Abstract
Foldable polymers with alternating single strand deoxyribonucleic acid (ssDNA) and planar fluorescent organic chromophores can self-organize into folded nanostructures and hence are hybrid foldamers with biological sequences and synthetic properties. The biological sequence provides highly specific molecular recognition properties while the physical properties of synthetic chromophores offer sensitive fluorescence detection. In this paper, we describe that rational designed hybrid foldamers exhibit potential in the detection of polynucleotides. Under strictly controlled laboratory conditions, fluorescence measurements indicate that configuration change due to binding of polynucleotides with one or two mismatched bases can be readily distinguished. These results shed light on the design and construction of nanostructured foldamers with actuator and sensory properties, which may find important applications as biological probes.
Keywords: DNA synthesis, fluorescence, self-assembly, foldable polymer, foldamer, FRET, folding
Detection of DNA has attracted great interest recently, especially in the area of applying emerging nanotechnology (1-3) to biotechnology (4-6). The advantages of integrating nano-bio-technology are that biological macromolecules such as DNA offer specific interaction whereas nanoparticles provide means of amplification of such molecular recognition events. Typically, a DNA macromolecule binds to its complementary strand in a subtle different way than that of mismatched strands (7,8). When such subtle discrimination is amplified, a potential viable DNA detection mechanism is developed. One way to amplify such molecular recognition events is to use the size-dependent dielectric properties of gold nanoparticles (9,10). Another is utilization of fluorescent properties of either nanoparticles or organic chromophores and their sensitivity to distance and size (11). Herein, we demonstrate that folding and unfolding of a hybrid polymer containing alternating biological sequences and synthetic chromophores with orthogonal fluorescent emissions can be applied to DNA detection.
Our general strategy for developing foldable polymers is to alternate a hydrophobic sequence and a hydrophilic sequence in a controlled placement and orientation with a single molecular weight distribution. Highly fluorescent chromophores are embedded in the hydrophobic sequences; quantum interactions between these chromophores, such as π-orbital overlap and resonance energy transfer, are very sensitive to their distance separation, thus providing a means in gauging folding and unfolding events. Fluorescence resonance energy transfer (FRET) between chromophores in the processes of folding or unfolding is particularly interesting because it can be amplified by distance change, even single molecular events could be monitored with fluorescence (12-16). The typical requirements for the hydrophilic sequences in foldable polymers are flexibility and solubility. Both characteristics are important because flexibility provides hinge-like function, allowing the hydrophobic chromophores to fold while solubility is needed for the studies of individual folded nanostructures in solution. Yet, more importantly if the flexible sequences exhibit exquisite molecular recognition properties then molecular recognition-induced folding and unfolding events can trigger a sensitive change in the quantum interactions among chromophores (17-19). An ideal hydrophilic sequence that satisfies all these requirements is the macromolecule used for genetic codes, single strand DNA.
Experimental Procedures
Synthesis of DDP
