Abstract
The World Wide Web has revolutionized how researchers from various disciplines collaborate over long distances. This is nowhere more important than in the Life Sciences, where interdisciplinary approaches are becoming increasingly powerful as a driver of both integration and discovery. Data access, data quality, identity, and provenance are all critical ingredients to facilitate and accelerate these collaborative enterprises and it is here where Semantic Web technologies promise to have a profound impact. This paper reviews the need for, and explores advantages of as well as challenges with these novel Internet information tools as illustrated with examples from the biomedical community.
Keywords: AJAX, OWL, RDF, Semantic Web, SPARQL, Web 2.0
Introduction
Over the last decade, the World Wide Web has forever changed the way people communicate, do business, and retrieve information (Berners-Lee and Hendler, 2001; http://www.w3.org/TR/webarch/). This is a process that has already and will continue to have a profound impact on the Life Sciences. There are, however, significant challenges related to the use of much of the data found on the web. Foremost is the issue that often the quality of that data itself has to be questioned. For example, it was widely reported (http://news.cnet.com/2100-1038_3-5997332.html) that Wikipedia published an average of 3.86 factual inaccuracies per article compared to Encyclopedia Britannica's 2.92 errors. Although measures have been taken to improve the editorial process, accuracy and completeness remain valid concerns. Perhaps the issue is one in which it has become difficult to establish exactly what has actually been peer reviewed and what has not, given that the low cost of digital publishing on the web has led to an explosive amount of publishing and republishing to the point where it is overwhelming.
Digital science is no different to science in the non-digital world where it is especially important to know not just the product of data but also its origin. The latter often has a significant bearing on the former. Unfortunately, simply posting one's methods along with the findings is only a partial solution, because of the problems of incomplete data provenance. If someone finds a statement in an article to be particularly interesting, they are likely to reference it in their own work. Like the physical world, the original author has little control over whether or not the context leading up to his quotable statement ever reaches an audience when it is referenced elsewhere. From the publisher's point of view, there are no successful mechanisms available to comprehensively protect or track the use of the content they create. This has not been as much of a problem in the world of printed academic journals, where writers honor copyrights and citation policies at the risk of legal penalties and academic infamy.
However, the viral nature of digital information is unchecked on the web. Seemingly unlimited copies of information can spawn, evolve, and propagate around the globe almost instantly, as the music and motion picture industries have already discovered. In the world of Science and dissemination of facts, the problem can often be much more subtle and virtual provenance remains a largely unsolved issue. It is worth noting that this is perhaps more of a social and historical problem than technical one. On a technical level, publication data can be stored as metadata (data about data, such as the author's contact details or the date of publication). In addition, certain document formats can be digitally locked to prevent copying or digitally signed to show evidence of tampering. Furthermore, the web hyperlink is a very simple technical mechanism that can provide instant references and form a chain of provenance—but only if a re-publisher of information chooses to use it. Much of the time though, these technical means are ignored, perhaps due to habit or simply that the available tools do not yet make their usage effortless enough for everyday use. Although the origin of data may not matter for someone leisurely surfing the web, it is crucial that scientists and researchers know that the data they gather online are legitimate.
In spite of these problems, researchers find enormous utility in continuing to use the web in their collaborations, information discovery, and dissemination processes. The fact that the spread of digital information is viral in nature and that it does tend to rapidly mirror itself and diverge rapidly across the globe is still clearly far more of a benefit than a problem. As research teams grow, so will the number of communication channels between team members (Wuchty et al, 2007). More and more scientific work requires the sharing of information and knowledge in meaningful ways beyond the simple transfer of unassociated pieces of data (Neumann, 2005). Increasingly it makes sense that the data they exchange need to be in standard formats and data representations that everyone can easily work with (Piwowar et al, 2007). Over the last decades, a number of fundamental technology standards (examples include TCP/IP, HTTP, HTML, URI, JPEG, XML, Web Services, etc.) have made it easier for important domain-specific data standards, and de facto standards, to emerge (examples include caBIG, SBML, CellML, MAGE-ML, PEDRo, and LSID; see Table I) in facilitating these exchanges. The Semantic Web concepts and related standards that have emerged from the World Wide Web Consortium over the last few years have the potential to improve this transfer of information to a whole new level.
