Abstract
There has been much work devoted to the mapping, alignment, and linking of ontologies (MALO), but little has been published about how to evaluate systems that do this. A fault model for conducting fine-grained evaluations of MALO systems is proposed, and its application to the system described in Johnson et al. [15] is illustrated. Two judges categorized errors according to the model, and inter-judge agreement was calculated by error category. Overall inter-judge agreement was 98% after dispute resolution, suggesting that the model is consistently applicable. The results of applying the model to the system described in [15] reveal the reason for a puzzling set of results in that paper, and also suggest a number of avenues and techniques for improving the state of the art in MALO, including the development of biomedical domain specific language processing tools, filtering of high frequency matching results, and word sense disambiguation.
1. Introduction
The mapping, alignment, and/or linking of ontologies (MALO) has been an area of active research in recent years [4,28]. Much of that work has been groundbreaking, and has therefore been characterized by the lack of standardized evaluation metrics that is typical for exploratory work in a novel domain. In particular, this work has generally reported coarse metrics, accompanied by small numbers of error exemplars. However, in similar NLP domains finer-grained analyses provide system builders with insight into how to improve their systems, and users with information that is crucial for interpreting their results [23,14,8]. MALO is a critical aspect of the National Center for Biomedical Ontology/Open Biomedical Ontologies strategy of constructing multiple orthogonal ontologies, but such endeavors have proven surprisingly difficult–Table 1 shows the results of a representative linking system, which ranged as low as 60.8% overall when aligning the BRENDA Tissue ontology with the Gene Ontology [15].
Table 1.
Correctness rates for the ontology linking system described in Johnson et al. (2006). The three OBO ontologies listed in the left column were linked to the GO via the three lexical methods in the right columns.
| Type of linking method | ||||
|---|---|---|---|---|
| Ontology | Overall | Exact | Synonyms | Stemming |
| ChEBI | 84.2% | 98.3% (650/661) | 60.0% (180/300) | 73.5%(147/200) |
| Cell Type | 92.9% | 99.3% (431/434) | 73.0% (65/89) | 83.8% (88/105) |
| BRENDA | 60.8% | 84.5% (169/200) | 76.0% (152/200) | 11.0% (22/200) |
This paper proposes a fault model for evaluating lexical techniques in MALO systems, and applies it to the output of the system described in Johnson et al. [15]. The resulting analysis illuminates reasons for differences in performance of both the lexical linking techniques and the ontologies used. We suggest concrete methods for correcting errors and advancing the state of the art in the mapping, alignment, and/or linking of ontologies. Because many techniques used in MALO include some that are also applied in text categorization and information retrieval, the findings are also useful to researchers in those areas.
Previous lexical ontology integration research deals with false positive error analysis by briefly mentioning causes of those errors, as well as some illustrative examples, but provides no further analysis. Bodenreider et al. mention some false positive alignments but offer no evaluations [3]. Burgun et al. assert that including synonyms of under three characters, substring matching, and case insensitive matching are contributors to false positive rates and thus are not used in their linking system [5]. They report that term polysemy from different ontologies contributes to false positive rates, but do not explain the magnitude of the problem. Zhang et al. report a multi-part alignment system but do not discuss errors from the lexical system at all [29]. Lambrix et al. report precision from 0.285-0.875 on a small test set for their merging system, SAMBO, which uses n-grams, edit distance, WordNet, and string matching. WordNet polysemy and the N-gram matching method apparently produce 12.5% and 24.3% false positive rates, respectively [17,16]. Lambrix and Tan state that the same alignment systems produce different results depending on the ontology used; they give numbers of wrong suggestions but little analysis [18]. For a linking system that matches entities with and without normalization of punctuation, capitalization, stop words, and genitive markers, Sarkar et al. report without examples a 4-5% false positive rate [26]. Luger et al. present a structurally verified lexical mapping system in which contradictory mappings occur at certain thresholds, but no examples or analyses are given [20]. Mork et al. introduce an alignment system with a lexical component but do not detail its performance [22]. Johnson et al. provide error counts sorted by search type and ontology but provide no further analysis [15]. Their system’s performance for matching BRENDA terms to GO is particularly puzzling because correctness rates of up to 100% are seen with some ontologies, but correctness for matching BRENDA is as low as 7% (see Table 1).