DDP was synthesized by condensing 2, 6-dimethyl-4-(dicyanomethylene) pyran with slightly excess 4-N-n-butyl-N-[2-[2-[2-(2-hydroxyethoxy) ethoxy] ethoxy] ethyl] aminobenzaldehyde in the presence of piperidine catalyst and purified using chromatography. The purified DDP was monotritylated by reacting with three equivalent of dimethoxy trityl chloride at room temperature. The monotritylated product was purified on silica column and verified with 1H NMR and mass spectrometry. 1H NMR (CDCl3) δ (ppm): 7.49-7.38 (m, 8H, aromatic rings and double bond trans-linkage), 7.34 (dt, 4H, J1 = 2.7Hz, J2 = 9Hz, methoxylbenzene ring), 7.31-7.15 (m, 3H, benzene ring), 6.81 (dt, 4H, J1 = 2.7Hz, J2 = 9Hz, methoxylbenzene ring), 6.69 (d, 2H, J = 8.7Hz, benzene ring), 6.66 (d, 2H, J = 9.0Hz, benzene ring), 6.54 (d, 1H, J = 2.1Hz, pyran ring), 6.52 (d, 1H, J = 2.1Hz, pyran ring), 6.48 (d, 1H, J = 15.9Hz, double bond trans-linkage), 6.46 (d, 1H, J = 15.9Hz, double bond trans-linkage), 3.77 (s, 6H, CH3O), 3.56-3.50 (m, 30H, tetraethylene glycol chain), 3.44-3.31 (m, 4H, n-butyl chain), 3.22 (t, 2H, J = 5.1Hz, tetraethylene glycol chain), 1.69-1.49 (m, 4H, n-butyl chain), 1.48-1.42 (m, 4H, n-butyl chain), 0.97 (t, 3H, J = 7.5Hz, n-butyl chain), 0.96 (t, 3H, J = 7.5Hz, n-butyl chain). 13C NMR (CDC13) δ (ppm): 159.5, 158.4, 156.2, 149.8, 145.1, 138.2, 136.4, 130.2, 129.9, 128.3, 127.8, 126.7, 122.2, 122.1, 116.5, 113.1, 112.94, 112.88, 111.8, 111.7, 105.4, 86.1, 72.7, 71.0, 70.92, 70.84, 70.76, 70.5, 68.6, 63.3, 61.9, 56.1, 55.4, 51.4, 50.6, 29.4, 20.5, 14.3. MS (ESI): m/z 1145.6 [M]+. The characterized monotritylated product was treated with slight excess chloro-N,N-diisopropylaminocyanoethoxyphosphane to yield the desired phosphoramidite of DDP.
Synthesis of HSB
HSB was synthesized by reacting a Wittig reagent tetraethyl 1,4-xylylenediphosphonate (1.05 g, 2.78 mmol) with 3 equivalent of 4-[2-[2-[2-(2-hydroxyethoxy)ethoxy]ethoxy]ethoxy]-benzaldehyde. The purified HSB was monotritylated using procedure similar to that of DDP and the tritylated product was purified with silica column and extensively characterized with NMR. 1H NMR (CDCl3) δ (ppm): 7.46 (s, 4H, aromatic ring of styryl benzene), 7.49-7.38 (m, 6H, aromatic rings of styryl benzene and DMTr), 7.34 (dt, 4H, J1 = 8.7Hz, J2 = 2.3Hz, methoxylbenzene ring), 7.31-7.15 (m, 3H, benzene ring of DMTr), 7.06 (d, 1H, J = 16.2Hz, double bond trans-linkage), 7.05 (d, 1H, J = 16.2Hz, double bond trans-linkage), 6.96 (d, 1H, J = 16.2Hz, double bond trans-linkage), 6.95 (d, 1H, J = 16.2Hz, double bond trans-linkage), 6.94-6.84 (m, 4 H, aromatic ring of styryl benzene), 6.81 (dt, 4H, J1 = 8.7Hz, J2 = 2.3Hz, methoxylbenzene ring), 4.19-4.08 (m, 4H, tetraethylene glycol chain), 3.89-3.82 (m, 4H, tetraethylene glycol chain), 3.80-3.58 (m, 28H, CH3O and tetraethylene glycol chain), 3.22 (t, 2H, J = 5.1Hz, DMTrOCH2); 13C NMR (CDC13) δ (ppm): 158.57, 158.55, 158.43, 145.2, 136.74, 136.70, 136.4, 130.47, 130.40, 130.2, 128.3, 128.00, 127.96, 127.87, 127.79, 126.77, 126.67, 126.44, 126.38, 115.0, 113.2, 86.1, 72.7, 71.09, 71.04, 70.98, 70.95, 70.88, 70.81, 70.56, 69.95, 67.64, 63.4, 62.0, 55.4. The phosphoramidite was obtained by reacting the monotritylated product with ∼2 equivalent chloro-N,N-diisopropylaminocyanoethoxyphosphane and used immediately on the solid state synthesizer.
Synthesis and Purification of Hybrid Foldermers with Alternating Fluorescent Chromophores and Oligonucleotides DNA
Since the coupling reactions of the fluorescent emitters on the automated DNA synthesizer had been optimized and were compatible with phosphoamidite chemistry used in DNA synthesis, the insertion of the above two chromophores via tetraethylene glycol spacer phosphoramidites into the designed DNA sequences during the DNA chain growing was successfully completed on a Expedite™ 8900 Nucleic Acid Synthesis System.
