images correspond to abstract sentences, and 66.5% of abstract sentences correspond to images that appear in the full-text articles. Yu and Lee further designed a user-interface BioEx in which the associations between images and abstract sentences are visualized. BioEx provides access to images through the associated abstract sentences. Those 119 scientists who annotated their articles were invited to evaluate the BioEx interface to compare it with two other baseline interfaces in which images cannot be accessed through abstract sentences. Forty-one scientists participated in the evaluation and 36 (87.8%) preferred the BioEx user-interface. The association of images and abstract sentences in Yu and Lee is achieved using hierarchical clustering algorithms based on the word level similarity between abstract sentences and image captions. One of the systems achieved a precision of 72% that corresponded to a recall of 33%.
Somewhat related to images by their nonlinear nature are chemical compound descriptions. Rhodes et al. [69] describe a molecular similarity search engine for identifying similar chemical compounds in a patent corpus. The system first identifies chemical names in text, converts the names to corresponding compound structures, and then presents each structure as a IUPAC International Chemical Identifier (InChI) code. Features are extracted from the InChI codes and the text-based Vector Space Model is then applied to index and retrieve relevant chemical compounds. Evaluation found that the similarity search outperformed a text-based search.
Outside the biological domain, systems have mainly been developed to retrieve medical images from databases. ImageCLEFmed [76] is a medical image retrieval task as a part of CLEF (Cross Language Evaluation Forum) since 2004. 12 groups participated in 2006 and IPAL [77] achieved the highest mean average precision (MAP: 0.3095) for automatic medical image retrieval. IPAL incorporated the UMLS as a knowledge base and found that it enhanced both text-based and visual retrieval.
Question answering
Question answering can be approached as a special case of high accuracy information retrieval. Rather than returning a list of documents from large text collections, question answering attempts to provide short, specific answers to questions and put them in context by providing supporting information and linking to original source documents [78]. Question answering has been initially addressed as an open-domain application, and more recently in restricted domains [79]. The clinical domain has seen active research earlier, hence it is covered in this section, while genomics has only been tackled more recently [80].
Question answering systems typically incorporate components of question analysis, query formulation, information retrieval, answer extraction, summarization and presentation. For question-answering in the biomedical domain, Zweigenbaum [81] is the most accessible introduction.
Although the needs for answering clinical questions have been widely recognized (e.g. [82]), medical question answering is a relatively new field. Jacquemart and Zweigenbaum [83] conducted a feasibility study for answering clinical questions in French. Huang et al. [84] mapped clinical questions based on Problem/Population, Intervention, Comparison and Outcome (PICO). Demner-Fushman and Lin [85] then identified and extracted the PICO texts to answer clinical questions; they also found that domain-specific knowledge (e.g. journal impact and MeSH terms) enhanced information retrieval [86]. Yu et al. [87] implemented a medical question answering system and conducted a usability study to compare the question answering system with other information retrieval systems (e.g. PubMed).
The Text Retrieval Conference (TREC, Section ‘Annotated text collections and large scale evaluation’) Genomics Track has been a driving force for question answering in the genomics domain. In 2006, the Genomics track single task focused on retrieval of short passages that specifically answer biological questions (e.g. ‘What is the role of PrnP in mad cow disease?’) [80]. Thirty-one groups participated in the Genomics Track, and obtained the following mean average precision scores: 0.0198–0.5439 (median: 0.3083) for document retrieval, 0.0007–0.1486 (median: 0.0345) for passage retrieval and 0.011–0.4411 (median 0.1581) for aspect retrieval.
One of the best-performing systems [88] integrated rule-based, dictionary and statistical methods for recognizing term variations, synonyms, hypernyms and hyponyms and other related terms, and found they greatly enhanced the performance of question answering. Another highly-performing system [89] combined the results of four independent information retrieval systems (Essie, EasyIR, SMART and Theme) and found that the fusion significantly outperformed individual systems. Advanced information retrieval models have been explored by many groups. For example, Jiang et al. [90] explored language models and relevance feedback; Caporaso et al. [91] explored Latent Semantic Analysis; Divoli et al. [92] took into consideration the structure of the questions and of the full-text documents; however, those models did not enhance the passage retrieval performance. Zheng et al. [93] selected sentences based on their syntactic tree-structure similarity with the question and found that shallow parsing enhanced the performance for answer extraction.
Literature-based discovery
An exciting usage of information extracted from the scientific literature by the various text-mining methods outlined above consists in trying to uncover ‘hidden’, indirect links: this is often called ‘literature-based discovery’ [10]. These links can be proposed as potential scientific hypotheses, the prototypical example being that between fish oil and Raynaud's disease, hypothesized by Swanson in his seminal paper [94]. Since then, few methods and systems have been designed to help such discovery. Given initial user-specified targets, they compute and traverse association links, and propose the highest-ranked associations to the user.
Some researchers [95] find NLP to be computationally too expensive for practical use in literature-based discovery, and fold back to using the manually assigned MeSH terms available in MEDLINE. Nevertheless, methods generally rely on some amount of natural language processing to obtain the basic facts: Jelier et al. [96] use named entity recognition; to perform NER, Seki et al. [97] extend terms with words of their definitions in an IR-style query-expansion mode; Pospisil et al. [98] use the NER facilities of the LSGraph system; Palakal et al. [99] start with simple co-occurrences to obtain associations, then learn patterns to identify the direction of associations; Rzhetsky et al. [100] exploit the full parsing done in their GeneWays project. Full parsing is more computationally demanding, so that Hristovski et al. [101] envisage its integration with less demanding co-occurrence-based methods. It may be made practical, though, by running systems on powerful computer clusters (see, e.g. Fundel et al. [30] above in section ‘Identifying relations between biomedical entities’).
Progress in literature-based discovery takes the form of advances in methods, a greater number of integrated systems (LitMiner [102], BBP [103], Arrowsmith [104]; see Section ‘Understanding user needs’), and more examples of actual usage of these systems to propose ‘discoveries’ for further biological experimentation [105].
A strand of research is akin to the distributional analysis commonly performed now in corpus-based semantics: two words are semantically similar if they occur in the same contexts (e.g. [106, 107]). Here, two biological entities are related if they occur in the same contexts in the literature. Co-occurrence-based methods, as discussed above in section ‘Identifying relations between biomedical entities’, are based on direct (‘first-order’) co-occurrence between biological elements. In literature-based discovery, ‘second-order’ relations are explored by looking for the shared co-occurrents of two biological terms.