There has been no comprehensive evaluation of errors in lexical MALO systems. This leaves unaddressed a number of questions with real consequences for MALO system builders: What types of errors contribute to reduced performance? How much do they contribute to error rates? Are there scalable techniques for reducing errors without adversely impacting recall? Here we address these questions by proposing a fault model for false-positive errors in MALO systems, providing an evaluation of the errors produced by a biomedical ontology linking system, and suggesting methods to reduce errors in MALO.
2. Methods
2.1. The ontology linking method in Johnson et al. (2006)
Since understanding the methodology employed in Johnson et al. is important to understanding the analysis of its errors, we review that methodology briefly here. Their system models inter-ontology relationship detection as an information retrieval task, where relationship is defined as any direct or indirect association between two ontological concepts. Three OBO ontologies’ terms (BRENDA Tissue, ChEBI, and Cell Type) are searched for in GO terms [9,27,11,1]. Three types of searches are performed: (a) exact match to OBO term, (b) OBO term and its synonyms, and (c) stemmed OBO term. The stemmer used in (c) was an implementation of the Porter Stemmer provided with the Lucene IR library [13,25]. Besides stemming, this implementation also reduces characters to lower case, tokenizes on whitespace, punctuation and digits (removing the latter two), and removes a set of General English stop words. The output of the system is pairs of concepts: one GO concept and one OBO concept.
To determine the correctness of the proposed relationships, a random sample of the output (2,389 pairs) was evaluated by two domain experts who answered the question: Is this OBO term the concept that is being referred to in this GO term/definition? Inter-annotator agreement after dispute resolution was 98.2% (393/400). The experts deemed 481 relations to be incorrect, making for an overall estimated system error rate of 20%. All of the system outputs (correct, incorrect, and unjudged) were made publicly available at compbio.uchsc.edu/dependencies.
2.2. The fault model
In software testing, a fault model is an explicit hypothesis about potential sources of errors in a system [2,8]. We propose a fault model, comprising three broad classes of errors (see Table 2), for the lexical components of MALO systems. The three classes of errors are distinguished by whether they are due to inherent properties of the ontologies themselves, are due to the processing techniques that the system builders apply, or are due to including inappropriate metadata in the data that is considered for locating relationships. The three broad classes are further divided into more specific error types, as described below. Errors in the lexical ambiguity class arise because of the inherent polysemy of terms in multiple ontologies (and in natural language in general) and from ambiguous abbreviations (typically listed as synonyms in an ontology). Errors in the text processing class come from manipulations performed by the system, such as the removal of punctuation, digits, or stop words, or from stemming. Errors in metadata matching occur when elements in one ontology matched metadata in another ontology, e.g. references to sources that are found at the end of GO definitions.
Table 2.
The fault model and results of its application to Johnson et al.’s erroneous outputs. The rows in bold are the subtotaled percentages of the broad categories of errors in relation to all errors. The non-bolded rows indicate the percentages of the subtypes of errors in relation to the broad category that they belong to. The counts for the subtypes of text processing errors exceed the total text processing count because multiple types of text processing errors can contribute to one erroneously matched relationship.
| Inter-judge agreement | ||||
|---|---|---|---|---|
| Type of error | Percent | Count | pre-resolution | post-resolution |
| Lexical ambiguity errors | ||||
| biological polysemy | 56% | (105/186) | 86% | 98% |
| ambiguous abbreviation | 44% | (81/186) | 96% | 99% |
| Lexical Ambiguity Total | 38% | (186/481) | ||
|
| ||||
| Text processing errors | ||||
| Stemming | 6% | (29/449) | 100% | 100% |
| digit removal | 51% | (231/449) | 100% | 100% |
| punctuation removal | 27% | (123/449) | 100% | 100% |
| stop word removal | 14% | (65/449) | 99% | 100% |
| Text Processing Total | 60% | (290/481) | ||
|
| ||||
| Matched Metadata Total | 1% | (5/481) | 100% | 100% |
| Total | 99% | (481/481) | 95% | 99% |
To evaluate whether or not the fault model is consistently applicable, two authors independently classified the 481 incorrect relationships from the Johnson et al. system into nine fine-grained error categories (the seven categories in the model proposed here, plus two additional categories, discussed below, that were rejected). The model allows for assignment of multiple categories to a single output. For instance, the judges determined that CH:29356 oxide(2-) erroneously matched to GO:0019417 sulfuroxidation due to both character removal during tokenization ((2-) was deleted) and to stemming (the remaining oxide and oxidation both stemmed to oxid ). Detailed explanations of the seven error categories, along with examples of each, are given belowa.
