Abstract
A growing number of modules including FYVE domains target key signaling proteins to membranes through specific recognition of lipid headgroups and hydrophobic insertion into bilayers. Despite the critical role of membrane insertion in the function of these modules, the structural mechanism of membrane docking and penetration remains unclear. In particular, the three dimensional orientation of the inserted proteins with respect to the membrane surface is difficult to define quantitatively. Here, we determined the geometry of the micelle penetration of early endosome antigen 1 (EEA1) FYVE domain by obtaining NMR-derived restraints that correlate with the distances between protein backbone amides and spin labeled probes. The 5- and 14-doxyl-phosphatidylcholine spin labels were incorporated into dodecylphosphocholine (DPC) micelles, and the reduction of amide signal intensities of the FYVE domain due to paramagnetic relaxation enhancement was measured. The vector of the FYVE domain insertion was estimated relative to the molecular axis by minimizing the paramagnetic restraints obtained in phosphatidylinositol 3-phosphate (PI3P)-enriched micelles containing only DPC or mixed with phosphatidylserine (PS). Additional distance restraints were obtained using a novel spin label mimetic of PI(3)P that contains a nitroxyl radical near the threitol group of the lipid. Conformational changes indicative of elongation of the membrane insertion loop (MIL) were detected upon micelle interaction, in which the hydrophobic residues of the loop tend to move deeper into the non-polar core of micelles. The micelle insertion mechanism of the FYVE domain defined in this study is consistent with mutagenesis data and chemical shift perturbations and demonstrates the advantage of using the spin label NMR approach for investigating the binding geometry by peripheral membrane proteins.
A diverse array of signaling and membrane trafficking proteins are transiently recruited to intracellular membranes by their phosphoinositide (PI)-binding domains (reviewed in (1-3). In addition to specifically recognizing the PI headgroups, these modules often insert into the interfacial and hydrocarbon layers of the membrane. They comprise a substantial fraction of the so-called peripheral proteins, which includes the ENTH (4, 5), FYVE (6-8) and PX domains (9, 10), as well as the FERM-homologous domain of Talin (11, 12) and the vinculin tail (13, 14). The majority of these proteins appear to insert relatively deeply into the bilayer interacting with both the polar headgroups and the hydrocarbon core. Multivalent anchoring resulting from electrostatic and polar interactions with multiple headgroups and insertion of set of aliphatic and aromatic residues provides the strength and selectivity that are necessary for the proper protein function and localization, as has been demonstrated for the EEA1 FYVE domain (15). Thus PI-recognizing domains bind membrane-embedded PIs several orders of magnitude more tightly than soluble lipids or isolated inositol headgroups (16-18). Mutations of the membrane-inserting residues of the EEA1, Hrs and Vps27p FYVE domains (7, 19), Epsin ENTH domain (4, 5) and p40phox PX domain (10) abolish or significantly decrease the membrane association and lead to the disruption of the normal biological activities of these proteins.
Although it is becoming evident that membrane penetration is fundamentally important for the function of many proteins, their orientations in membranes and insertion interfaces remain challenging to characterize. The most common methods of studying membrane properties of peripheral proteins include electron paramagnetic resonance (EPR) (20-22), tryptophan fluorescence (21-24), monolayer penetration (5, 7, 10, 25), computational modeling (8), solid state NMR (26) and solution NMR spectroscopy utilizing chemical shift (6, 27), nuclear Overhauser effect (NOE) (28) and spin label (29-32) data. While providing important insights, these approaches suffer several limitations. The EPR method requires design and purification of a large number of cysteine mutants and subsequent attachment of spin-labeled paramagnetic probes to the mutant proteins. NMR resonance perturbation analysis offers qualitative mapping of the affected residues, but does not differentiate between changes due to direct binding and indirect effects. Spin diffusion along the lipid side chains results in overestimation of the interproton distances by nuclear Overhauser effect spectroscopy (NOESY). Fluorescence spectroscopy and monolayer penetration provide qualitative analysis and often require covalent attachment of fluorescent groups to various positions of the protein or mutations of residues. The inevitable effects of these modifications, which can alter the protein's structure, dynamics and interactions, indicate a need to develop new methods that provide accurate measurements of membrane insertion by proteins in their native, unmodified states.
Here we develop a theoretical basis and a general NMR-based approach for determining the geometry of protein insertion into membrane-mimetic micelle systems using spin label probes.
EXPERIMENTAL PROCEDURES
Experimental Methods
Protein Expression and Purification
The FYVE domain of human EEA1 (residues 1325-1410) was cloned into a pGEX-KG vector (Amersham) and expressed in E. coli BL21 (DE3) pLysS in minimal media supplemented with zinc sulfate and 15NH4Cl (Cambridge Isotope). Bacteria were harvested by centrifugation after induction with IPTG (0.5 mM), lysed by sonication or by French press. The 15N-uniformly labeled glutathione S-transferase (GST)-fusion protein was purified on a glutathione sepharose 4B column (Amersham) as previously described (6, 19). The GST tag was cleaved with thrombin (Sigma). The protein was concentrated in Millipore concentrators (Millipore) into 20 mM d11-Tris, pH 6.8 in the presence of 200 mM KCl, 20 mM perdeuterated dithiothreitol, 50 μM 4-amidinophenylmethane sulfonyl fluoride, 1 mM NaN3, and 7% 2H2O. The identity and purity of the protein was analyzed by FPLC, SDS-PAGE and 1H NMR.