In this line of research, Jelier et al. [96] aim at identifying genes which are functionally similar by comparing their distributional profiles. They use statistics based on the co-occurrence of concepts in MEDLINE abstracts, as defined by MeSH terms and a combination of genetic databases: this produces concept profiles where each identified co-occurring concept is assigned a strength of association with the source gene, using the log likelihood ratio. Identified concepts are restricted to those having prespecified UMLS semantic types. Gene concept profiles are then subjected to hierarchical clustering. In a given cluster, the concepts which contribute most to cluster similarity identify the shared functions.
Another series of second-order association research, in the line of Swanson's investigations, relies on ‘B’ elements (e.g. blood viscosity) found in the same ‘literatures’ (sets of papers) as ‘C’ (e.g. Raynaud's disease) and ‘A’ terms (e.g. Fish oil) [A more detailed introduction can be found in (10)]. Second-order relations are explored by looking for shared co-occurrents of ‘C’ and ‘A’ terms: they provide the hypothesized uncovered links. Additionally, whereas corpus-based semantics focuses on finding synonyms (or also hypernyms, translations, etc.) through tightly controlled co-occurrence (short-distance or syntactic dependencies), literature-based discovery is interested in more varied associative relations (e.g. ‘causes’, ‘treats’). Yetisgen-Yildiz and Pratt [95] implement an ‘open-discovery’ approach, in the sense that a starting term ‘C’ is specified, but target terms ‘A’ are left open. Their LitLinker system looks for co-occurrents of ‘C’ (linking terms ‘B’), then for co-occurrents of these linking terms (target terms ‘A’). LitLinker differs from BITOLA ([108], see below) in the statistical processing it performs. Documents are represented by their indexing MeSH terms; the co-occurrence of terms is weighted by their z-score, and a predefined threshold keeps the most associated terms. Too general and too similar terms are pruned with the help of the MeSH hierarchy; co-occurring terms are also filtered on their semantic groups, with different constraints on linking terms (Chemicals and drugs, Disorders, etc.) and target terms (Chemicals and drugs or Genes and molecular sequence). The obtained target terms are ranked according to the number of linking terms that connect that target term to the original starting term.
Hristovski et al. [101] help provide more precise information about the ‘B’ terms, leveraging the ‘semantic predications’ extracted by BioMedLEE [45] and SemRep [109] for B- or C-related literature. This refines the search done using the BITOLA literature-based discovery system [108]. They focus on the ‘treats’ predication, with the following discovery pattern: looking for a new drug treatment of Disease C, find a Substance B changed (e.g. increased or decreased) in Disease C, then a Drug A which provokes the opposite change or another Disease C2 which provokes the same change and which is treated by a Drug A. Instead of leaving it to the user to read relevant C-B and B-A MEDLINE citations and find out what is ‘increased’ in relation to a disease and what can be used to ‘decrease’ it, this information is obtained from the semantic predications produced by NLP systems run on this literature. For instance, Eicosapentaenoic acid (A, found in fish oil) is proposed to reduce blood viscosity (B) and treat Raynaud's disease (C); and since insulin (B) is decreased in Huntington disease (C) as in diabetes mellitus (C2), insulin treatment (A) is proposed to treat Huntington disease.
Another strand of research explores the transitivity of labeled relations extracted from the literature. Individual relations are collected into large interaction networks whose paths can reveal indirect relationships. Palakal et al. [99] built a directed relationship graph from the individual directional relationships they collected by text processing. The user can then formulate queries to look for genes, cells, molecules, proteins or diseases associated with the presence or absence of given biological entities: e.g. “Find all the cells that are present in inflammation but not in multiple sclerosis and experimental allergic encephalomyelitis.”
Seki et al. [97] adapt an information retrieval model called an ‘inference network’ [110] to the search of indirect gene–disease associations. In their network, the disease is the query, genes are documents, and intermediate nodes are gene functions (GO terms) and phenotypes (MeSH C terms, i.e. diseases). Conditional probabilities in this network are estimated from co-occurrence in MEDLINE: between MeSH terms (disease and phenotypes) and between MeSH terms and cross-referenced Entrez Gene entries (phenotypes and gene functions). The latter is complemented by taking into account textual co-occurrence in MEDLINE abstracts, which improved system prediction by 4.6% (area under the ROC curve or AUC) on known gene–disease associations from the genetic association database (GAD). Overall, AUC values ranged from 0.623 to 0.786 depending on the domain of the disease. An additional preliminary experiment, with the full text of papers showed another increase of 5.1% in AUC.
Besides the literature-based discovery work described here, let us note that statistics of co-occurrence over MEDLINE abstracts are widely used in other biomedical text mining work. For instance, validation and improvement of existing semi-automatic methods for functional annotation of genes was developed by Aubry et al. [105]. Evaluation of this method on over 7000 genes showed that combining evidence from the Gene Ontology with co-occurrence statistics of gene and GO terms in MEDLINE citations provides more information about gene function than either approach alone.
Evaluation of literature-based discovery systems is not easy, since for a true discovery, there is no immediately available ground truth which could be used as a gold standard. A classical test consists in replicating known discoveries: typically, Swanson's Raynaud–fish oil or migraine–magnesium links. Another test [95] consists in dividing MEDLINE publications into two sets separated by a cutoff date: literature-based discovery proceeds on the older set and results are tested against the more recent set. Precision and recall measures can then be computed on all generated discoveries. Most recently, Torvik and Smalheiser [111] have made available a gold standard for evaluating the sets of terms that are typically a product of literature-based discovery tools (‘A’, ‘B’ and ‘C’ terms above).
ASSESSMENT AND USER-FOCUSED SYSTEMS
The biomedical text mining community has made large strides in the development of materials and infrastructure for large-scale comparative evaluations of text mining systems (in the broader sense of that term) in the recent past. These advances include both the development of a large set of annotated textual resources (known as corpora), and an infrastructure for conducting shared tasks. Along with this attention to principled, comparative system evaluation, there has recently been some movement away from the development of systems based on long-accepted categories of NLP applications, and towards the development of systems based on carefully assessed user needs. The shared tasks themselves have been carefully constructed to target the actual workflow of biomedical researchers, and an additional small body of very recent work has investigated specialized user communities.