3. Results
Table 2 displays the counts and percentages of each type of error, with inter-judge agreement (IJA) for each category. Section 3.1 discusses inter-judge agreement and the implications that low IJA has for the fault model. Sections 3.2-3.3 explain and exemplify the categories of the fault model, and 3.4 describes the distribution of error types across orthogonal ontologies.
3.1. Inter-judge agreement
Inter-judge agreement with respect to the seven final error categories in the fault model is shown in Table 2. Overall IJA was 95% before dispute resolution and 99% after resolution. In the 1% of cases where the judges did not agree after resolution, the judge who was most familiar with the data assigned the categories. The initial fault model had two error categories that were eliminated from the final model because of low IJA. The first category, tokenization, had an abysmal 27% agreement rate even after dispute resolution. The second eliminated category, general English polysemy, had 80% pre-resolution agreement and 94% post-resolution agreement, with only 10 total errors assigned to this category. Both judges felt that all errors in this category could justifiably be assigned to the biological polysemy category; therefore, this category is not included in the final fault model.
3.2. Lexical ambiguity errors
Lexical ambiguity refers to words that denote more than one concept. It is a serious issue when looking for relationships between domain-distinct ontologies [10:1429]. Lexical ambiguity accounted for 38% of all errors.
Biological polysemy is when a term that is present in two ontologies denotes distinct biological concepts. It accounted for 56% of all lexical ambiguity errors. Examples of biological polysemy include (1–3) below. Example (1) shows a polysemous string that is present in two ontologies.
| (1) | BTO | 0000280: | cone |
| def: | A mass of ovule-bearing or pollen-bearing scales or bracts in trees of the pine family or in cycads that are arranged usually on a somewhat elongated axis. | ||
| GO | 0042676: | cone cell fate commitment | |
| def: | The process by which a cell becomes committed to become a cone cell. |
OBO terms have synonyms, some of which polysemously denote concepts that are more general than the OBO term itself, and hence match GO concepts that are not the same as the OBO term. Examples (2) and (3) show lexical ambiguity arising because of the OBO synonyms.
| (2) | BTO | 0000131: | blood plasma |
| synonym: | plasma | ||
| def: | The fluid portion of the blood in which the particulate components are suspended. | ||
| GO | 0046759: | lytic plasma membrane viral budding | |
| def: | A form of viral release in which the nucleocapsid evaginates from the host nuclear membrane system, resulting in envelopment of the virus and cell lysis. | ||
| (3) | CH | 17997: | dinitrogen |
| synonym: | nitrogen | ||
| GO | 0035243: | protein-arginine omega-N symmetric methyltransferase activity | |
| def: | … Methylation is on the terminal nitrogen (omega nitrogen) … |
Example (4) shows that by the same synonymy mechanism, terms from different taxa match erroneously.
| (4) | CL | 0000338: | neuroblast (sensu Nematoda and Protostomia) |
| synonym: | neuroblast | ||
| GO | 0043350: | neuroblast proliferation (sensu Vertebrata) |
Ambiguous abbreviation errors happen when an abbreviation in one ontology matches text in another that does not denote the same concept. The ambiguity of abbreviations is a well-known problem in biomedical text [7,6]. In the output of [15] it is the cause of 43% of all lexical ambiguity errors. The chemical ontology includes many one- and two-character symbols for elements (e.g. C for carbon, T for thymine, As for arsenic, and At for astatine). Some abbreviations are overloaded even within the chemical domain. For example, in ChEBI C is listed as a synonym for three chemical entities besides carbon, viz. L-cysteine, L-cysteine residue, and cytosine. So, single-character symbols match many GO terms, but with a high error rate. Examples (5) and (6) illustrate such errors.