Synthesis of a spin-labeled derivative of PI(3)P, 1{(2R,3R)-4-[(2-(3-carboxy-PROXYL)amino)ethoxyphosphoryloxy]-2,3-di-O-palmitoylbutoxyphosphoryloxy}-3-myo-phosphate [PROXYL-Pea-PI(3)P]
A solution of PROXYL- N-hydroxysuccinimidyl-3-carboxylate ester (NHS) (0.4 mg, 1.26 μmol) in 250 μL of DMF was added to a solution of 1-[(2R,3R)-4-(2-aminoethoxyphosphoryloxy)-2,3-di-O-palmitoylbutoxyphosphoryloxy]-3-myo-phosphate (1.2 mg, 1.05 μmol) in 0.5 M TEAB (250 μL, pH 7.5). The mixture was stirred at room temperature for 18 h, and solvents were then removed in vacuo. The residue was washed with acetone (5×1.5 mL) and purified on DEAE-cellulose column with a step gradient (0 to 2 M) of triethylammonium bicarbonate (TEAB). The desired fraction was lyophilized yielding the proxylated product as the triethylammonium salt (1.0 mg, 59%). MALDI-MS m/z = 971.9 [M – C15H35CO, free acid]
NMR spectroscopy and titration of the paramagnetic spin-label probes
NMR spectra were recorded at 25°C on Varian INOVA 500 and 600 MHz spectrometers. The 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectra of 0.2-0.3 mM uniformly 15N-labeled FYVE domain containing 0-1.5 mM dibutanoyl (C4)- or dipalmitoyl (C16)-PI(3)P (Echelon Biosciences Inc.), 250 mM (4.5 mM micellar) d38-dodecyl phosphocholine (DPC) (Cambridge Isotopes), and 0-10% w/w 1,2-dicaproyl-sn-glycero-3-[phospho-L-serine] (PS) (Avanti) were collected using 1024 t1 increments of 2048 data points, 96 number of increments and spectral widths of 7500 and 1367 Hz in the 1H and 15N dimensions, respectively, with a relaxation delay time of 1.5 and 2 s. The paramagnetic spin-label probes, 5- and 14-doxyl derivatives of 1-palmitoyl-2-stearoyl-sn-glycero-3-phosphocholines (Avanti) and PROXYL-Pea-PI(3)P (up to 5.8 mM) were added stepwise as powder or as methanol-d4 solutions. The ratio of spin-label to micelle was 0.7 to 1.3 and therefore the interaction between radicals can be neglected. The 1H-15N HSQC spectra were collected before and after addition of the spin labeled probes, and intensities of the backbone amide resonances of the protein were monitored and compared. Significant levels of cross peak intensity reduction were judged to be greater than the average plus one standard deviation. The average level of intensity reduction was validated by comparing it to the average line broadening in similar HSQC experiments using the spin labels when PI(3)P was absent. The spin labels did not alter the FYVE domain structure based on the lack of chemical shift perturbations in the NMR spectra of the protein.
Intramolecular NOEs were obtained from 15N-edited NOESY-HSQC (33) spectra (τm=135 ms) collected on the 1 mM FYVE domain sample in the presence and the absence of 5 mM C4-PI(3)P and 250 mM d38-DPC.
Theoretical Methods
Definition of the molecular axis
The long molecular axis of the FYVE domain M⃗ was obtained by minimizing the sum of the distances between atoms and the axis.
First, the origin of the coordinate system was moved to the non-weighted center of the protein
Where xi, yi, zi are the coordinates of each atom i of the PI(3)P-bound FYVE domain (PDB 1HYI).
The molecular axis is then given by a vector r = [xa,yaza] in a form
The distance between atom i and the axis is defined as di = |ri| cos(γi), where ri is a distance from the center of micelle and γ is the angle between the two vectors ra and ri calculated as
Vector ra is obtained by minimizing the sum of di′s.
Direction of the insertion
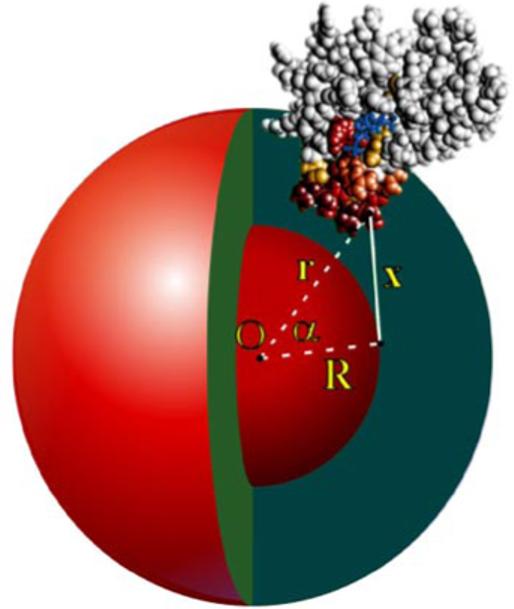
The doxyl radical is assumed to be randomly distributed inside the spherical micelle over an inner sphere of radius R (Figure 1). Significant attenuation of the signal intensities of each backbone amide nitrogen atom i due to paramagnetic line broadening in the presence of spin-label probes is denoted by a set of experimental observables . For any backbone nitrogen atom i located at a distance ri from the center of micelle O (xo, yo, zo) the attenuation effect is inversely proportional to the sixth power of the distance (x) between the atom and the radical located on the inner sphere of radius R (34). The effect is then integrated over the surface of the inner sphere.
FIGURE 1.

Model of the FYVE domain insertion into a spherical micelle. The FYVE domain is shown in a space-filling form with the residues perturbed by the 5-doxyl spin label probe colored in gradations of red. The inositol group of the bound PI(3)P is shown as a stick model and colored blue. For any FYVE domain backbone atom i located at a distance ri from the center of micelle O, the attenuation effect is proportional to x-6, where x is a distance between the atom and the spin label located at a distance R from the micelle center.