Annotated text collections and large-scale evaluation
Evaluation is an essential tool that allows determining whether a given BioNLP method or system effectively achieves its stated objectives and the extent to which it succeeds in performing a task and achieving the anticipated results. As in any other field, BioNLP researchers are concerned with repeatability, comparability and viability of their experimental results. A methodology that addresses these concerns was pioneered by the KDD Cup [112] and continues to be actively researched within TREC [52]. This evaluation methodology involves creation of test collections and development of reliable and valid evaluation measures [113]. The GENIA corpus [40] has marked the start of such test collections in the biomedical domain. Recent developments in creation of such collections and metrics for biomedical text processing, address both the methodological and practical issues.
Wilbur et al. [114] explored methodological issues of finding and annotating general text properties for text mining. They identify the following dimensions to characterize information-bearing fragments of scientific text: (i) Focus (scientific, generic or methodology); (ii) Polarity (positive, negative, lack of knowledge); (iii) Certainty (degree ranging from 0 to 3); (iv) Evidence (absence, reference to, or presence in the fragment); and (v) Direction/trend (high/low level or an increase/decrease in a finding). Based on good agreement in annotation of 101 sentences extracted from biomedical publications, the authors express hope that they defined an executable, reproducible and machine-learnable practical task. Annotation of a large collection using the above methodology is underway.
Such annotation requires domain knowledge and significant time: annotation of 1100 sentences in the BioInfer collection reported by Pyysalo et al. [115] was started in 2001. This collection builds upon entity annotation of the GENIA corpus and includes annotation for relationships, named entities and syntactic dependencies. Information about these and other test collections and their availability can be found at the ‘Corpora for biomedical natural language processing’ website. (http://compbio.uchsc.edu/ccp/corpora/pubs.shtml)
Several ongoing large-scale evaluations not only generate reusable test collections, but also provide a platform for exchange of ideas, fast adoption of best practices and technology transfer.
With the goal of bringing together bioinformatics and information retrieval researchers, a Genomics track was started within TREC in 2002. The 2006 Genomics track task [80] was to extract passages (paragraphs) providing answers and context for 28 questions collected from biomedical researchers. The document collection consists of 162 259 full text documents subdivided into 12 641 127 paragraphs. Content experts determined the relevance of passages to each question and grouped them into aspects identified by one or more MeSH terms. Document relevance was defined by the presence of one or more relevant aspects. Thus the collection provides relevance judgments at the passage, aspect and document level.
The goals of the second BioCreAtIvE evaluation were finding mentions of genes in the text, normalization of gene names and extraction of protein– protein interactions. Morgan et al. [116] analyze issues involved in organizing the evaluation and preparing the text collection on the example of the BioCreAtIvE task of finding EntrezGene identifiers for all human genes and proteins mentioned in a MEDLINE abstract.
Although the large-scale evaluation tasks are modeled using some practical tasks and real user needs, an in-depth principled study of information needs of biologists would provide further insights for conducting evaluations. Similarly, although some discussion of the reliability and validity of the evaluation measures is taking place within the community-wide evaluations, the community would greatly benefit from a principled analysis of the currently used metrics.
Understanding user needs
Studies of user needs, behavior and interactions with tools are an effective way to determine which bioinformatics tools and services are needed, and whether they will be useful. Unfortunately, this area of research has mostly been neglected in BioNLP, although this has changed somewhat in the recent past. Recent efforts primarily focus on the application of natural language processing methods to support advanced functionality of tools for researchers and database curators, taking into account user needs. The systems are mostly developed to address a specific task and/or user group, e.g. a specific organism database curation or creation of a personal digital library of scientific publications.
Iterative development based on user observation and user's feedback was applied in the implementation of a tool for FlyBase curation [117]. Natural language processing integrated in this tool includes recognition of mentions of genes and related noun phrases. The tool provides capabilities to navigate to listed mentions and visual cues that help identify related entities. A pilot evaluation of the tool helped to identify additional desirable features, such as highlighting tables and captions, and keeping track of users' actions.
Similarly to the FlyBase curation tool, LitMiner [102] was developed to enable biologists' analysis of published articles. The LitMiner application is a suite of tools for searching the biomedical literature via PubMed and for manipulating the results. The results could be manipulated as follows: (i) clustered into a hierarchical subject list based on keywords extracted from the titles and abstracts of the articles; (ii) saved and shared with collaborators; (iii) gene co-occurrences could be compared and the relationships between genes could be visualized using a network graph. Aliases used to refer to genes in searching could be tuned using a thesaurus. In a case study, increased access to publications (measured by the numbers of orders) was observed after introduction of this customized service.
The Brucella Bioinformatics Portal (BBP) provides integrated access to information available for the Brucella genome and possibilities for research, text mining and Brucella database curation [103]. The BBP text processing pipeline uses TextPresso [118] to extract Brucella-related information from MEDLINE/PubMed citations into the database.
Although there has been some recent research on user needs, we hope to see more studies and systems grounded in real-life tasks. It would be interesting to see a systematic approach of user observations and dialog with intended users, and whether such approach will improve the initial system design. As a number of systems and services are becoming fairly mature, we might see more user-centered rigorous evaluations in the future.
Additionally, the TREC Genomics track (Section ‘Annotated text collections and large-scale evaluation’) has made serious efforts to focus its recent question-answering evaluations on the actual information needs of a range of types of bioscientists. Specifically, the track has engaged in a concerted effort to collect actual questions from working scientists with a broad range of backgrounds and from a wide range of working environments [119, 120]. The BioCreative shared task (Section ‘Annotated text collections and large-scale evaluation’) has made concerted efforts to focus its tasks around applications of actual use to biological researchers, especially database curators. BioCreative's approach to this has centered on aggressively pursuing and maintaining collaboration with biologists at CNIO, Mouse Genome Informatics, InterAct, MINbT and EBI, both in defining tasks and in evaluating system outputs [121, 122].
OUTLOOK FOR THE FUTURE: WHAT ARE THE ‘NEW FRONTIERS’ FOR BIOMEDICAL TEXT MINING?
As we have seen, there has been significant progress in a number of areas of biomedical text mining research. Nonetheless, there are significant unsolved problems—both ones that have thus far resisted our attempts to solve them, and ones that have only barely been attempted.
Hunter and Cohen [2] identify some encouraging trends, which include the following:
The increasing sophistication of knowledge representations, both in terms of semantic resources such as proposition banks [123, 124] that take us beyond the binary representations that have characterized most of the early work on biomedical relation extraction [125] and in terms of work that targets binary relations of increasing granularity [126].
Increasing awareness of the importance of being able to map from strings in text to the things in the world (unique identifiers) or the concepts in ontologies to which they refer [127–129, 116].
The increasing availability of tools that are actually being used by working scientists [130–132, 118].