| (5) | CH | 17821: | thymine |
| synonym: | T | ||
| GO | 0043377: | negative regulation of CD8-positive T cell differentiation |
One- and two-character abbreviations sometimes also match closed-class or function words, such as a or in, as illustrated in example (6).
| (6) | CH | 30430: | indium |
| synonym: | In | ||
| GO | 0046465: | dolichyl diphosphate metabolism | |
| def: | … In eukaryotes, these function as carriers of … |
3.3. Text processing errors
As previously mentioned, Johnson et al.’s system uses a stemmer that requires lower-case text input. The system performs this transformation with a Lucene analyzer that splits tokens on non-alphabetic characters, then removes digits and punctuation, and removes stop words. This transformed text is then sent to the stemmer. Example (7) illustrates a ChEBI term and a GO term, and the search and match strings that are produced by the stemming device.
| (7) | Original text | Tokenized/stemmed text | ||
| CH | 32443: | L-cysteinate(2-) | l cystein | |
| GO | 0018118: | peptidyl-L-cysteine … | peptidyl l cystein … |
Errors arise from the removal of digits and punctuation, the removal of stop words, and the stemming process itself (see Table 2). These are illustrated in examples (8–16). Few errors resulting from text processing can be attributed to a single mechanism.
Digit removal is the largest contributor among the text processing error types, constituting 51% of the errors. Punctuation removal is responsible for 27% of the errors. These are illustrated in examples (8–10).
| (8) | CL | 0000624: | CD4 positive T cell |
| GO | 0043378: | positive regulation of CD8-positive T cell differentiation | |
| (9) | CH | 20400: | 4-hydroxybutanal |
| GO | 0004409: | homoaconitate hydratase activity | |
| def: | Catalysis of the reaction: 2-hydroxybutane-1,2,4-tri … | ||
| (10) | CH | 30509: | carbon(1+) |
| GO | 0018492: | carbon-monoxide dehydrogenase (acceptor) activity |
Six percent of the errors involve the stemming mechanism. (This is somewhat surprising, since the Porter stemmer has been independently characterized as being only moderately aggressive [12].)
Of the 580 evaluated relationships that were processed by the stemming mechanism in the original linking system, 43% (253/580) match because of the stemming applied. Of those, 73% (185/253) are correct relationships; 27% (68/253) are incorrect. Table 3 displays a list of all suffixes that were removed during stemming and the counts of how many times their removal resulted in a correct or an incorrect match. Examples (11–13) display errors due to stemming:
Table 3.
Counts of correct and incorrect relationships that resulted after the stemming mechanism was applied.
| Matches | -al | -ate | -ation | -e | -ed | -ic | -ing | -ize | -ous | -s |
|---|---|---|---|---|---|---|---|---|---|---|
| Correct | 19 | 1 | 2 | 12 | 0 | 11 | 0 | 0 | 2 | 157 |
| Incorrect | 1 | 17 | 3 | 26 | 3 | 2 | 4 | 1 | 0 | 39 |
| (11) | CH | 25741: | oxides |
| GO | 0016623: | oxidoreductase activity, acting on the aldehyde or oxo … | |
| def: | Catalysis of an oxidation-reduction (redox) reaction … | ||
| (12) | CH | 25382: | monocarboxylates |
| GO | 0015718: | monocarboxylic acid transport | |
| def: | The directed movement of monocarboxylic acids into … | ||
| (13) | CH | 32530: | histidinate(2-) |
| GO | 0019558: | histidine catabolism to 2-oxoglutarate |
While stemming works most of the time to improve recall—the count of correct matches in Table 3 is more than double the count of incorrect matches (204 versus 96)—an analysis of the errors shows that in this data, there is a subset of suffixes that do not stem well from biomedical terms, at least in these domains. Removal of -e results in incorrect matches far more often than it results in correct matches, and removal of -ate almost never results in a correct match. These findings illustrate the need for a domain-specific stemmer for biomedical text.