Detailed protocol of solid phase synthesis of foldamers was reported in the previous report17 and here we simply summarize the key parameters. The DDP and HSB phosphoramidite reagents were prepared in dichloromethane because they have low solubility in the “standard” solvent of the synthesizer acentonitrile. The concentration of the DDP and HSB phosphoramidite was ∼100 mg/mL and their coupling reaction time on the solid phase columns were extended to about 30 min in order to achieve high coupling yields. Reaction parameters of other steps (oxidation and detritylation) on the solid phase columns are similar to what was used for DNA synthesis.
The Oligonucleotide Purification Cartridge (ABI Masterpiece™) was used to purify the obtained crude oligonucleotides with the terminal DMTr group on. With high coupling yield during foldamer chain assembly (>80-99% based on trityl monitor) and good overall yield, the OPC (Oligonucleotide Purification Cartridge) purified foldamers could be used for DNA detection after desalting. A typical procedure is as follows.
After solid-state synthesis, the column containing 0.2 μmol of the designed foldamer is removed from the DNA synthesizer. A syringe is attached to each end of the column, one syringe is containing 1 mL of fresh ammonium hydroxide (NH4OH, 29% w), and the other is empty. The ammonium hydroxide solution is injected through one end of the column and forced through the column by depressing the syringe, and simultaneously collected in the empty syringe attached on the opposite end of the column. After passing all ammonium hydroxide solution through, the solution collected in the opposite syringe is then forced back through the column. Again the solution is collected in the original syringe. This “extraction” process is repeated 3-4 times to ensure that the support in the column is fully saturated. The column/syringe assembly is allowed to rest at RT for 1 h, and the extraction process is repeated another 3-4 times. After allowing the column/syringe assembly to rest once more at RT for another 1 h, the solution is drawn completely into one syringe, which is then carefully removed from the column. The solution is then delivered into a 10-mL flask with a sealed septum and a stir bar. After stirring for 48 hours at RT, 20 μL of the solution is diluted with 3000 μL distilled water, and the optical density unit (ODU) value, which is equivalent to absorbance unit, of this solution is measured at 260 nm. Based on the ODU value, the original ammonium hydroxide solution is diluted so that the final ODU value is below 10. The foldamer purification is then carried out according to the protocol provided by ABI Masterpiece™ (2 mL diluted solution with ODU ≤ 10 for each OPC column). The OPC purified trityl-off foldamers are stored as a dry solid at -80 °C. According to the desalting protocol provided by ABI Masterpiece™, the trityl-off foldamers are dissolved in water (1 mL), and the resulting solution is passed through another activated OPC column, at last collected with 1 mL of 50% acetonitrile-water solution. The obtained acetonitrile-water solution is then concentrated to dryness in vacuum and the salt-free foldamer is obtained as a solid, which can be stored at -20 °C for further studies. This completes the crude separation of foldamers and they are further purified with HPLC as described below.
HPLC Purification of Oligo DNA and the Hybrid Foldamers
The oligo DNA 18-mers were purified using ion-exchange column (Zorbax Oligo; 5 μm). The mobile phase consists of two buffers and a linear salt gradient is used to elute out the DNA samples. The first buffer contains 20% acetonitrile and 80% water (v:v) with 0.01 M NaH2PO4 and 0.01 M Na2HPO4 to ensure pH = 7. The second buffer has 2M NaCl dissolved in the first buffer. The desired oligo DNA is eluted by increasing the salt gradient and the samples are >95% pure.
The hybrid foldamer is purified using a Genesis 300 C4 (4 μm) reverse phase column. A linear gradient is used with an increasing organic component to elute out the desired hybrid foldamer. The aqueous buffer contains 0.1 mM of triethyl ammonium acetate (TEAA) in filtered milli-Q water. The organic buffer contains 0.1 mM of triethyl ammonium acetate (TEAA) in HPLC grade acetonitrile. A typical run will start with high aqueous buffer composition and slowly ramp up the organic buffer to elute out the desired foldamer. The final foldamer is >95% pure.
Fluorescence Measurements
Fluorescence measurements were carried out on SPEX Fluorolog-3-21. A typical experiment uses 10 nM solution of the hybrid foldamer in a 1-mL quartz cell and the sample is excited at 365 nm. Sequences of DNA with the complementary structure or various mismatches are then added to the sample cell after initial reading of the fluorescence intensity. The fluorescence spectra of various samples with reduced fluorescence resonance energy transfer (FRET) efficiency are collected.