Despite these encouraging signs of progress, Zweigenbaum et al. [133] were able to identify six areas that truly constitute ‘new frontiers’ in biomedical text mining: question-answering; summarization; mining data from full-text journal articles; co-reference resolution and normalization; user-driven systems, including assessment of user needs and of user interfaces; and evaluation. Furthermore, we can add to this list: quality assurance and robustness remain mostly ignored in biomedical text mining, and there is a clear need for portable systems, as well as for methodologies for assessing the utility and impact of text mining technologies for a range of users encompassing biologists, clinicians and hospital billing departments [134]. In this review, we have been able to report recent work, and in some cases encouraging progress, in a number of these areas. However, others of these areas remain neglected. Much work remains to be done; happily, biomedical text mining is an extremely active area of research at this time [135], and the likelihood of continued progress seems high.
Key Points.
Text mining often relies on information extraction technology, including NER and relation extraction, which show continued progress.
Beyond information extraction, methods such as summarization, question answering and the exploitation of figures provide new ways to offer easier access to information contained in scientific articles.
Progress in literature-based discovery has taken the form of advances in methods, a greater number of integrated systems, and more examples of actual usage of these systems to propose avenues for further biological experimentation.
The biomedical text mining community has made large strides in the development of materials and infrastructure for large-scale comparative evaluations of text mining systems.
Figure 2.
Image category defined in [67]: Gel, from cover page of [71].
Figure 3.
Image category defined in [67]: Graph, from cover page of [72].
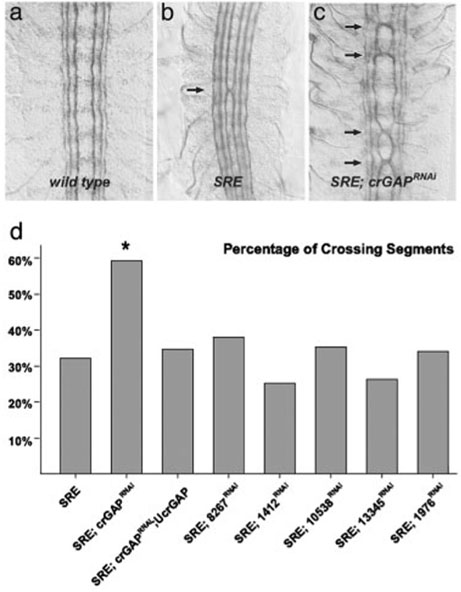
Figure 4.
Image category defined in [67]: Model, from cover page of [73].
Acknowledgements
DDF was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and Lister Hill National Center for Biomedical Communication (LHNCBC). HY was supported by a Research Committee Award, a Research Growth Initiative grant, and an MiTAG award from the University of Wisconsin-Milwaukee, as well as NIH grant R01-LM009836-01A1. KBC was supported by NIH grants ‘Construction of a Full Text Corpus for Biomedical Text Mining’ (#1G08LM009639-01) and ‘Technology Development for a Molecular Biology Knowledge-base’ (#5R01 LM008111-03). We wish to thank the journal's anonymous reviewers, whose insightful comments helped significantly improve this article.
References
S indicates other surveys of the domain
*indicates papers of particular interest published within the period of this review
**indicates papers of extreme interest published within the period of this review
- 1.Aronson AR, Bodenreider O, Demner-Fushman D . Biological, Translational, and Clinical Language Processing. Prague, Czech Republic: Association for Computational Linguistics; 2007. From indexing the biomedical literature to coding clinical text: experience with MTI and machine learning approaches; pp. 105–12. [Google Scholar]
- 2S.Hunter L, Cohen KB. Biomedical language processing: what's beyond PubMed? Mol Cell. 2006;21:589–94. doi: 10.1016/j.molcel.2006.02.012. [indicates other surveys of the domain] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SK. Integrating text mining with data mining. In: Ananiadou S, McNaugh J, editors. Text Mining for Biology and Biomedicine. Norwood, Massachussets: Artech House Publishers; 2006. [Google Scholar]
- 4.Baumgartner WA, Jr., Cohen KB, Fox L, et al. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics (ISMB proceedings issue) 2007;23(13):i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5S.Cohen AM, Hersh WA. Survey of current work in biomedical text mining. Brief Bioinform. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 6S.Spasic I, Ananiadou S, McNaught J, et al. Text mining and ontologies in biomedicine making sense of raw text. Brief Bioinform. 2005;6(3):239–51. doi: 10.1093/bib/6.3.239. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 7S.Ananiadou S, Kell DBB, Tsujii JII. Text mining and its potential applications in systems biology. Trends Biotechnol. 2006;24(12):571–579. doi: 10.1016/j.tibtech.2006.10.002. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 8S.de Bruijn B, Martin J. Getting to the (c)ore of knowledge: mining biomedical literature. Int J Med Inform. 2002;67:7–18. doi: 10.1016/s1386-5056(02)00050-3. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 9S.Cohen KB, Hunter L. Natural language processing and systems biology. In: Dubitzky W, Azuaje F, editors. Artificial Intelligence Methods and Tools for Systems Biology. Heidelberg: Springer; 2004. pp. 147–74. [indicates other surveys of the domain] [Google Scholar]
- 10S.Weeber M, Kors JA, Mons B. Online tools to support literature-based discovery in the life sciences. Brief Bioinform. 2005;6(3):277–86. doi: 10.1093/bib/6.3.277. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 11S.Shatkay H. Hairpins in bookstacks: information retrieval from biomedical text. Brief Bioinform. 2005;6(3):222–38. doi: 10.1093/bib/6.3.222. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 12S.Krallinger M, Valencia A. Text-mining and information retrieval services for molecular biology. Genome Bioly. 2005;6:224. doi: 10.1186/gb-2005-6-7-224. [indicates other surveys of the domain] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13S.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet. 2006;7:119–29. doi: 10.1038/nrg1768. [indicates other surveys of the domain] [DOI] [PubMed] [Google Scholar]
- 14S.Ananiadou S, McNaught J. Text Mining for Biology and Biomedicine. Norwood, Massachussets: Artech House Publishers; 2006. [indicates other surveys of the domain] [Google Scholar]