Finally, stop word removal contributed 14% of the error rate. Examples like (14–16) are characteristic:
| (14) | CL | 0000197: | receptor cell |
| GO | 0030152: | bacteriocin biosynthesis | |
| def: | … at specific receptors on the cell surface … | ||
| (15) | CH | 25051: | lipid As |
| GO | 0046834: | lipid phosphorylation | |
| (16) | CH | 29155: | His-tRNA(His) |
| GO | 0050562: | lysine-tRNA(Pyl) ligase activity |
3.4. Applying the fault model to orthogonal ontologies
The fault model that this paper proposes explains the patterns observed in the Johnson et al. work. They report an uneven distribution of accuracy rates across the ontologies (see Table 1); Table 4 shows that this corresponds to an uneven distribution of the error types across ontologies. Most striking is that ChEBI is especially prone to ambiguous abbreviation errors, which were entirely absent with the other two ontologies. BRENDA is prone to deletion-related errors — in fact, over half of the errors in the text processing error category are due to a specific type of term in BRENDA (169/290). These terms have the structure X cell, where X is any combination of capital letters, digits, and punctuation, such as B5/589 cell, T-24 cell, and 697 cell. The search strings rendered from these after the deletions—B cell, T cell, and cell, respectively—match promiscuously to GO (see Figure 1).
Table 4.
Distribution of error types across ontologies
| Deletion of: | |||||||
|---|---|---|---|---|---|---|---|
| Ontology | Biological polysemy | Abbreviation ambiguity | digit | punct. | stopword | Stemming | Totals |
| BRENDA | 84 | 0 | 187 | 89 | 54 | 2 | 416 |
| Cell Type | 29 | 0 | 9 | 0 | 7 | 0 | 45 |
| ChEBI | 26 | 81 | 35 | 34 | 4 | 27 | 207 |
Figure 1.
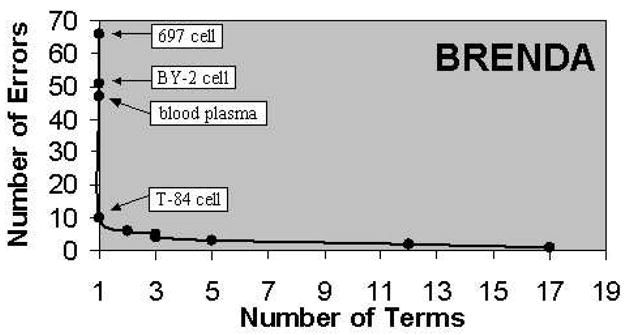
A few terms from BRENDA caused a large number of errors.
Biological polysemy errors are a problem in all three ontologies. Sixty-four percent of the errors for Cell Type were related to polysemy, 20% in BRENDA, and 12% in ChEBI. Dealing with word sense disambiguation could yield a huge improvement in performance for these ontologies.
None of this error type distribution is apparent from the original data reported in [15], and all of it suggests specific ways of addressing the errors in aligning these ontologies with GO.
4. Fault-driven analysis suggests techniques for improving MALO
Part of the value of the fault model is that it suggests scalable methods for reducing the false positive error rate in MALO without adversely affecting recall. We describe some of them here.
4.1. Error reduction techniques related to text processing
Johnson et al. reported exceptionally low accuracy for BRENDA relationships based on stemming: only 7-15% correctness. Our investigation suggests that this low accuracy is due to a misapplication of an out-of-the-box Lucene implementation of the Porter stemmer: it deletes all digits, which occur in BRENDA cell line names, leading to many false-positive matches against GO concepts containing the word cell. Similarly, bad matches between ChEBI chemicals and the GO (73-74% correctness rate) occur because of digit and punctuation removal. This suggests that a simple change to the text processing procedures could lower the error rate dramatically.
4.2. Error reduction techniques related to ambiguity
For ontologies with error patterns like ChEBI and BRENDA, excluding synonyms shorter than three characters would be beneficial. For example, Bodenreider and Burgun excluded synonyms shorter than three characters [5]. Length-based filtering of search candidates has been found useful for other tasks in this domain, such as entity identification and normalization of Drosophila genes in text [21].