For kinetics experiments, the fluorescence intensity versus time curve is recorded and the folded concentration is calculated using the fluorescence intensity of the FRET peak. The data are then fitted into a first, second, or third order kinetics equation and the rate constants are obtained from the slope of the straight lines. All samples used in kinetics measurements are purified with HPLC and used immediately after dissolving the purified sample in autoclaved water.
Results and Discussions
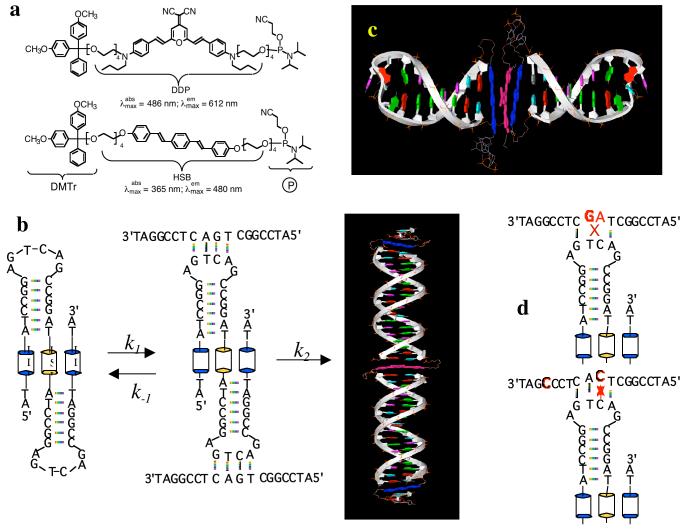
To construct foldable polymers with alternating hydrophobic and hydrophilic structures, we employed solid-phase synthesis (20,21), which requires that the fluorescent building blocks have an activated functional group for attachment to the growing end of the DNA polymer chain and a removable protecting group so that it can be removed to permit further chain extension (22). Two fluorescent chromophores are synthesized: one emits in the yellow with a λmax ∼ 612 nm and the other emits in the blue with an emission maximum at 480 nm in organic solvents. In addition to being orthogonal in fluorescent emissions, the absorption maximum of the yellow chromophore (486 nm) overlaps perfectly with the fluorescent emission of the blue chromophore. The yellow emitter is based on the 2,6-diaminostyryl 4-dicyanomethylene-4H-pyran or DDP structure (Figure 1a). Flexible tetra ethylene glycol (TEG) linkers (23) were attached to both amino groups of DDP, yielding two hydroxyl groups at either end of the chromophore. One hydroxyl group was protected with a removable blocker, dimethoxy trityl (DMTr) group and the other was activated with a phosphoramidite group. The blue emitter is 1,4-(bis-4, 4′-(2-(2-(2-(2-hydroxyl ethoxy) ethoxy) ethoxy) ethoxy) styryl) benzene (HSB) and it is similarly functionalized with a removable DMTr protection and an active phosphoramidite for efficient coupling to hydroxyl groups.
Figure 1.
(a). Chemical structures of the two fluorescent chromophores (HSB and DDP), modified with a removable DMTr protecting group and an activated phosphoramidite group. (b). Foldamer BYB sequence and its reaction with the complementary DNA sequence. (c). Model of folded molecular structure. (d). Mis-matched sequences do not form supramolecular assemblies.
Solid phase DNA synthesis is well developed (21). Synthesis of hybrid oligonucleotides containing stilbene units is also reported with a satisfactory yield (24,25). However, general incorporation of organic chromophores into DNA main chains is still not trivial (26, 27) and frequently requires special manipulation (28, 29). Rarely, more than two chromophores are incorporated into DNA main chains due to limited synthetic yields. Herein, the incorporation of two HSB and one DDP chromophores to DNA backbone is achieved in high yield by repeating the coupling reaction. The foldable polymers synthesized on solid supports are purified using HPLC (Genesis 300, C4 column) to yield > 95% pure samples. The foldable polymer for this study has a sequence of DNA-HSB-DNA- DDP-DNA- HSB-DNA, where HSB and DDP are the two fluorescent segments and DNA has a base sequence of 5′-ATC-CGG-AGT-CAG-CCG-GAT-3′ except the terminal DNA sequences which has been truncated after two AT bases (Figure 1b & c). The above foldamer is named as BYB because of an alternating blue-yellow-blue fluorescent sequence.