Table 1.
A table of commonly used bioinformatics data standards
| Examples of Web 2.0 applications data standards and semantic web projects | |
|---|---|
| Web 2.0 | |
| OpenWetWare (http://openwetware.org) | A wiki for the synthetic biology and biological engineering community |
| YeastPheromoneModel.org (http://yeastpheromonemodel.org/wiki/Main_Page) | A wiki used as a modeling resource for the Saccharomyces cerevisiae mating pathway |
| UsefulChem (http://usefulchem.wikispaces.com) | An example of OpenNotebook science |
| Nature Network (http://network.nature.com) | A web 2.0 platform facilitating scientific networking |
| SciVee (http://scivee.tv) | A site for sharing science videos |
| Standards | |
| BioPax (http://biopax.org) | Open source data exchange ontology and format for biological pathways |
| CellML (http://www.cellml.org/) | XML-based open standard for storing and exchanging computer-based mathematical models; utilizes MathML and RDF |
| FuGE (http://fuge.sf.net) | XML-based model for shared components in different functional genomics domains |
| LSID (http://lsids.sf.net) | Life Science Identifier is a unique and persistent ID for naming documents and objects |
| Mage-ML (http://www.mged.org/Workgroups/MAGE/mage.html) | Language based on XML for representing gene expression |
| mzXML (http://tools.proteomecenter.org/mzXMLschema.php) | Mass spectrometry representation based on XML |
| SBML (http://sbml.org) | Machine-readable format for representing quantitative models |
| Semantic Web | |
| YeastHub (http://yeasthub.gersteinlab.org) | Integration of diverse types of biological data stored in a variety of formats |
| CViT (http://www.cvit.org) | Computational and mathematical cancer modeling community; includes a semantic RDF-based repository |
| WikiProfessional (http://www.wikiprofessional.org) | Web-based environment that combines wiki functionalities with semantic navigation capabilities |
| Freebase (http://www.freebase.com) | Free online database for structured information |
Semantic Web and data integration
A significant issue facing the web in its current form is that the textual and other unstructured forms of information found on most web pages today are transmitted across the internet as a stream of character bytes intended simply to facilitate the display of the contained information on the screen of a computer for a human to interpret. Beyond the formatting and rendering instructions contained in these byte streams, all the remaining information contained therein is for the most part entirely unintelligible to the computers facilitating them today. The fact that these characters can be displayed on a computer to form words and that these words have meaning that can be accumulated and acted upon is something that we humans recognize while computers cannot (Seringhaus and Gerstein, 2007). The goal of the Semantic Web is to provide a way for computers to be able to process the data they make available for the user and to allow programs to amass, contextualize, and reason over this information and even act upon it automatically. The Semantic Web, as proposed by Tim Berners-Lee, is intended to allow meaning (i.e. semantics) to be associated with information on the web through a universal mechanism in a machine-interpretable way (Berners-Lee and Hendler, 2001; Hendler, 2003; Neumann and Quan, 2006). The Semantic Web makes information essentially self-describing through the adoption of the Resource Description Framework (RDF) standard (http://www.w3.org/RDF/FAQ). RDF is designed to provide a common machine-readable data representation that maps to most other data representations by making statements in the form of a subject–predicate–object. These statements are called triples. Unique identity is an essential concept in RDF and is implemented through the use of Uniform Resource Identifiers (URIs) (http://www.w3.org/TR/REC-rdf-syntax/).