Integration details
The distance from atom i to the radical on the surface defined by R can be expressed in terms of r, R and the angle α (the angle between the vectors r and R) as
The distance dependence integrated over the inner sphere is then:
Where the numerator has 2πRsin(α), i.e. a circumference of a circle produced by rotating vector x around r, and the denominator is x6.
Substituting d(α)sin(α) by d(cos(α))=dy we obtain:
It further follows that value F(r,R) proportional to the predicted effect at atom i, can be expressed as a function of ri and R:
It was shown that spin labels are broadly distributed in z-direction of membranes (35, 36). In our model we approximated such distribution with a normalized Gaussian function of R, so that the ith atomic contribution of one spherical slice of radius Rj is
where R0 is an average radial position of the spin label in the micelle, and σ is the Gaussian width parameter. Both parameters are systematically varied in the optimization runs, 0 < R0 < Rmicelle; σ = 1, 2 or 3 Å
Value F(ri,Rj) is then summed, or numerically integrated, over 1 < j < M Gaussian-weighted spherical slices.
To avoid singularity the summation is carried out for |ri – Rj| ≥ d, where d = 1, 2 or 3 Å is one of the optimization parameters.
The protein's position relative to the micelle is then optimized to maximize correlation between the set of predicted effects {Fi } and the set of experimental values . Two sets of initial protein orientations were used to avoid local minima.
The insertion angle is defined as the angle between the vector M⃗ (protein's long axis) and the vector O⃗P (center of micelle to the non-weighted center of the protein). The insertion angles, calculated with various combinations of the optimized parameters, were ranked by the correlation values and a weighted average is obtained for the top 40 structures (listed with weighted standard deviations).
RESULTS
Paramagnetic spin-label probes define the micelle insertion interface of the FYVE domain
To determine the precise micelle orientation of the FYVE domain, NMR experiments with the lipid spin label probes were carried out. The paramagnetic 5- and 14-doxyl-phosphatidylcholine (PC) carry a nitroxyl radical at positions 5 and 14 of the stearoyl side chain. They readily incorporate into DPC micelles and induce the paramagnetic relaxation enhancements of nearby nuclei. Thus the 5-doxyl probe causes the line broadening of NMR resonances of nuclei located in close proximity to the micelle surface, i.e. 1-3 bonds away from the polar head groups. In contrast, 14-doxyl PC broadens the resonances in the micelle center, that is 10-12 bonds away from the polar head groups, as was previously measured by NMR and EPR studies (29-31). Consequently, protein residues buried in the micelle hydrophobic core and residues located at the level of micelle phosphates are expected to experience selective line broadening upon addition of 14- or 5-doxyl probes, respectively.
When the 5-doxyl probe was added to the 15N-labeled FYVE domain that was pre-bound to C16-PI(3)P and DPC micelles, a substantial reduction of the Asp1351, Asn1352, Val1354, Phe1364, Ser1365, Val1366, Thr1367, Val1368, Arg1369, Arg1370, Cys1381, Ala1382 and Arg1399 amide signal intensities was observed (Table 1). These residues are located in and around the MIL and form a continuous surface, which was used in this study to estimate the volume and angle of the FYVE domain insertion into the micelle interior. In contrast, addition of the 14-doxyl probe caused line broadening of only Val1366, Thr1367 and Val1368 amide resonances (Table 1). These three residues are located at the tip of the MIL and were previously found to be buried deeply in the micelle core (15). A variety of micelle systems including diheptanoyl phosphatidylcholine, cyclohexylbutylphosphocholine and mixed micelles were tested with similar results (15). DPC was used here since it mimics PC, the predominant phospholipid in mammalian cell membranes and forms small micelles suitable for NMR studies. It is also well characterized and widely used for solution NMR studies being readily available in a perdeuterated form. The size limit in solution NMR precludes the use of actual membranes or liposomes. Although in contrast to planar membranes micelles possess a curved surface, they are commonly used for studying structures and functions of membrane proteins, and for modeling of the lipid environments (27, 28, 37).
Table 1.
Experimental restraints.