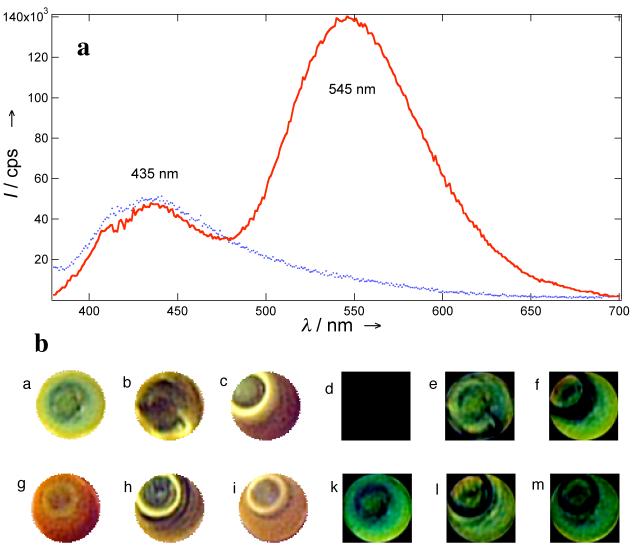
In aqueous solutions (pH ∼5), HSB and DDP in BYB fluoresce at 435 and 545 nm, respectively. The shifts in fluorescence emissions are attributed to the change of solvent and pH effects on chromophores. The purified BYB sequence forms the predominately folded structure, in which the two chromophores are in approximately juxtaposed positions. Since the emission bands of HSB overlap strongly with the absorption band of the DDP, fluorescence resonance energy transfer (FRET) will quench the blue emitter and yellow emission will dominate the fluorescence spectrum. Indeed, this behavior was observed (Figure 2a & 2b image g).
Figure 2.
(a). Florescence spectra show that folded BYB (10 nM) emit dominantly at 545 nm (line). Upon binding to the complementary strand, the fluorescence resonance energy transfer efficiency drops significantly between HSB and DDP chromophores and the unfolded BYB fluoresce at 435 nm (dots). (b). Comparison of fluorescence colors of foldamers binding to the perfect match (a), one-mismatch (OM1: b and OM2: c), two-mismatch (TM2: h and TM1: i), and blank (g). The perfect match generates a green spot (a) whereas pure foldamer yields an orange spot (g). The difference of the colors is presented between perfect match with perfect match (d), OM1 (e), OM2 (f), blank (k), TM1 (m), and TM2 (l).
Subsequently, BYB foldamer was exposed to various oligonucleotides, having either the complementary sequence or various mismatched base pairs to the DNA in BYB (Table 1). To evaluate the feasibility that foldamer can discriminate perfect match strand from those mismatched strands, 2 μL (∼9.2 pmole) of reaction solution was condensed on glass slides to dryness. The resulting fluorescent spots on glass have enough color differences to allow visual identification of the perfect match from those having one or two mismatched bases.
Table 1.
Various DNA sequences used to probe BYB foldamer responses
| Name | DNA Sequences |
|---|---|
| Complementary | 3′-TAG-GCC-TCA-GTC-GGC-CTA-5′ |
| OM1 | 3′-TAG-GCC-TCT-GTC-GGC-CTA-5′ |
| OM2 | 3′-TAC-GCC-TCA-GTC-GGC-CTA-5′ |
| TM1 | 3′-TAG-CCC-TCA-CTC-GGC-CTA-5′ |
| TM2 | 3′-TAG-GCC-TCG-ATC-GGC-CTA-5′ |
Two sequences with one-base mismatch were studied and the mismatched base was introduced either in the middle of (OM1) or toward the end of the oligonucleotide (OM2). Both sequences with one base mismatch react with BYB, yielding yellow or yellow green fluorescent spots (Figure 2b images b & c). The reaction between BYB and its complementary sequence yields green fluorescent spots (Figure 2b image a). The color difference between perfect match and the two one-mismatched sequences is distinguishably green (Figure 2b images e & f), whereas perfect match against perfect match in control experiments yields black background (Figure 2b image d).