- 15S.Shatkay H, Craven M. Biomedical Text Mining. Cambridge, Massachussets: MIT Press; 2007. [indicates other surveys of the domain] [Google Scholar]
- 16.Jackson P, Moulinier I. Natural Language Processing for Online Applications: Text Retrieval, Extraction, and Categorization. Amsterdam: John Benjamins Publishing Company; 2002. [Google Scholar]
- 17.Hearst MA. What is text mining? http://www.ischool.berkeley.edu/~hearst/text-mining.html, (October 2003 date last accessed)
- 18.Fukuda K, Tamura A, Tsunoda T, et al. Pac Symp Biocomput. Maui: Hawaii; 1998. Toward information extraction: identifying protein names from biological papers; pp. 707–18. [PubMed] [Google Scholar]
- 19.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields. BMC Bioinform. 2005;6(Suppl):1–S6. doi: 10.1186/1471-2105-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, McDonald R, Lerman K, et al. Automated recognition of malignancy mentions in biomedical literature. BMC Bioinform. 2006;7:492. doi: 10.1186/1471-2105-7-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Yeh A, Morgan A, Colosimo M, et al. BioCreAtIvE task 1A: gene mention finding evaluation. BMC Bioinform. 2005;6(Suppl):1. doi: 10.1186/1471-2105-6-S1-S2. [indicates papers of particular interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson F, Eriksson G, Franzén K, et al. Notions of correctness when evaluating protein name taggers; Proceedings of the 19th international conference on computational linguistics (COLING2002):Taipei, Taiwan 2002; pp. 765–71. [Google Scholar]
- 23.Dingare S, Nissim M, Finkel J, et al. A system for identifying named entities in biomedical text: how results from two evaluations reflect on both the system and the evaluations: Conference papers. Comp Funct Genomics. 2005;6(1–2):77–85. doi: 10.1002/cfg.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler T, Schein A, Ungar L. Automatic term list generation for entity tagging. Bioinformatics. 2006;22(6):651–7. doi: 10.1093/bioinformatics/bti733. [DOI] [PubMed] [Google Scholar]
- 25.Tanabe L, Thom L, Matten W, et al. AMIA Annu Symp Proc. Washington, DC: 2006. SemCat: semantically categorized entities for genomics; pp. 754–8. [PMC free article] [PubMed] [Google Scholar]
- 26.Tanabe L, Wilbur W. A priority model for named entities. BioNLP 2006.
- 27.Maguitman AG, Rechtsteiner A, Verspoor K, et al. Pac Symp Biocomput 11. Maui: Hawaii; 2006. Large-scale testing of bibliome informatics using Pfam protein families; pp. 76–87. http://psb.stanford.edu/psb-online/proceedings/psb06/maguitman.pdf. [PubMed] [Google Scholar]
- 28.Bunescu R, Mooney R, Ramani A, et al. BioNLP 2006. Integrating co-occurrence statistics with information extraction for robust retrieval of protein interactions from MEDLINE. [Google Scholar]
- 29.Curran JR, Moens M. Scaling context space; Proceedings of the 38th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA. ACL 2002; pp. 231–8. [Google Scholar]
- 30*.Fundel K, Küffner R, Zimmer R. RelEx—relation extraction using dependency parse trees. Bioinformatics. 2007;23:365–71. doi: 10.1093/bioinformatics/btl616. http://bioinformatics.oxfordjournals.org/cgi/content/full/23/3/365. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 31.Hanisch D, Fundel K, Mevissen HT, et al. Prominer: rule-based protein and gene entity recognition. BMCBioinforma. 2005;6(Suppl 1):S14. doi: 10.1186/1471-2105-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nédellec C. Proceedings of the ICML05 workshop: Learning Language in Logic (LLL05) Bonn: Germany; 2005. Learning language in logic—genic interaction extraction challenge. [Google Scholar]
- 33.Rinaldi F, Schneider G, Kaljurand K, et al. Mining of relations between proteins over biomedical scientific literature using a deep-linguistic approach. Artif Intell Med. 2007;39(2):127–36. doi: 10.1016/j.artmed.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Rinaldi F, Schneider G, Kaljurand K, et al. An environment for relation mining over richly annotated corpora: the case of GENIA. BMC Bioinforma. 2006;7(Suppl 3):S3. doi: 10.1186/1471-2105-7-S3-S3. http://www.biomedcentral.com/content/pdf/1471-2105-7-S3-S3.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyao Y, Ohta T, Masuda K, et al. Semantic retrieval for the accurate identification of relational concepts in massive textbases; Proceedings of the 21st International Conference on Computational Linguistics and 44th Annual Meeting of the Association for Computational Linguistics (COLING/ACL 2006), Sydney, Australia 2006; pp. 1017–24. [Google Scholar]
- 36.Ninomiya T, Tsuruoka Y, Miyao Y, et al. Fast and scalable HPSG parsing. TAL 2005. 2007;46(2) [Google Scholar]
- 37.Ohta T, Miyao Y, Ninomiya T, et al. An intelligent search engine and GUI-based efficient MEDLINE search tool based on deep syntactic parsing; Proceedings of the 21st International Conference on Computational Linguistics and 44th Annual Meeting of the Association for Computational Linguistics (COLING/ACL 2006), Interactive Presentation Sessions, Sydney, Australia 2006; pp. 17–20. [Google Scholar]
- 38.Tsai TH, Chou WC, Lin YC, et al. BIOSMILE: Adapting semantic role labeling for biomedical verbs: an exponential model coupled with automatically generated template features. BioNLP. 2006. [DOI] [PMC free article] [PubMed]