| Residuea | 5-doxyl C16- PI(3)P (%) |
5-doxyl C4-PI(3)P (%) |
14-doxyl C16- PI(3)P (%) |
14-doxyl C4-PI(3)P (%) |
5-doxyl C16-PI(3)P C6-PSb(%) |
|---|---|---|---|---|---|
| LYS1347 | 0 | 0 | 6.1 | 0 | 7.4 |
| TRP1348 | 31.7 | 20.1 | 1.7 | 0 | 7.7 |
| ALA1349 | 37.1 | 14.7 | 9.7 | 0 | 12.7 |
| GLU1350 | 2.0 | 9.5 | 21.1 | 21.4 | 0 |
| ASP1351 | 39.4 | 2.6 | 11.9 | 19.4 | 12.4 |
| ASN1352 | 46.1 | 39.3 | 14.7 | 2.3 | 23.1 |
| GLU1353 | 25.7 | 23.2 | 17.4 | 25.3 | 22.8 |
| VAL1354 | 50.9 | 9.2 | 10.1 | 5.1 | 13.6 |
| GLN1355 | 33.3 | 12.5 | 7.7 | 8.4 | 0 |
| ASN1356 | 26.9 | 23.2 | 5.9 | 9.1 | 10.6 |
| CYS1357 | 18.7 | 28.2 | 1.7 | 5.6 | 3.5 |
| MET1358 | 20.6 | 9.8 | 13.3 | 19.1 | 20.8 |
| ALA1359 | 12.6 | 9.2 | 4.2 | 7.5 | 9.5 |
| CYS1360 | 4.3 | 20.1 | 9.2 | 4.7 | 9.5 |
| GLY1361 | 5.3 | 16.8 | 8.9 | 9.0 | 7.9 |
| LYS1362 | 30.4 | 24.3 | 15.4 | 1.3 | 7.8 |
| GLY1363 | 36.1 | 30.1 | 12.2 | 17.0 | 3.9 |
| PHE1364 | 41.3 | 29.4 | 11.0 | 24.9 | 15.7 |
| SER1365 | 39.6 | 44.6 | 18.0 | 23.2 | 31.3 |
| VAL1366 | 72.4 | 76.7 | 57.3 | 49.0 | 49.2 |
| THR1367 | 75.4 | 68.1 | 54.1 | 55.5 | 43.6 |
| VAL1368 | 75.9 | 52.8 | 40.3 | 47.1 | 47.3 |
| ARG1369 | 51.9 | 43.2 | 26.9 | 20.4 | 35.8 |
| ARG1370 | 43.8 | 36.7 | 6.4 | 3.1 | 45.0 |
| HIS1371 | 12.9 | 43.0 | 8.9 | 0 | 31.4 |
| HIS1372 | 0 | 10.8 | 26.1 | 14.3 | 18.2 |
| CYS1373 | 0 | 0 | 8.3 | 0 | 30 |
| ARG1374 | 2.0 | 29.2 | 6.9 | 24.8 | 3.5 |
| GLN1375 | 17.3 | 18.6 | 9.6 | 5.7 | 20.7 |
| CYS1376 | 38.5 | 12.0 | 11.7 | 21.9 | 18.7 |
| GLY1377 | 37.1 | 8.3 | 15.6 | 24.0 | 25.5 |
| ASN1378 | 22.9 | 18.3 | 2.3 | 0 | 10.7 |
| ILE1379 | 12.9 | 20.9 | 27.6 | 20.8 | 21.6 |
| PHE1380 | 25.4 | 8.4 | 7.8 | 20.6 | 17.9 |
| CYS1381 | 48.8 | 21.6 | 2.8 | 7.1 | 24.6 |
| ALA1382 | 38.9 | 44.5 | 13.5 | 24.8 | 20.1 |
| GLU1383 | 37.1 | 35.0 | 12.9 | 12.3 | 16.8 |
| CYS1384 | 20.4 | 22.2 | 15.2 | 17.6 | 25.1 |
| SER1385 | 31.1 | 0.4 | 18.7 | 5.3 | 22.0 |
| ALA1386 | 9.4 | 30.4 | 6.5 | 11.0 | 7.1 |
| LYS1387 | 26.3 | 29.3 | 12.7 | 21.4 | 23.8 |
| ASN1388 | 32.7 | 24.4 | 7.4 | 9.2 | 0.3 |
| ALA1389 | 32.7 | 15.4 | 15.2 | 7.3 | 26.6 |
| LEU1390 | 27.9 | 22.7 | 6.9 | 2.7 | 6.0 |
| THR1391 | 17.0 | 19.4 | 2.7 | 11.0 | 12.5 |
| SER1393 | 18.9 | 13.8 | 0 | 12.4 | 12.6 |
| SER1394 | 30.6 | 9.6 | 5.9 | 13.5 | 11.3 |
| LYS1396 | 28.4 | 19.7 | 1.7 | 1.8 | 5.1 |
| VAL1398 | 26.2 | 8.6 | 13.8 | 6.3 | 3.5 |
| ARG1399 | 38.8 | 17.9 | 21.9 | 12.7 | 24.6 |
| VAL1400 | 30.6 | 33.1 | 11.6 | 3.1 | 22.1 |
| CYS1401 | 24.1 | 24.2 | 10 | 12.5 | 24.3 |
| ASP1402 | 30.4 | 8.9 | 16.6 | 3.5 | 5.0 |
| ALA1403 | 27.9 | 16.7 | 12.2 | 21.6 | 9.7 |
| CYS1404 | 32.3 | 28.2 | 9.9 | 14.8 | 24.6 |
| PHE1405 | 27.1 | 23.2 | 27.0 | 15.0 | 6.6 |
| ASN1406 | 15.8 | 26.9 | 19.1 | 2.8 | 9.4 |
| ASP1407 | 30.3 | 27.7 | 10.7 | 5.4 | 1.5 |
| LEU1408 | 24.7 | 6.3 | 12.0 | 3.3 | 10.3 |
| GLN1409 | 18.5 | 20.3 | 4.8 | 3.6 | 2.2 |
| GLY1410 | 17.6 | 23.4 | 4.4 | 13.5 | 3.0 |
Intensity reduction of the amide resonances of the FYVE domain pre-bound to C16- or C4-PI(3)P and DPC induced by the addition of indicated spin label probes, 5-doxyl or 14-doxyl PC as % of the initial signal intensity.
DPC micelles contain 10%, w/w of C6-PS.
To quantitatively define insertion of the FYVE domain, the intensity reduction due to paramagnetic line broadening was used in a Raphson-Newton minimization approach. The angles of insertion into micelles were estimated by optimizing an average position of the 5-doxyl radical relative to the PI3P/DPC-bound FYVE domain to satisfy the experimental paramagnetic restraints (see Theoretical Methods for more details). The vector of FYVE domain insertion into PI(3)P-containing DPC micelles (O⃗P) diverges from the molecular axis of the FYVE domain (M⃗) by an angle of 48+/-14°. This angle changed to 67°+/-10° when dicaproyl PS (10% of DPC) was added to the micelles, revealing a critical role of the non-specific electrostatic interactions between the solvent-exposed basic patches of the FYVE domain and the acidic PS headgroups (Figure 2 and Table 1). These results are consistent with the chemical shift (6) and mutagenesis data (19) and yield a model of the inserted state of the FYVE domain in which the hydrophobic protuberance and the surrounding exposed polar residues maximize complementary contacts with the interior and interfacial zones of the micelle, respectively.