Numerous techniques have been proposed for resolving word sense ambiguities [24]. The OBO definitions may prove to be useful resources for knowledge-based ontology term disambiguation [19].
4.3. Error reduction by filtering high error contributors
The Zipf-like distribution of error counts across terms (see Figure 1) suggests that filtering a small number of terms would have a beneficial effect on the error rates due to both text processing and ambiguity-related errors. This filtering could be carried out in post-processing, by setting a threshold for matching frequency or for matching rank. Alternatively, it could be carried out in a pre-processing step by including high-frequency tokens in the stop list. This analysis would need to be done on an ontology-by-ontology basis, but neither method requires expert knowledge to execute the filtering process. As an example of the first procedure, removing the top contributors to false-positive matches in each ontology would yield the results in Table 5.
Table 5.
Effect of filtering high-frequency match terms.
| Ontology | Terms removed | Increase in correctness | Decrease in matches |
|---|---|---|---|
| BRENDA | 697 cell, BY-2 cell, blood plasma, T-84 cell | 27% | 41% |
| Cell Type | band form neutrophil, neuroblast | 4% | 3% |
| ChEBI | iodine, L-isoleucine residue, groups | 2% | 2% |
5. Conclusion
The analysis presented in this paper supports the hypotheses that it is possible to build a principled, data-driven fault model for MALO systems; that the model proposed can be applied consistently; that such a model reveals previously unknown sources of system errors; and that it can lead directly to concrete suggestions for improving the state of the art in ontology alignment. Although the fault model was applied to the output of only one linking system, that system included linking data between four orthogonal ontologies. The model proved effective at elucidating the distinct causes of errors in linking the different ontologies, as well as the puzzling case of BRENDA. A weakness of the model is that it addresses only false-positive errors; evaluating failures of recall is a thorny problem that deserves further attention.
Based on the descriptions of systems and false positive outputs of related work, it seems that the fault model presented in this work could be applied to the output of many other systems, including at least [3,5,16,17,18,26,20,22,29]. Note that in the data that was examined in this paper, the distribution of error types was quite different across not just lexical techniques, but across ontologies, as well. This reminds us that specific categories in the model may not be represented in the output of all systems applied to all possible pairs of ontologies, and that there may be other categories of errors that were not reflected in the data that was available to us. For example, the authors of the papers cited above have reported errors due to case folding, spelling normalization, and word order alternations that were not detected in the output of Johnson et al.’s system. However, the methodology that the present paper illustrates—i.e., combining the software testing technique of fault modelling with an awareness of linguistic factors—should be equally applicable to any lexically-based MALO system. Many of the systems mentioned in this paper also employ structural techniques for MALO. These techniques are complementary to, not competitive with, lexical ones. The lexical techniques can be evaluated independently of the structural ones; a similar combination of the software testing approach with awareness of ontological/structural issues may be applicable to structural techniques. We suggest that the quality of future publications in MALO can be improved by discussing error analyses with reference to this model or very similar ones derived via the same techniques.
Acknowledgments
The authors gratefully acknowledge the insightful comments of the three anonymous PSB reviewers, and thank Michael Bada for helpful discussion and Todd A. Gibson and Sonia Leach for editorial assistance. This work was supported by NIH grant R01-LM008111 (LH).
Footnotes
In all paired concepts in our examples, BTO=BRENDA Tissue Ontology, CH=ChEBI Ontology, CL=Cell Type Ontology, and GO=Gene Ontology. Underlining indicates the portion of GO and OBO text that matches, thereby causing the linking system to propose that a relationship exists between the pair.