- 39.Kim JD, Ohta T, Tateisi Y, et al. Genia corpus— a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(Suppl 1):180–2. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 40.Masseroli M, Kilicoglu H, Lang FM, et al. Argument-predicate distance as a filter for enhancing precision in extracting predications on the genetic etiology of disease. BMC Bioinform. 2006;7(291) doi: 10.1186/1471-2105-7-291. http://www.biomedcentral.com/content/pdf/1471-2105-7-291.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Rodriguez-Esteban R, Iossifov I, Rzhetsky A. Imitating manual curation of text-mined facts in biomedicine. PLoS Comput Biol. 2006;2(9) doi: 10.1371/journal.pcbi.0020118. [indicates papers of extreme interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee LC, Horn F, Cohen FE. Automatic extraction of protein point mutations using a graph bigram association. PLoS Comput Biol. 2007;3(2) doi: 10.1371/journal.pcbi.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang DTH, Weng YZ, Lin JH, et al. Protemot: prediction of protein binding sites with automatically extracted geometrical templates. Nucleic Acids Res. 2006;34:W303–9. doi: 10.1093/nar/gkl344. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1538868&blobtype=pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun HW, Tsuruoka Y, Kim JD, et al. Pac Symp Biocomput. Vol. 11. Maui: Hawaii; 2006. Extraction of gene-disease relations from Medline using domain dictionaries and machine learning; pp. 4–15. http://psb.stanford.edu/psb-online/proceedings/psb06/chun.pdf. [PubMed] [Google Scholar]
- 45.Lussier Y, Borlawsky T, Rappaport D, et al. Pac Symp Biocomput. Vol. 11. Maui: Hawaii; 2006. PhenoGO: assigning phenotypic context to Gene Ontology annotations with natural language processing; pp. 64–75. http://psb.stanford.edu/psb-online/proceedings/psb06/lussier.pdf. [PMC free article] [PubMed] [Google Scholar]
- 46.Ahlers CB, Fiszman M, Demner-Fushman D, et al. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Extracting semantic predications from MEDLINE citations for pharmacogenomics; pp. 209–20. [PubMed] [Google Scholar]
- 47.Baker CJO, Witte R. Mutation mining: a prospector's tale. J Inform Syst Front. 2006;8(1):47–57. [Google Scholar]
- 48*.Kim JJ, Zhang Z, Park JC, et al. BioContrasts: extracting and exploiting protein-protein contrastive relations from biomedical literature. Bioinformatics. 2006;22:597–605. doi: 10.1093/bioinformatics/btk016. http://bioinformatics.oxfordjournals.org/cgi/reprint/22/5/597.pdf. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 49.DUC task, documents, and measures. 2006 http://duc.nist.gov/duc2006/tasks.html (September 2007, date last accessed)
- 50.Demner-Fushman D, Lin J. Answer extraction, semantic clustering and extractive summarization for clinical question answering; Proceedings of the 21st International Conference on Computational Linguistics and 44th Annual Meeting of the Association for Computational Linguistics (COLING/ACL 2006), Sydney, Australia 2006; pp. 841–8. [Google Scholar]
- 51*.Ling X, Jiang J, He X, et al. Pac Symp Biocomput 11. Maui: Hawaii; 2006. Automatically generating gene summaries from biomedical literature; pp. 40–51. http://psb.stanford.edu/psb-online/proceedings/psb06/ling.pdf. [indicates papers of particular interest published within the period of this review] [PubMed] [Google Scholar]
- 52.Hersh W, Bhupatiraju RT. The twelfth Text Retrieval Conference, TREC 2003. National Institute of Standards and Technology. Gaithersburg: Maryland; 2003. TREC Genomics track overview; pp. 14–23. [Google Scholar]
- 53.Lu Z, Cohen KB, Hunter L. Pac Symp Biocomput 11. Maui: Hawaii; 2006. Finding GeneRIFs via Gene Ontology annotations; pp. 52–63. http://psb.stanford.edu/psb-online/proceedings/psb06/lu.pdf. [PMC free article] [PubMed] [Google Scholar]
- 54.Rebholz-Schuhmann D, Kirsch H, Arregui M, et al. EBIMed-text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–44. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 55.Chiang JH, Shin JW, Liu HH, et al. GeneLibrarian: an effective gene-information summarization and visualization system. BMC Bioinform. 2006;7(392) doi: 10.1186/1471-2105-7-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernández J, Hoffmann R, Valencia A. iHOP Web services. Nucleic Acids Res. 2007;35:W21–6. doi: 10.1093/nar/gkm298. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanabe L, Scherf U, Smith LH, et al. Medminer: an Internet text-mining tool for biomedical information, with application to gene expression profiling. BioTechniques. 1999;27(6):1210–4. doi: 10.2144/99276bc03. 1216–7. [DOI] [PubMed] [Google Scholar]
- 58.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinform. 2004;5(147) doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Névéol A, Shooshan SE, Humphrey SM, et al. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Multiple approaches to fine-grained indexing of the biomedical literature; pp. 292–303. [PubMed] [Google Scholar]
- 60.Stoica E, Hearst M. Pacific Symposium on Biocomputing 11, 2006. Maui: Hawaii; 2006. Predicting gene functions from text using a cross-species approach; pp. 88–99. http://psb.stanford.edu/psb-online/proceedings/psb06/stoica.pdf. PSB. [PubMed] [Google Scholar]
- 61.Fyshe A, Szafron D. Term generalization and synonym resolution for biological abstracts: using the gene ontology as a source of expert knowledge. BioNLP. 2006.
- 62.Höglund A, Blum T, Brady S, et al. Pac Symp Biocomput 11. Maui: Hawaii; 2006. Significantly improved prediction of subcellular localization by integrating text and protein sequence data; pp. 16–27. http://psb.stanford.edu/psb-online/proceedings/psb06/hoglund.pdf. [PubMed] [Google Scholar]
- 63.Murphy RF, Velliste M, Yao J, et al. Searching online journals for fluorescence microscope images depicting protein subcellular location patterns. IEEE International Symposium on BioInformatics and Biomedical Engineering, Rockville, Maryland, USA 2001. pp. 119–28.
- 64.Kou Z, Cohen W, Murphy R. ACM SIGKDD Workshop on Data Mining in Bioinformatics (BIOKDD) Washington, DC: 2003. Extracting information from text and images for location proteomics; pp. 2–9. [Google Scholar]
- 65**.Shatkay H, Chen N, Blostein D. Integrating image data into biomedical text categorization. Bioinformatics. 2006;22:e446–53. doi: 10.1093/bioinformatics/btl235. [indicates papers of extreme interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 66*.Yu H, Lee M. Accessing bioscience images from abstract sentences. Bioinformatics. 2006;22:e547–56. doi: 10.1093/bioinformatics/btl261. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 67.Rafkind B, Lee M, Chang S, et al. Exploring text and image features to classify images in bioscience literature. BioNLP, NewYork, USA 2006. pp. 73–80.