FIGURE 2.
The insertion interface of the FYVE domain. The three orthogonal views of the FYVE domain bound to PI(3)P- and PS- enriched DPC micelles are shown. Residues that exhibit significant signal intensity reduction upon addition of 5-doxyl spin label probe are colored red, orange and yellow for large, medium and small changes, respectively. The micelle surface is depicted as a green mesh and indicates the position of the phosphate group of DPC.
While changing the distance dependence of the spin label restraints from 1/r6 to 1/r2 generally resulted in similar angles of insertion, the 1/r6 model appears to describe the experimental data sets significantly better, i.e. the correlation coefficients were consistently (by 0.03-0.09) higher than for the 1/r2 model.
Novel PROXYL-Pea- derivative of PI(3)P
To obtain additional restraints and to determine the position of glycerol moiety of the ligand, a novel two-headed hybrid phosphoinoside-phosphatidylethanolamine spin label mimetic of PI(3)P was synthesized (Figure 3). This Pea- PI(3)P analog carries a proxyl moiety attached to the reactive amine of the phosphatidylethanolamine head group at the C1 position of the lipid's threitol backbone. Addition of PROXYL-Pea-PI(3)P to the FYVE domain, which was pre-bound to C16-PI(3)P and PS-containing DPC micelles, resulted in line broadening of several amide resonances, most significantly those of the Val1366-Cys1373 sequence and Arg1399-Val1400 (Figure 3b,c). Mapping these residues onto the surface of the FYVE domain revealed that the proxyl group is on average positioned near the inositol ring and the MIL. Moreover, line broadening was observed for the residues located predominantly on one side of the bound PI(3)P, indicating that the radical is restrained by the bound inositol ring and also by hydrophobic contacts of the ligand's acyl chains with the MIL. Consequently, this novel spin-label derivative of PI(3)P offers an effective way for characterizing the interactions between the lipid ligand and the micelle inserted protein. Furthermore, it complements another spin labeled PI3P derivative with a proxyl radical attached to the acyl chain that was previously used to map interactions with the FYVE domain (15).
FIGURE 3.

PROXYL-Pea- derivative of PI(3)P. a, Synthesis of the PROXYL-Pea- PI(3)P is shown as described in the methods. b, The histogram displays loss of amide signal intensities in the HSQC spectra caused by the addition of PROXYL-Pea- PI(3)P to the 15N-labeled FYVE domain in the presence of C16-PI(3)P and d38-DPC micelles. The colored bars indicate significant changes, being greater than the average plus one standard deviation. c, Residues that exhibit significant intensity reduction in (b) are labeled and colored in red, orange and yellow on the FYVE domain surface. The proxyl lipid is shown as ball-and-stick model with C, O, N and H atoms colored green, red, blue and light gray, respectively.
Conformational change in the MIL
The structural changes induced in the C4-PI(3)P-bound FYVE domain due to the insertion into DPC micelles were investigated by NMR. The experimental set of restraints obtained using spin labels and the predicted restraints derived from the theoretical model are presented as a bar graph in Figure 4. Comparison of the two data sets reveals that the theoretical model of the FYVE domain insertion underestimates line broadening of Val1354, Val1366, Thr1367, Val1368 and Cys1381 residues, while overestimating the effect on Ser1365, Phe1364 and Arg1370. The most significant deviations are observed for the Val1368 and Ser1365 residues. Both residues are located in the MIL, on the opposite sides of the loop (Figure 5b). Based on these deviations we predict that, as a result of insertion, the predominantly hydrophobic residues including Val1354, Val1366, Val1367 and Val1368 would move deeper into the hydrophobic core of micelles, while hydrophilic Ser1365 and Arg1370 would shift toward the aqueous interface, thus causing the MIL to become elongated (Figure 5b).
FIGURE 4.

Comparison of the spin label experimental data with those predicted by the derived model. The histogram displays the significant amide signal intensity reductions observed in HSQC spectra (light gray) and a theoretical model (dark gray) obtained from the titration of 5-doxyl PC (5 mM) into the FYVE domain (0.2 mM) bound to C4-PI(3)P (1 mM) and micelles formed by DPC (250 mM).
FIGURE 5.
Conformational changes in the MIL of the FYVE domain upon insertion into micelles. a, The corresponding stripes derived from 3D 15N-edited NOESY spectra of 1 mM FYVE domain recorded in the absence and presence of 5 mM PI(3)P and 250 mM d38-DPC. Upon insertion the intensities of a number of interresidue NOEs within the MIL are perturbed, which are highlighted by green and red boxes in (a) and indicated as green and red lines in (b) for the weakened and strengthened NOEs, respectively. b, The MIL region of the FYVE domain bound to PI(3)P (only 1-phosphate group is shown for clarity) is depicted as a gray ribbon. The MIL residues are labeled and shown as ball-and-stick models. Residues that move towards the hydrophobic core of micelles or the aqueous interface are colored orange and green, respectively.