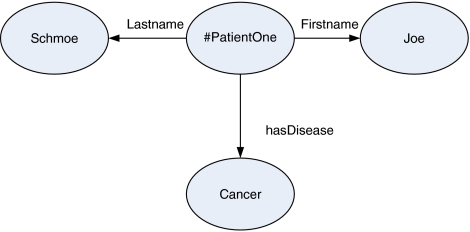
Here is an example of a simple triple representing the fact that Patient One suffers from cancer (also see Figure 1)—http://www.example.org/identity#PatientOne, http://www.example.org/diagnosis#SuffersFrom, and http://www.example.org/hcls#Cancer, which are respectively the subject, predicate, and object of this triple. In this example, ‘http://www.example.org/diagnosis#SuffersFrom' is a unique name for a previously defined relationship concept between two elements and http://www.example.org/hcls#Cancer is the unique name for a previously defined ailment concept, whereas http://www.example.org/identity#PatientOne is the unique identifier for Patient One. URIs such as these make it possible to make further unambiguous statements about all three elements in this statement using additional triples with the appropriate URI as the subject or object. The meaning of biological concepts, in this case http://www.example.org/hlcs#Cancer, as well as applicable relationships to cancer could be stored on servers available for information retrieval (Mukherjea, 2005). A particular shared URI for an object listed in two different RDF documents containing triples is what ties the information in those two documents together and indicates that the concept that links them is one and the same. In the Life Sciences, one form of domain-specific URI is the LSID (Life Science Identifier), which is an established data identifier standard representing unique, immutable data objects with links to metadata and dynamic relations that are created and versioned by different authorities (Clark et al, 2004).
Figure 1.
This graph illustrates three RDF relations: #PatientOne Lastname ‘Schmoe'; #PatientOne Firstname ‘Joe'; #PatientOne hasDisease Cancer. An English interpretation would be Patient One is named Joe Schmoe and Joe Schmoe has been diagnosed with cancer.
In addition to being able to point to another uniquely identified concept (e.g. another URI) to indicate a relationship, the object portion of a triple has an alternative form in which it can contain a literal value storing information about the uniquely named node that forms the subject of the triple. For example, with the triple http://www.example.org/identity#PatientOne, http://www.example.org/measures#hasMetricWeight, and ‘73kg', we are recording the fact that Patient One currently weighs 73 kg using a previously defined #hasMetricWeight concept. We now know two things about Patient One and how additional triples using this simple three-part structure could be added to tell us (and a machine) a great deal more about Patient One. In this manner, a collection of triples can form networks of interconnected logical graphs that describe information nodes with individual node properties and their interrelationships with other nodes. These directed, labeled, interconnected graphs can grow arbitrarily large and complex, and can contain many millions of triples describing even the most complex data structures at whatever level of granularity it is useful to represent. Triples form graphs that can span many documents and because these graphs have no set order, merging data from multiple sources using RDF becomes trivial (Feigenbaum et al, 2007). RDF is expressive enough to encapsulate nearly any sort of data model and is therefore a reasonable way to represent the Semantic Web in a form that is machine readable. RDF has a number of serialization (storage) formats including an XML representation.
Two other key ingredients of the Semantic Web are the Web Ontology Language (OWL), which defines the types of objects, their vocabulary, and their relations in an RDF document (http://www.w3.org/TR/owl-features/), and RDF Schema (RDFS), which defines how to use RDF to define vocabularies by specifying a standardized way to describe resources. Life Sciences research can benefit from RDF, RDFS, and OWL, which can be explicitly defined and used to integrate genomic, proteomic, cellular, physiological, biochemical, and indeed almost any other kind of information even when all the contributory data exist in different databases under different interpretation schemas. This exposition and linkage of data from many sources in a standard manner comprehensible to software programs is what makes up the Semantic Web. It is important to note that the underlying data need not be stored as RDF but can be converted dynamically as needed for this common representation, just as the OWL ontology describing these data and their relationships can also be added long after the original database was created.
Another key standard recently emerged from the World Wide Web Consortium. This is the SPARQL standard, the query language for RDF and the first standardized distributed query language and access protocol. The SPARQL language is used to express single queries simultaneously across diverse RDF and non-RDF sources of data and integrate the results. It is likely to become a critically important standard for achieving data integration and significantly leverages the standards described earlier. There are already a good number of standards-compliant implementations available for early adopters to experiment with.