- 68.Kou Z, Cohen W, Murphy R. A stacked graphical model for associating sub-images with sub-captions. Pacific Symposium on Biocomputing 12, 2007. pp. 257–68. [PMC free article] [PubMed]
- 69.Rhodes J, Boyer S, Kreulen J, et al. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Mining patents using molecular similarity search; pp. 304–15. [PubMed] [Google Scholar]
- 70.Claudinot S, Nicolas M, Oshima H, et al. Long-term renewal of hair follicles from clonogenic multipotent stem cells. PNAS. 2005;102(41):14677–82. doi: 10.1073/pnas.0507250102. Image on cover page. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan SL, Nakao H, He Y, et al. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. PNAS. 1999;96(10):5533–8. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schuler B, Lipman E, Steinbach P, et al. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. PNAS. 2005;102(8):2754–9. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kihara D, Lu H, Kolinski A, et al. TOUCHSTONE: an ab initio protein structure prediction method that uses threading-based tertiary restraints. PNAS. 2001;98(18):10125–30. doi: 10.1073/pnas.181328398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu H, Li M, Labrador J, et al. Cross GTPase-activating protein (CrossGAP)/Vilse links the Roundabout receptor to Rac to regulate midline repulsion. PNAS. 2005;102(12):4613–8. doi: 10.1073/pnas.0409325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H. AMIA Annu Symp Proc. Washington, DC: 2006. Towards answering biological questions with experimental evidence: Automatically identifying text that summarize image content in full-text articles; pp. 834–8. [PMC free article] [PubMed] [Google Scholar]
- 76.Muller H, Deselaers T, Lehmann T, et al. Overview of the ImageCLEFmed 2006 medical retrieval and annotation tasks. CLEF 2006 Working Notes, Alicante, Spain. 2006.
- 77*.Lacoste C, Chevallet JP, Lim J, et al. IPAL knowledge-based medical image retrieval in ImageCLEFmed 2006. CLEF 2006 Working Notes, Alicante, Spain. 2006. [indicates papers of particular interest published within the period of this review]
- 78.Voorhees EM, Tice DM. The TREC-8 question answering track evaluation. In: Voorhees EM, Harman D, editors. Proceedings of the Eigth Text REtrieval Conference (TREC-8). NIST, 2000. Gaithersburg: Maryland; 2000. [Google Scholar]
- 79.Mollá D, Vicedo J. Question answering in restricted domains: An overview. Comput Linguist. 2007;33(1):41–61. [Google Scholar]
- 80.Hersh W, Cohen AM, Roberts P, et al. The FifteenthText Retrieval Conference—TREC 2006. NIST. Gaithersburg: Maryland; 2006. TREC 2006 genomics track overview. [Google Scholar]
- 81.Zweigenbaum P. Question answering in biomedicine. In: de Rijke M, Webber B, editors. Proc Workshop on Natural Language Processing for Question Answering, EACL 2003. Budapest: ACL; 2003. 2003. pp. 1–4. [Google Scholar]
- 82.Ely J, Osheroff J, Chambliss M, et al. Answering physicians' clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005;12:217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacquemart P, Zweigenbaum P. Stud Health Technol Inform. Vol. 95. Amsterdam: IOS Press; 2003. Towards a medical question-answering system: a feasibility study; pp. 463–8. [PubMed] [Google Scholar]
- 84.Huang X, Lin J, Demner-Fushman D. AMIA Annu Symp Proc. Washington, DC: 2006. Evaluation of PICO as a knowledge representation for clinical questions; pp. 359–63. [PMC free article] [PubMed] [Google Scholar]
- 85*.Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques. Comput Linguist. 2007;33:63–103. [indicates papers of particular interest published within the period of this review] [Google Scholar]
- 86.Lin J, Demner-Fushman D. 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR) Washington: Seattle; 2006. The role of knowledge in conceptual retrieval: A study in the domain of clinical medicine; pp. 469–76. [Google Scholar]
- 87.Yu H, Lee M, Kaufman D, et al. Development, implementation, and a cognitive evaluation of a definitional question answering system for physicians. J Biomed Inform. 2007;40(3):236–51. doi: 10.1016/j.jbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 88.Zhou W, Yu C, Torvik V, et al. A concept-based framework for passage retrieval in Genomics; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006. [Google Scholar]
- 89*.Demner-Fushman D, Humphrey S, Ide N, et al. Finding relevant passages in scientific articles: fusion of automatic approaches vs. an interactive team effort; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006; [indicates papers of particular interest published within the period of this review] [Google Scholar]
- 90.Jiang J, He X, Zhai C. Robust pseudo feedback estimation and HMM passage extraction: UIUC at TREC 2006 Genomics Track; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006. [Google Scholar]
- 91.Caporaso J, Baumgartner W, Kim H, et al. Concept recognition, information retrieval, and machine learning in genomics question-answering; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006. [Google Scholar]
- 92.Divoli A, Hearst M, Nakov P, et al. BioText team report for the TREC 2006 Genomics Track; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006. [Google Scholar]
- 93.Zheng H, Lin C, Huang L, et al. Using profile matching and text categorization for answer extraction in TREC Genomics; Proceedings of Fifteenth Text REtrieval Conference, Gaithersburg. 2006. [Google Scholar]
- 94.Swanson DR. Fish oil, Raynaud's syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986;30:7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 95*.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. J Biomed Inform. 2006;39(6):600–11. doi: 10.1016/j.jbi.2005.11.010. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 96.Jelier R, Jenster G, Dorssers LC, et al. Text-derived concept profiles support assessment of DNA microarray data for acute myeloid leukemia and for androgen receptor stimulation. BMC Bioinform. 2007;8:14. doi: 10.1186/1471-2105-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97*.Seki K, Mostafa J. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Discovering implicit associations between genes and hereditary diseases; pp. 316–27. [indicates papers of particular interest published within the period of this review] [PubMed] [Google Scholar]
- 98.Pospisil P, Iyer LK, Adelstein SJ, et al. A combined approach to data mining of textual and structured data to identify cancer-related targets. BMC Bioinform. 2006;7(354) doi: 10.1186/1471-2105-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99*.Palakal M, Bright J, Sebastian T, et al. A comparative study of cells in inflammation, EAE and MS using biomedical literature data mining. J Biomed Sci. 2007;14(1):67–85. doi: 10.1007/s11373-006-9120-8. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed] [Google Scholar]
- 100**.Rzhetsky A, Iossifov I, Loh JM, et al. Microparadigms: chains of collective reasoning in publications about molecular interactions. PNAS. 2006;103:4940–5. doi: 10.1073/pnas.0600591103. [indicates papers of extreme interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101**.Hristovski D, Friedman C, Rindflesch T, et al. AMIA Annu Symp Proc. Washington, DC: 2006. Exploiting semantic relations for literature-based discovery; pp. 349–53. [indicates papers of extreme interest published within the period of this review] [PMC free article] [PubMed] [Google Scholar]