To test whether such conformational changes occur upon insertion of the FYVE domain into micelles, the patterns of NOEs within the MIL were compared. Figure 5a displays corresponding strips derived from 15N-edited NOESY spectra, which were collected in the absence and presence of PI(3)P and DPC micelles. Upon addition of the ligands, several NOE crosspeaks became weaker or disappeared and, conversely, some NOEs appeared or became stronger. The perturbed NOEs are indicated in Figure 5a by green (decreased intensities) and red (increased intensities) boxes and by green and red lines in Figure 5b. In particular, the HβSer1365-HNVal1366, HNSer1365-HNThr1367 and HNSer1365-HNVal1368 NOE intensities were significantly decreased, whereas the HεPhe1364-HNSer1365 and HβVal1368-HNArg1369 interproton contacts became stronger (Figure 5). Thus, the changes in NOE patterns for the MIL residues upon binding to PI(3)P-containing DPC micelles support the lengthwise conformational extension, in which the Val1366-Val1368 sequence moves closer to the core of micelles, while the polar and charged residues are pulled in the opposite direction, toward the polar interface.
DISCUSSION
Paramagnetic relaxation enhancement of NMR signals by spin label compounds has been used to study peptide orientations in micelles for many years (ref. (29)). It was first applied for short peptides of an average size of 35 residues (29-31, 38-41), and has only recently been extended onto larger biological molecules (15). Yet the geometric interpretation of the line broadening data and elucidation of the protein's position in micelles remain limited and qualitative.
In this study we develop a quantitative NMR-based approach for determining the geometry of protein insertion into membrane mimetic micelle systems. At the core of this approach is the utilization of lipids with spin label probes attached at various depths of the acyl chain. Such spin label amphiphiles incorporate into micelles and attenuate resonance signals of protein nuclei in a distance-dependent fashion. The protein backbone resonances are monitored by recording 1H,15N HSQC spectra before and after addition of the spin-label probes, and the signal intensities are compared. The intensity attenuation due to paramagnetic line broadening is related to the distance between the spin label and each affected protein backbone atom. A set of such paramagnetic restraints is used in a Raphson-Newton minimization. The attenuation effect is inversely proportional to the sixth power of the distance between the atom and the spin label. The projected combined effect of the spin labels, broadly distributed in the micelle, on each backbone nucleus is obtained via integration over a sphere corresponding to the given depth of the label insertion and over a radial distribution of the labels. Correlation of the observed and theoretical values allows us to quantitatively describe various critical geometrical parameters including the vector of the protein insertion (Figure 1). This straightforward approach has a number of advantages over the previously reported methods. It offers a rapid mapping of the membrane binding regions of macromolecules providing many complementary distance restraints, using only HSQC spectra, 200 micromolar 15N labeled protein (less with a cryogenic probe), under a mg of a spin label, and does not require mutations or other modifications of the proteins. Furthermore, this approach provides a nearly complete set of paramagnetic restrains for large complexes, such as the PI3P/DPC-bound FYVE domain, in which severe line broadening precludes obtaining the restraints by other commonly used methods including analysis of R1 relaxation rates.
The FYVE domain has been chosen here as a model because this stable protein specifically binds a single ligand while inserting a single loop into the membranes. This small cysteine-rich module targets PI(3)P-containing endosomes (42-44) and is distinguished from other RING zinc fingers by two unique structural elements, the basic PI binding pocket and the adjacent MIL. Three different membrane orientations have been proposed for the Vps27p (45), Hrs (46) and EEA1 FYVE (6, 17) domains based on crystal and solution structures of the proteins. In two models, described for the Vps27p and EEA1 proteins, the hydrophobic loop inserts such that the primary (long) molecular axis of the FYVE domain is either almost perpendicular (6, 45) or parallel (17) to the membrane surface. The third model is based on a crystallized dimer (46) and positions the hydrophobic loop away from the surface and instead predicts membrane interactions with the second β sheet.
We found that the FYVE domain orientation relative to the micelle surface can be altered by varying the lipid composition of the micelles. The vector of FYVE domain insertion into PI(3)P-containing DPC micelles (O⃗P) differs from the protein primary molecular axis (M⃗) by an angle of ∼50°. This tilted orientation places the FYVE domain neither perpendicular nor parallel to the micelle surface, but at an intermediate angle that favors complementary interactions between polar residues located around the MIL and the zwitterionic PC headgroups. However when an acidic phospholipid such as PS was added to the PI(3)P-enriched DPC micelles, the micelle insertion angle of the FYVE domain was changed to ∼70° (Figure 2). The estimated ∼20° difference in the insertion angle is most likely due to the non-specific electrostatic interactions between basic regions of the FYVE domain and the acidic PS headgroups (15). This orientation tips the FYVE domain such that the basic residues located near the PI(3)P binding site can better interact with the PS headgroups. The PS-stabilized orientation is almost parallel to the micelle surface, and is similar to that suggested by the crystal structure of the EEA1 dimer (17) and by computer modeling (8).
Additional intermolecular restraints were obtained using a novel two-headed mimetic of PI(3)P which contains a proxyl radical near the glycerol moiety (Figure 3). This class of hybrid lipids was first designed for the corresponding PI(4,5)P2 and PI(3,4,5)P3 head groups (reviewed in (47, 48)). The residues most perturbed upon addition of PROXYL-Pea-PI(3)P in the FYVE domain are located near the inositol ring's binding pocket and the MIL (Figure 3). Interestingly only one side of the FYVE domain is affected by the spin label indicating that the PI(3)P analog does not freely rotate but rather is restrained by the interaction of the acyl chains of the lipid with the hydrophobic MIL. The lack of discernible NOEs between the FYVE domain and the PI(3)P threitol moiety or acyl chains calls for alternative approaches. The method based on spin-label restraints represents such an alternative for characterization of interactions between the ligand and the micelle inserted protein.