Similarly, two sequences with two-base mismatch were also investigated; the first sequence has a mismatch in the middle and another towards the end of the sequence (TM1) while the second sequence has two positionally switched bases in the middle. Both TM1 and TM2 have considerable yellow fluorescent emissions (Figure 2b images i and h). Therefore, the difference in color between the perfect match and TM1 and TM2 are vividly green (image m & l). These results indicate that sequences with greater than one base mismatch (TM1, & TM2) can be readily distinguished.
Quantitative fluorescence changes due to unfolding of the BYB foldamer are listed in Table 2 for complementary target DNA along with the mismatched sequences. At 30 min of reaction time, the perfect match reduces the fluorescence intensity at 545 nm by more than 20% while the mismatched sequences reduce by approximately 1-7%. At 60 min, the reaction of the complementary sequence is about half way to completion while the mismatched sequences only finish 3-11%. These results are in agreement with fluorescent color observed in Figure 2.
Table 2.
Normalized fluorescence intensity of yellow fluorescence band at 545 nm at various reaction times. The complementary strand is very efficient at unfolding the BYB foldamer and hence produces the largest reduction of the yellow fluorescent band.
| Reaction Time (min) | Samples | ||||
|---|---|---|---|---|---|
| Complementary | OM1 | OM2 | TM1 | TM2 | |
| 30 | 0.79 | 0.93 | 0.99 | 0.97 | 0.99 |
| 60 | 0.55 | 0.89 | 0.96 | 0.92 | 0.97 |
| 90 | 0.41 | 0.83 | 0.95 | - | - |
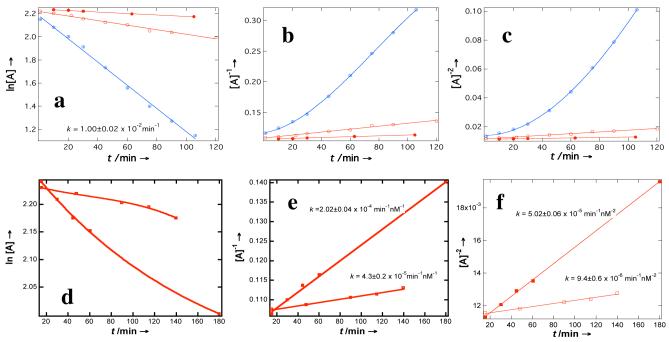
The origin of fluorescence color identification comes from differences in reactivity between the complementary strand and mismatched strands, which manifest in the kinetic studies. Figure 3 plots the kinetic data of the reaction between BYB complementary oligonucleotide along with sequences having mismatched bases. Although hybridization of complementary strands is not a unimolecular reaction, the rate law surprisingly fit better to first order reaction than those of second or third order reactions with a first-order rate constant of k(1) = 1.00±0.02 × 10-2 min-1. This behavior is unexpected, indicating that the reaction pathway must have undergone through a supramolecular intermediate species and the rate-determining step must be the unfolding of the supramolecular assembly (Figure 1b).
Figure 3.
(a). Kinetics measurements of the rate constant for the reaction of BYB with the complementary oligonucleotide (circles with plus) yield a first-order reaction, whereas one-mismatched sequences (OM1: open circles; OM2: solid circles) fit first-, second- (b), and third-order reactions (c). Two-mismatched sequences (TM1: open squares; TM2: solid squares), however, no longer fit to the first order reactions (d); rather they fit to the second- (e), and third-order reactions (f).