Naturally, the Semantic Web idea has its complications. The most often raised criticism centers around the notion of standardization, based on the belief that absolute ‘top to bottom' homogeny is required for the Semantic Web to work and that it is nearly impossible to achieve this kind of widespread agreements (Wang et al, 2005). This notion points to a common misunderstanding of the technology. It is true that the benefits of the Semantic Web are generally easier to achieve if everyone concerned agrees to use common standards for intra-domain naming and organizing their information by sharing the same URI-defined concepts, specific vocabularies, and specific OWL ontologies describing their data's interrelationships. However, this level of accord is by no means essential for success nor can reasonably be expected in reality. Communities of information providers generally grow independently, with separate requirements and information collection objectives. Information collected by different communities may logically only overlap in a few areas. This is precisely where the semantic technologies become useful. Provided information producers make use of just the Semantic Web's lowest level standards for their data representations (i.e. RDF, RDFS, and OWL), or if they can convert to these representations dynamically, it need not matter at all that the RDFS and OWL descriptions of each community are different and that the URIs used are initially unrelated. The design of the Semantic Web makes it possible that by using these same underlying standards, third parties may create and share bridging ontologies for performing data integration between information architectures that have evolved entirely independently all the while creating their own sets of ontologies and URI named concepts (Lam et al, 2006). Bridging separately evolved information can often have extremely synergistic results as the two or more data sets exploring different aspects of a knowledge domain are merged revealing new information and connections and improving our overall understanding.
A closely related criticism is that some information is going to be difficult to classify in any system. Smith et al (2007) state that ‘there are many synonyms for the same underlying entity caused by independent naming, e.g. structural genomics centers assigning their own protein identifiers in addition to UniProt's.' There can also be lexical variants. ‘We have to make sure that the knowledge captured in statements in one language is changed as little as possible when transforming them into statements of another language. Hence, the semantics of one language needs to be reconciled with the semantics of the other' (Aranguren et al, 2007). Again OWL and RDFS can play a reconciliatory role to bridge differing naming schemes and vocabularies.
In both cases, by identifying equivalent classes, attributes, and properties (including deterministically transformable properties), isomorphic structures, and defaults for non-overlapping parts of the data models, ontology maps can be created. Armed with such a map, a program employing forward-chaining reasoning can automatically instantiate new data that allow all the existing data to be viewed in a consistent model. Similarly, a backward-chaining reasoner combined with a query engine can answer queries posed against the two (or more) disparate data sets as if they were a single data model.
Awareness of Semantic Web standards and technologies is growing and accelerating together with the list of both commercial and open source software tools, libraries, SPARQL implementations, and applications needed to begin enabling the transformation to, storage and query of, and reasoning over semantically enabled data. The World Wide Web Consortium has established an active special interest group dedicated to pioneering the use of Semantics standards in Health Care and Life Sciences (Ruttenberg et al, 2007). With data available in a semantic format, a variety of new IT capabilities will become possible. Because the data are self-describing, this can change the fundamental nature of the applications that access them, with the potential for much greater flexibility in designing, modifying, and changing semantic-based applications far more quickly and perhaps even by their end users. This will help address several major challenges facing Life Science and Healthcare research and practice today where often the discovery of new knowledge outstrips the ability of applications to represent, store, integrate, share, and otherwise utilize it. For example, developing applications that support sophisticated event-based processes, captured perhaps during the course of a simulation, an experiment, or a diagnosis, becomes much more realistic along with far more advanced and widespread uses of rules based systems than we see today. Capturing and acting on data in ‘real time' and understanding their content and context becomes more practical, which in turn leads to more automated actions. As touched on above, integration of services and data, so crucial for advancing biomedical research, becomes more straightforward because all information is presented to the application developer or end user in a single common data representation through which that information can be merged. It is highly likely that significant inclusion of semantic standards-based technologies will be essential to achieve widespread adoption and the full promise of the Service Oriented Architectures (SOA). This especially includes the automated discovery, mediation, and adaptation of reusable services in applications that the IT industry is driving toward. Currently, even discovery is an issue for large-scale SOA implementations not yet using semantics. Self-describing data do much to address the Life Sciences' emerging data explosion problem where enormous data sets are generated, as data that are self-describing can be more readily searched, sorted, filtered for relevance, and exchanged.