Insertion into membranes induces transformations in proteins and peptides ranging from local conformational changes to global structural rearrangements such as α-helix or β-strand formation (49). Our findings indicate that the MIL of the FYVE domain becomes elongated when the protein inserts into micelles, while a global fold of the FYVE module remains unchanged. Comparison of the experimental data obtained from the titration of the 5-doxyl probe and the theoretical model derived from this experimental data suggests rearrangement in the MIL conformation. In particular, our model predicts that upon insertion the hydrophobic residues of the MIL move closer to the micelle's hydrophobic core whereas hydrophilic residues move toward the aqueous interface, thus stretching the MIL (Figure 5b). Such conformational changes were corroborated by the altered NOEs observed for the MIL residues in the ligand-free and the micelle-bound FYVE domain. Specifically, upon micelle insertion the interproton distances between Ser1365 and the Val1368-Thr1367 sequence became significantly longer whereas the distance between Ser1365 and the buried Phe1364 residue became shorter, indicating the local rearrangements within the MIL (Figure 5).
Strong conservation of the MIL residues among all but one of the 31 human FYVE domains suggests similar site of insertion for other FYVE proteins. The FSVTV sequence of EEA1 is replaced by the FTFTN and FSLLN sequences in Hrs and Vps27 proteins, and these exposed loops assume similar conformations (6, 17, 45, 46). Mutations of the MIL residues of EEA1, Hrs, and Vps27p reveal their critical role in membrane association and function of these proteins. Thus the subcellular localization of EEA1 is completely lost when the Val1366 and Thr1367 residues are mutated to Gly or Glu (50). Similarly, replacement of corresponding hydrophobic residues of the Vps27p or Hrs FYVE domains results in a seven to twenty fold reduction of affinity for the membrane-bound PI(3)P (51). Taken together the mutagenesis data, the chemical shift perturbations in the FYVE domain occurred upon binding to micelles (6) and the conserved hydrophobic nature of MIL we suggest a general model of the FYVE insertion in which the elongated MIL is positioned to make complementary hydrophobic and electrostatic contacts with the interior and interfacial zones of the micelle.
In conclusion, we have developed a theoretical basis for quantitative determination of the geometry of micelle insertion by peripheral proteins, and have demonstrated a general application of this approach by defining the micelle orientation of the FYVE domain.
For Table of Contents Use Only
ACKNOWLEDGMENT
We thank S. Emr and A. Sorkin for discussions, and R. Muhandiram and L. E. Kay for NMR pulse sequences. This work was supported by grants from the National Institutes of Health (NS29632 to G.D.P., GM067655 to A.G.K., CA085716 to T.G.K. and GM089074 to Echelon Biosciences), Wellcome Trust (M.O.), the American Cancer Society (RSG0513601 to T.G.K.) and in part by Cancer League of Colorado (T.G.K.).
Abbreviations
- NMR
nuclear magnetic resonance
- EEA1
early endosome antigen 1
- DPC
dodecylphosphocholine
- PS
phosphatidylserine
- PI(3)P
phosphatidylinositol 3-phosphate
- MIL
membrane insertion loop
- PI
phosphoinositide
- HSQC
heteronuclear single quantum coherence
- NOE
nuclear Overhauser effect
- 5- and 14-doxyl PC
5- and 14-doxyl derivatives of 1-palmitoyl-2-stearoyl-sn-glycero-3-phosphocholine
REFERENCES
- 1.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–13. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Misra S. Signaling and subcellular targeting by membrane-binding domains. Annual Review of Biophysics & Biomolecular Structure. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overduin M, Cheever ML, Kutateladze TG. Signaling with phosphoinositides: better than binary. Molecular interventions. 2001;1:150–9. [PubMed] [Google Scholar]
- 4.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin.[comment] Nature. 2002;419:361–6. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 5.Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisma of AP180 N-terminal homology (ANTH) and Epsin N-terminal homology (ENTH) domains. Journal of Biological Chemistry. 2003;278:28993–9. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 6.Kutateladze TG, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 7.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 2002;277:26379–88. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 8.Diraviyam K, Stahelin RV, Cho W, Murray D. Computer modeling of the membrane interaction of FYVE domains. J. Mol. Biol. 2003;328:721–36. doi: 10.1016/s0022-2836(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 9.Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–8. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 10.Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. Journal of Biological Chemistry. 2003;278:14469–79. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 11.Isenberg G, Niggli V, Pieper U, Kaufmann S, Goldmann WH. Probing phosphatidylinositolphosphates and adenosinenucleotides on talin nucleated actin polymerization. FEBS Letters. 1996;397:316–20. doi: 10.1016/s0014-5793(96)01203-3. [DOI] [PubMed] [Google Scholar]
- 12.Seelig A, Blatter XL, Frentzel A, Isenberg G. Phospholipid binding of synthetic talin peptides provides evidence for an intrinsic membrane anchor of talin. J. Biol. Chem. 2000;275:17954–61. doi: 10.1074/jbc.M002264200. [DOI] [PubMed] [Google Scholar]
- 13.Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–13. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RP, Niggli V, Durrer P, Craig SW. A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry. 1998;37:10211–22. doi: 10.1021/bi9727242. [DOI] [PubMed] [Google Scholar]
- 15.Kutateladze TG, Capelluto DGS, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–7. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 16.Gaullier JM, Ronning E, Gillooly DJ, Stenmark H. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 2000;275:24595–600. doi: 10.1074/jbc.M906554199. [DOI] [PubMed] [Google Scholar]