For one-mismatched sequences, kinetic data indicate the formation of a supramolecular complex is not the dominating reactions pathway, whereas a supramolecular self-assembly is no longer the major reaction pathway for two-mismatched sequences (Figure 3d) because fit to the first order is not a straight line. Second feature is that the reaction between complementary sequences is much faster than those of mismatched sequences. For OM1 and OM2 sequences, the rate constants for the first, second, and third order reactions are k(1) = 1.99±0.09 × 10-3 min-1, 6.3±0.3 × 10-4 min-1; k(2) = 2.40±0.01 × 10-4 min-1 nM-1, 6.9±0.3 × 10-5 min-1 nM-1; k(3) = 5.9±0.2 × 10-5 min-1 nM-2, 1.53±0.05 × 10-5 min-1 nM-2, respectively. For TM1 and TM2 sequences, the rate constants for the second and third order reactions are k(2) = 4.3±0.2 × 10-5 min-1 nM-1, 2.02±0.04 × 10-4 min-1 nM-1 and k(3) = 9.4±0.6 × 10-6 min-1 nM-2, 5.02±0.06 × 10-5 min-1 nM-2, respectively. All these rate constants are much smaller than that of the complementary strand and therefore the complementary polynucleotide yields more green to blue fluorescent spots than those of mismatched sequences.
In summary, we have demonstrated detection of single nucleotide polymorphorism (SNP) using hybrid foldamer at 1-10 nM concentration range. These results suggest foldamers with optimized structures should have potential application in DNA detection based on the sensitive switching of fluorescent colors. The simplicity of visual color change under UV-radiation makes foldamers attractive as DNA probes.
ACKNOWLEDGMENT
We acknowledge the supports of National Institute of General Medicine Science (NIGMS 065306) and Arnold and Mabel Beckman Foundation. ADQL is a Beckman Young Investigator (BYI).
ABBREVIATIONS
- DNA
deoxyribonucleic acid
- FRET
fluorescence resonance energy transfer
- DMTr
dimethoxy trityl
- OPG
oligonucleotide purification cartridge
- ODU
optical density unit, which is equivalent to absorbance unit (AU)
- RT
room temperature
- HPLC
High performance liquid chromatography
- TEAA
triethyl ammonium acetate
- TEG
tetra ethylene glycol
- DDP
2,6-diaminostyryl 4-dicyanomethylene-4H-pyran
- HSB
1,4-(bis- 4, 4′-(2-(2-(2-(2-hydroxyl ethoxy) ethoxy) ethoxy) ethoxy) styryl) benzene
- BYB
blue-yellow-blue [foldamer]
- OM1
one-mismatched [sequence] 1
- OM2
one-mismatched [sequence] 2
- TM1
two-mismatched [sequence] 1
- TM2
two-mismatched [sequence] 2
REFERENCES
- 1.Zhao XJ, Tapec-Dytioco R, Tan WH. Ultrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. J. Am. Chem. Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell DJ, Taylor JR, Nie SM. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J. Am. Chem. Soc. 2002;124:9606. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 4.Gaylord BS, Heeger AJ, Bazan GC. DNA detection using water-soluble conjugated polymers and peptide nucleic acid probes. Proc. Natl. Acad. Sci. 2002;99:10954–10957. doi: 10.1073/pnas.162375999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patolsky F, Lichtenstein A, Willner I. Electronic transduction of DNA sensing processes on surfaces: Amplification of DNA detection and analysis of single-base mismatches by tagged liposomes. J. Am. Chem Soc. 2001;123:5194–5205. doi: 10.1021/ja0036256. [DOI] [PubMed] [Google Scholar]
- 6.Dore K, Dubus S, Ho HA, Levesque I, Brunette M, Corbeil G, Boissinot M, Boivin G, Bergeron MG, Boudreau D, Leclerc M. Fluorescent polymeric transducer for the rapid, simple, and specific detection of nucleic acids at the zeptomole level. J. Am. Chem. Soc. 2004;126:4240–4244. doi: 10.1021/ja038900d. [DOI] [PubMed] [Google Scholar]
- 7.Ali MF, Kirby R, Goodey AP, Rodriguez MD, Ellington AD, Neikirk DP, McDevitt JT. DNA hybridization and discrimination of single-nucleotide mismatches using chip-based microbead arrays. Anal. Chem. 2003;75:4732–4739. doi: 10.1021/ac034106z. [DOI] [PubMed] [Google Scholar]