- 102*.Demaine J, Martin J, Wei L, et al. LitMiner: integration of library services within a bio-informatics application. Biomed Digit Libr. 2006;3(11) doi: 10.1186/1742-5581-3-11. doi:10.1186/1742-5581-3-11, http://www.bio-diglib.com/content/3/1/11, http://www.litminer.com/ [indicates papers of particular interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103*.Xiang Z, Zheng W, He Y. BBP: Brucella genome annotation with literature mining and curation. BMC Bioinform. 2006;7(347) doi: 10.1186/1471-2105-7-347. [indicates papers of particular interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smalheiser NR, Torvik VI, Bischoff-Grethe A, et al. Collaborative development of the Arrowsmith two node search interface designed for laboratory investigators. J Biomed Discov Collab. 2006;1(8) doi: 10.1186/1747-5333-1-8. http://www.j-biomeddiscovery.com/content/1/1/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aubry M, Monnier A, Chicault C, et al. Combining evidence, biomedical literature and statistical dependence: new insights for functional annotation of gene sets. BMC Bioinform. 2006;7:241. doi: 10.1186/1471-2105-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Firth JR. Papers in Linguistics. London: Oxford University Press; 1957. pp. 1934–1951. [Google Scholar]
- 107.Habert B, Zweigenbaum P. Contextual acquisition of information categories: what has been done and what can be done automatically? In: Nevin BE, Johnson SM, editors. The Legacy of Zellig Harris: Language and information into the 21st Century, Mathematics and computability of language. Vol. 2. Amsterdam: John Benjamins; 2002. pp. 203–31. [Google Scholar]
- 108.Hristovski D, Peterlin B, Mitchell J, et al. Using literature-based discovery to identify disease candidate genes. Int J Med Inform. 2005;74(2–4):289–98. doi: 10.1016/j.ijmedinf.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 109.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 110.Turtle HR, Croft WB. Evaluation of an inference network-based retrieval model. ACM T Inform Syst. 1991;9(3):187–222. [Google Scholar]
- 111*.Torvik VI, Smalheiser N. A quantitative model for linking two disparate sets of articles in MEDLINE. Bioinformatics. 2007. [indicates papers of particular interest published within the period of this review] [DOI] [PubMed]
- 112.Yeh AS, Hirschman L, Morgan AA. Evaluation of text data mining for database curation: lessons learned from the KDD Challenge Cup. Bioinformatics. 2003;19(Suppl 1):i331–9. doi: 10.1093/bioinformatics/btg1046. [DOI] [PubMed] [Google Scholar]
- 113.Voorhees E. TREC: Improving information access through evaluation. Bulletin of the American Society for Information Science and Technology. 2005;32(1) http://www.asis.org/Bulletin/Oct-05/voorhees.html. [Google Scholar]
- 114**.Wilbur WJ, Rzhetsky A, Shatkay H. New directions in biomedical text annotation: definitions, guidelines and corpus construction. BMC Bioinform. 2006;25(356) doi: 10.1186/1471-2105-7-356. [indicates papers of extreme interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pyysalo S, Ginter F, Heimonen J, et al. BioInfer: a corpus for information extraction in the biomedical domain. BMC Bioinform. 2007;8(50) doi: 10.1186/1471-2105-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116*.Morgan AA, Wellner B, Colombe JB, et al. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Evaluating the automatic mapping of human gene and protein mentions to unique identifiers; pp. 281–91. [indicates papers of particular interest published within the period of this review] [PubMed] [Google Scholar]
- 117*.Karamanis N, Lewin I, Sealy R, et al. Pac Symp Biocomput 12. Maui: Hawaii; 2007. Integrating natural language processing with Flybase curation; pp. 245–56. [indicates papers of particular interest published within the period of this review] [PubMed] [Google Scholar]
- 118.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2(11):e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hersh W, Cohen A, Yang J, et al. The Fourteenth Text Retrieval Conference—TREC 2005. Gaithersburg: Maryland; 2005. Trec 2005 genomics track overview. [Google Scholar]
- 120.Hirschman L, Yeh A, Blaschke C, et al. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinform. 2005;6 doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Camon EB, Barrell DG, Dimmer EC, et al. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA. BMC Bioinform. 2005;6(Suppl 1):S17. doi: 10.1186/1471-2105-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wattarujeekrit T, Shah PK, Collier N. PASBio: predicate-argument structures for event extraction in molecular biology. BMC Bioinform. 2004;5:155. doi: 10.1186/1471-2105-5-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chou WC, Tsai RTH, Su YS, et al. Proceedings of the workshop on frontiers in linguistically annotated corpora 2006. Association for Computational Linguistics; Australia, Sydney: 2006. A semi-automatic method for annotating a biomedical proposition bank; pp. 5–12. [Google Scholar]
- 124.Cohen KB, Hunter L. A critical review of PASBio's argument structures for biomedical verbs. BMC Bioinform. 2006;7(Suppl 3):S5. doi: 10.1186/1471-2105-7-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. Proceedings of ACL2004. 2004. pp. 430–7.
- 126*.Hirschman L, Colosimo M, Morgan A, et al. Overview of BioCreative Task 1B: normalized gene lists. BMCBioinform. 2005;6(Suppl 1):S11. doi: 10.1186/1471-2105-6-S1-S11. [indicates papers of particular interest published within the period of this review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cohen AM. Linking biological literature, ontologies and databases: mining biological semantics. Association for Computational Linguistics; Detroit, Michigan: 2005. Unsupervised gene/protein named entity normalization using automatically extracted dictionaries; pp. 17–24. [Google Scholar]
- 128.Fang HR, Murphy K, Jin Y, et al. Linking natural language processing and biology: towards deeper biological literature analysis. Association for Computational Linguistics; Brooklyn, New York: 2006. Human gene name normalization using text matching with automatically extracted synonym dictionaries; pp. 41–8. [Google Scholar]
- 129.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinform. 2004;5:1471–2105. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shah PK, Jensen LJ, Boue S, et al. Extraction of transcript diversity from scientific literature. PLoS Comput Biol. 2005;1(1):67–73. doi: 10.1371/journal.pcbi.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20(4):557–68. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- 132.Zweigenbaum P, Demner-Fushman D, Yu H, et al. Proc Pac Symp Biocomput 12. Maui: Hawaii; 2007. New frontiers in biomedical text mining; pp. 205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hirschman L, Bourne P, Cohen KB, et al. Translating Biology: text mining tools that work, 2007. http://psb.stanford.edu/cfp-nlp.html (September 2007, date last accessed) [PMC free article] [PubMed]
- 134.Verspoor K, Cohen KB, Mani I, et al. Linking natural language processing and biology: towards deeper biological literature analysis. Brooklyn, New York: Association for Computational Linguistics; 2006. Introduction to BioNLP'06; pp. 3–5. [Google Scholar]