- 17.Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright D. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 2001;8:947–58. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 18.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–7. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 19.Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol. Cell. 1999;3:805–11. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- 20.Ball A, Nielsen R, Gelb MH, Robinson BH. Interfacial membrane docking of cytosolic phospholipase A2 C2 domain using electrostatic potential-modulated spin relaxation magnetic resonance; Proceedings of the National Academy of Sciences of the United States of America; 1999. pp. 6637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling.[erratum appears in Biochemistry 2002 Jun 11;41(23):7528] Biochemistry. 2002;41:6282–92. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 22.Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ. C2 domains of protein kinase Cα: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–65. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman ER, Davis AF. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. Journal of Biological Chemistry. 1998;273:13995–4001. doi: 10.1074/jbc.273.22.13995. [DOI] [PubMed] [Google Scholar]
- 24.Perisic O, Paterson HF, Mosedale G, Lara-Gonzalez S, Williams RL. Mapping the phospholipid-binding surface and translocation determinants of the C2 domain from cytosolic phospholipase A2. J. Biol. Chem. 1999;274:14979–87. doi: 10.1074/jbc.274.21.14979. [DOI] [PubMed] [Google Scholar]
- 25.Medkova M, Cho W. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. Journal of Biological Chemistry. 1999;274:19852–61. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- 26.Mascioni A, Karim C, Barany G, Thomas DD, Veglia G. Structure and orientation of sarcolipin in lipid environments. Biochemistry. 2002;41:475–82. doi: 10.1021/bi011243m. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nature Structural Biology. 1997;4:717–24. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez C, Hilty C, Wider G, Wuthrich K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy; Proceedings of the National Academy of Sciences of the United States of America; 2002. pp. 13533–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papavoine CH, Konings RN, Hilbers CW, van de Ven FJ. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994;33:12990–7. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- 30.Jarvet J, Zdunek J, Damberg P, Graslund A. Three-dimensional structure and position of porcine motilin in sodium dodecyl sulfate micelles determined by 1H NMR. Biochemistry. 1997;36:8153–63. doi: 10.1021/bi970193b. [DOI] [PubMed] [Google Scholar]
- 31.Van Den Hooven HW, Spronk C, Van De Kamp M, Konings RNH, Hilbers CW, Van De Ven FJM. Surface location and orientation of the lantibiotic nisin bound to membrane-mimicking micelles of dodecylphosphocholine and of sodium dodecylsulphate. Eur. J. Biochem. 1996;235:394–403. doi: 10.1111/j.1432-1033.1996.00394.x. [DOI] [PubMed] [Google Scholar]
- 32.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–65. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 33.Marion D, Kay LE, Sparks SW, Torchia DA, Bax A. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J. Am. Chem. Soc. 1989;111:1515–7. [Google Scholar]
- 34.Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of an ensemble of structures. J Mol Biol. 1997;268:170–84. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 35.Vogel A, Scheidt HA, Huster D. The distribution of lipid attached spin probes in bilayers: application to membrane protein topology. Biophys J. 2003;85:1691–701. doi: 10.1016/S0006-3495(03)74599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellena JF, Archer SJ, Dominey RN, Hill BD, Cafiso DS. Localizing the nitroxide group of fatty acid and voltage-sensitive spin-labels in phospholipid bilayers. Biochim Biophys Acta. 1988;940:63–70. doi: 10.1016/0005-2736(88)90008-9. [DOI] [PubMed] [Google Scholar]
- 37.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. Journal of the American Chemical Society. 2002;124:2450–1. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 38.Shenkarev ZO, Balashova TA, Efremov RG, Yakimenko ZA, Ovchinnikova TV, Raap J, Arseniev AS. Spatial structure of zervamicin IIB bound to DPC micelles: implications for voltage-gating. Biophysical Journal. 2002;82:762–71. doi: 10.1016/S0006-3495(02)75438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindberg M, Jarvet J, Langel U, Graslund A. Secondary structure and position of the cell-penetrating peptide transportan in SDS micelles as determined by NMR. Biochemistry. 2001;40:3141–3149. doi: 10.1021/bi0008985. [DOI] [PubMed] [Google Scholar]
- 40.Ohman A, Lycksell PO, Jureus A, Langel U, Bartfai T, Graslund A. NMR study of the conformation and localization of porcine galanin in SDS micelles. Comparison with an inactive analog and a galanin receptor antagonist. Biochemistry. 1998;37:9169–78. doi: 10.1021/bi980153n. [DOI] [PubMed] [Google Scholar]
- 41.Lindberg M, Graslund A. The position of the cell penetrating peptide penetratin in SDS micelles determined by NMR. FEBS Letters. 2001;497:39–44. doi: 10.1016/s0014-5793(01)02433-4. [DOI] [PubMed] [Google Scholar]
- 42.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–62. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 43.Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P [letter; comment] Nature. 1998;394:432–3. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 44.Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif [letter] [see comments] Nature. 1998;394:433–4. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 45.Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–66. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 46.Mao Y, Nickitenko A, Duan X, Lloyd TE, Wu MN, Bellen H, Quiocho FA. Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell. 2000;100:447–56. doi: 10.1016/s0092-8674(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 47.Rzepecki PW, Prestwich GD. Synthesis of hybrid lipid probes: derivatives of phosphatidylethanolamine-extended phosphatidylinositol 4,5-bisphosphate (Pea-PIP(2)) J Org Chem. 2002;67:5454–60. doi: 10.1021/jo011185a. [DOI] [PubMed] [Google Scholar]
- 48.Prestwich GD. Phosphoinositide signaling; from affinity probes to pharmaceutical targets. Chem Biol. 2004;11:619–37. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 49.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–65. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 50.Kutateladze TG, Ogburn KD, Watson WT, de Beer T, Emr SD, Burd CG, Overduin M. Phosphatidylinositol 3-phosphate recognition by the FYVE domain. Mol Cell. 1999;3:805–11. doi: 10.1016/s1097-2765(01)80013-7. [DOI] [PubMed] [Google Scholar]
- 51.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–88. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]