This said, writing applications against semantic data and ontologies is difficult today and few good programming patterns and mature software toolkits currently exist that allow a developer to take full advantage of semantic information. All the flexibility that semantic data make possible requires far more complicated programming models to cope with and best utilize them. There are still too few tools and sophisticated libraries (i.e., middleware that supports scalable semantic application building), and especially no scalable frameworks that address issues related to semantically marked up data being far more voluminous and unwieldy from the point of view of efficient querying than data have been in the past. These problems coupled with a lack of experience of actually writing semantic applications are sizeable inhibitors for the majority of developers who rely on ‘best practices' and sophisticated software libraries. Without the experience of existing applications to guide them, early adopters find it difficult to know what general-purpose frameworks to build. This presents an interesting dilemma: the potential of semantic technology is great but should one seriously consider utilizing it now or wait until its value has been more clearly demonstrated and the means for implementing it made easier? Earliest uses of semantics, particularly in the areas of data federation and integration, are more mature than the applications that require significant use of semantics-based reasoning. Acting now one can jump start evaluations of a promising new technology and might allow the development of solutions that can provide great immediate advantage. The adoption of the appropriate World Wide Web Consortium (W3C) standards for representing data today helps to future-proof the usefulness of the data and makes it available to important emerging integrative technologies such as SPARQL. The most serious concern related to the emergence of the Semantic Web is that not enough immediate value is created by the individual adoption of the semantic technologies and World Wide Web Consortium semantic standards to get to a point where a network effect provides enormous general value. While the World Wide Web has very long since passed the point where people and businesses questioned whether it was worth getting online or establishing a web presence, the Semantic Web still has some way to go before reaching this tipping point. In Life Sciences, the early value that begins to justify investment is already being found (see examples below). It is a domain that has extraordinary data integration problems with high motivation to solve them. It is also one that lends itself fairly obviously to the semantic approach. Early adopters are starting to find value in utilizing the necessary standards and technologies to create even small, private Semantic Webs that perhaps only integrate a few databases to start with but that begin to establish that overall value network while solving more immediate problems. It therefore may well be that this domain blazes the trail for everyone else.
Finally, Web Services technology has opened up whole new avenues for Life Sciences research. A Web Service is an application whose input and output are provided by XML documents (Digiampietri et al, 2005) conforming to various schemas. The earliest ‘de facto' form of this was called XMLRPC, which is still in use today, but this is being superceded by the Simple Object Access Protocol (SOAP) from the XML Protocol Working Group at the World Wide Web Consortium. One web service-based system of note is BioMOBY, which is unique due to its classification of all valid data types in an ontology (Wilkinson et al, 2005). BioMOBY goes beyond SOAP's data types and structures by treating data as different semantically defined data types. By standardizing this ontology of data types, BioMOBY helps to ensure the robustness and interoperability of its web services even though those services are written by many different people who may never directly collaborate. There are many successful projects available in bioinformatics, which are built on this system, for example, for exploring and visualizing genomic data (Turinsky et al, 2005), for building workflows linking different services addressing specific biological problems (Garcia Castro et al, 2005), and for discovering available services (Carrere and Gouzy, 2006). Although BioMOBY is a noteworthy system for integrating ontologies into a web service platform, it is not the only one out there. The National Cancer Institute's caBIG consortium effort delivers a vast collection of SOAP-based web services and will be discussed in further detail toward the end of the paper.