References
- 1.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6(2) doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder RV. Testing Object-Oriented Systems: Models, Patterns, and Tools. Addison-Wesley Professional; October 1999 [Google Scholar]
- 3.Bodenreider O, Hayamizu TF, Ringwald M, De Coronado S, Zhang S. Of mice and men: aligning mouse and human anatomies. AMIA Annu Symp Proc. 2005:61–65. [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O, Mitchell JA, McCray AT. Biomedical ontologies: Session introduction. Pac Symp Biocomput. 2003, 2004, 2005 doi: 10.1142/9789812704856_0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgun A, Bodenreider O. An ontology of chemical entities helps identify dependence relations among Gene Ontology terms. Proc SMBM. 2005 [Google Scholar]
- 6.Chang J, Schüutze H. Abbreviations in biomedical text. In: Ananiadou S, McNaught J, editors. Text mining for biology and biomedicine. Artech House; 2006. pp. 99–119. [Google Scholar]
- 7.Chang JT, Schütze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J Am Med Inform Assoc. 2002;9(6):612–620. doi: 10.1197/jamia.M1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen KB, Tanabe L, Kinoshita S, Hunter L. A resource for constructing customized test suites for molecular biology entity identification systems. BioLINK 2004. 2004:1–8. [Google Scholar]
- 9.Consortium TGO. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium TGO. Creating the Gene Ontology resource: design and implementation. Genome Research. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degtyarenko K. Proc 2003 Itnl Chem Info Conf. 2003. Chemical vocabularies and ontologies for bioinformatics. [Google Scholar]
- 12.Harman D. How effective is suffixing? J Am Soc Info Sci. 1991;42(1):7–15. [Google Scholar]
- 13.Hatcher E, Gospodnetific O. Lucene in Action (In Action series) Manning Publications; 2004. [Google Scholar]
- 14.Hirschman L, Mani I. Evaluation. In: Mitkov R, editor. Oxford handbook of computational linguistics. Oxford University Press; 2003. pp. 414–429. [Google Scholar]
- 15.Johnson HL, Cohen KB, Baumgartner WA, Lu Z, Bada M, Kester T, Kim H, Hunter L. Evaluation of lexical methods for detecting relationships between concepts from multiple ontologies. Pac Symp Biocomput. 2006:28–39. [PMC free article] [PubMed] [Google Scholar]
- 16.Lambrix P, Edberg A. Evaluation of ontology merging tools in bioinfor-matics. Pac Symp Biocomput. 2003:589–600. doi: 10.1142/9789812776303_0055. [DOI] [PubMed] [Google Scholar]
- 17.Lambrix P, Edberg A, Manis C, Tan H. Merging DAML+OIL bio-ontologies. Description Logics. 2003 [Google Scholar]
- 18.Lambrix P, Tan H. A framework for aligning ontologies. PPSWR. 2005:17–31. [Google Scholar]
- 19.Lesk M. SIGDOC ‘86: Proceedings of the 5th annual international conference on systems documentation. New York, NY, USA: ACM Press; 1986. Automatic sense disambiguation using machine readable dictionaries: how to tell a pine cone from an ice cream cone; pp. 24–26. [Google Scholar]
- 20.Luger S, Aitken S, Webber B. Automated terminological and structural analysis of human-mouse anatomical ontology mappings. BMC Bioinformat-ics. 2005;6(Suppl 3) [Google Scholar]
- 21.Morgan AA, Hirschman L, Colosimo M, Yeh AS, Colombe JB. Gene name identification and normalization using a model organism database. J Biomedical Informatics. 2004;37(6):396–410. doi: 10.1016/j.jbi.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Mork P, Pottinger R, Bernstein PA. Challenges in precisely aligning models of human anatomy using generic schema matching. MedInfo. 2004;11(Pt 1):401–405. [PubMed] [Google Scholar]
- 23.Oepen S, Netter K, Klein J. Linguistic Databases. CSLI Publications; 1998. TSNLP - Test suites for natural language processing. [Google Scholar]
- 24.Pedersen T, Mihalcea R. Advances in word sense disambiguation. Tutorial, Conf of ACL. 2005 [Google Scholar]
- 25.Porter M. An algorithm for suffix stripping. Program. 1980;14:130–137. [Google Scholar]
- 26.Sarkar IN, Cantor MN, Gelman R, Hartel F, Lussier YA. Linking biomedical language information and knowledge resources: GO and UMLS. Pac Symp Biocomput. 2003:439–450. doi: 10.1142/9789812776303_0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32(Database issue) doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shvaiko P, Euzenat J. A survey of schema-based matching approaches. Journal on Data Semantics. 2005;4 [Google Scholar]
- 29.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. AMIA Annu Symp Proc. 2003:753–757. [PMC free article] [PubMed] [Google Scholar]