- 8.Gerion D, Chen FQ, Kannan B, Fu AH, Parak WJ, Chen DJ, Majumdar A, Alivisatos AP. Room-temperature single-nucleotide polymorphism and multiallele DNA detection using fluorescent nanocrystals and microarrays. Anal. Chem. 2003;75:4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- 9.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell GP, Mirkin CA, Letsinger RL. Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 1999;121:8122–8123. [Google Scholar]
- 11.Chakrabarti R, Klibanov AM. Nanocrystals modified with peptide nucleic acids (PNAs) for selective self-assembly and DNA detection. J. Am. Chem. Soc. 2003;125:12531–12540. doi: 10.1021/ja035399g. [DOI] [PubMed] [Google Scholar]
- 12.Tan X, Hu DH, Squire TC, Lu P. Probing nanosecond protein motions of calmodulin by single molecule fluorescence anisotropy. Biophys. J. 2004;86:475A–475A. [Google Scholar]
- 13.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc. Natl. Acad. Sci. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartley LE, Zhuang XW, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 15.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Luo GB, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun LY, Xie XS. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Wan W, Zhou H-Z, Niu S, Li ADQ. Alternating DNA and π-conjugated sequences. Thermophilic foldable polymers. J. Am. Chem. Soc. 2003;125:5248–5249. doi: 10.1021/ja0341900. [DOI] [PubMed] [Google Scholar]
- 18.Saghatelian A, Guckian KM, Thayer DA, Ghadiri MR. DNA detection and signal amplification via an engineered allosteric enzyme. J. Am. Chem. Soc. 2003;125:344–345. doi: 10.1021/ja027885u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince RB, Barnes SA, Moore JS. Foldamer-based molecular recognition. J. Am. Chem. Soc. 2000;122:2758–2762. [Google Scholar]
- 20.Wang W, Li LS, Helms G, Zhou HH, Li ADQ. To fold or to assemble? J. Am. Chem. Soc. 2003;125:1120–1121. doi: 10.1021/ja027186h. [DOI] [PubMed] [Google Scholar]
- 21.Gait MJ. Oligonucleotide Synthesis: A Practical Approach. IRL Press; Washington DC: 1984. [Google Scholar]
- 22.Hayakawa Y, Kawai R, Hirata A, Sugimoto J, Kataoka M, Sakakura A, Hirose M, Noyori R. Acid/azole complexes as highly effective promoters in the synthesis of DNA and RNA oligomers via the phosphoramidite method. J. Am. Chem. Soc. 2001;123:8165–8176. doi: 10.1021/ja010078v. [DOI] [PubMed] [Google Scholar]
- 23.Amabilino DB, Ashton PR, Brown CL, Cordova E, Godinez LA, Goodnow TT, Kaifer AE, Newton SP, Pietraszkiewicz M, Philp D, Raymo FM, Reder AS, Rutland MT, Slawin AMZ, Spencer N, Stoddart JF, Williams DJ. Molecular meccano. 2. Self-assembly of [ETA]catenanes. J. Am. Chem. Soc. 1995;117:1271–1293. [Google Scholar]
- 24.Letsinger RL, Wu TF. Use of a stilbenedicarboxamide bridge in stabilizing, monitoring, and photochemically altering folded conformations of oligonucleotides. J. Am. Chem. Soc. 1995;117:7323–7328. [Google Scholar]
- 25.Lewis FD, Wu TF, Burch EL, Bassani DM, Yang JS, Schneider S, Jager W, Letsinger RL. Hybrid oligonucleotides containing stilbene units - excimer fluorescence and photodimerization. J. Am. Chem. Soc. 1995;117:8785–8792. [Google Scholar]
- 26.Ren RXF, Chaudhuri NC, Paris PL, Rumney S, Kool ET. Naphthalene, phenanthrene, and pyrene as DNA base analogues: Synthesis, structure, and fluorescence in DNA. J. Am. Chem. Soc. 1996;118:7671–7678. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berlin K, Jain RK, Simon MD, Richert CA. A porphyrin embedded in DNA. J. Org. Chem. 1998;63:1527–1535. [Google Scholar]
- 28.Bevers S, O’Dea TP, McLaughlin LW. Perylene- and naphthalene-based linkers for duplex and triplex stabilization. J. Am. Chem. Soc. 1998;120:11004–5. [Google Scholar]
- 29.Rahe N, Rinn C, Carell T. Development of donor-acceptor modified DNA hairpins for the investigation of charge hopping kinetics in DNA. Chem. Comm. 2003;(17):2120–2121. [PubMed] [Google Scholar]