Other ‘ontologies exist to describe the anatomy, developmental processes, phenotypes and pathologies of several species, as well as those oriented toward the experimental and physical properties' (Cote et al, 2006). One of the best-known ontologies is GO, the Gene Ontology. GO has aimed to describe the role of genes and gene products throughout many organisms (Ashburner et al, 2000). Additionally, Protégé is an environment centered on the development of new ontologies. Protégé has a large community of users and contributing developers. The ontologies developed range from taxonomies and classifications to data base schemas and axiomatized theories. It is important to note that the ontologies Protégé creates are built for Semantic Web use (Noy et al, 2003). The aforementioned OWL is designed for use by applications that need to process the content of information instead of just presenting the information to humans. Also of note here is the Taverna project (Oinn et al, 2004), which is part of UK's myGrid. Taverna allows building up a workflow of BioMOBY services (Kawas et al, 2006). As complicated as this sounds, the interface is roughly that of a flow chart, that is, a user indicates what type of input data will be used, and is linked to a number of web service modules that accept the appropriate type of data. Taverna automatically generates provenance information in a semantic format that researchers have found great utility in mining (Zhao et al, 2007). Finally, MyExperiment is an exciting new online collaborative space that will allow researchers to share scientific workflows built by Taverna. In essence, MyExperiment is a social networking platform as a front-end to Taverna. It allows users to track each other's work on a per user level rather than a per workflow level.
Web 2.0 and data sharing
Over the past couple of years, the internet has been revolutionized by what is turning out to be more than just a new buzzword: Web 2.0. Despite what the term implies, there is no new client software to use or browser updates for a web user to download. Web 2.0 is not about any one major new technology taking over. It is about teaching the old web new tricks, using almost all the same old tools but with a new paradigm in mind.
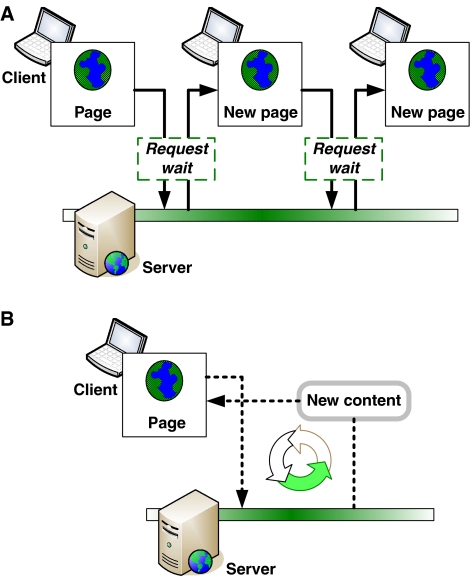
What makes Web 2.0 sites different from their predecessors is the sort of interaction between site and visitor (Kamel Boulos and Wheeler, 2007). Although older sites were dynamically generated, the pages they produced were largely static (Boulos et al, 2006). The adoption of AJAX (Asynchronous JavaScript and XML) has drastically increased interactivity in websites, a significant attribute of the Web 2.0 revolution. AJAX is not a new language or program (http://www.adaptivepath.com/ideas/essays/archives/000385.php), rather it is simply a new way to use JavaScript to pass messages in the background (Figure 2). What AJAX allows a site to do is transmit messages back and forth with the server, while the user continues to interact with the site without interruptions. This seems trivial at first glance—after all, the web is all about passing messages back and forth between server and browser. However, the difference with AJAX is that users can continue directly interacting with the web page while those transactions happen in the background. By making transactions between a site and its readers and publishers easier and more user-friendly, AJAX has effectively improved and accelerated information transfer. These days, posting updates to a site sometimes does not even require a publishing individual to leave his current web page. AJAX also has examples of use in Life Sciences applications and can advance current platforms of medical research, for example, enhancing user interaction with MEDLINE/PubMed (Muin and Fontelo, 2006), increasing usability and speed of protein–protein association requests (von Mering et al, 2007), improving presentation and visualization of cell signaling networks (Berger et al, 2007), and developing more rapid searching and browsing functions for biomedical ontologies (Cote et al, 2006; Beauheim et al, 2007). The popularity of AJAX will likely continue to grow, hence will improve the interactivity of the web in everyday use and Life Sciences applications alike (Figure 2).
Figure 2.
Traditional and AJAX page requests. AJAX allows a page to fetch content from the server without making the user wait for a page to load. New content is dynamically injected into an already loaded web page without disrupting the user's experience.
One piece of technology that has greatly influenced the Web 2.0 revolution is the wiki. Wikis are websites in which content is both created and edited by users in a manner akin to using a word processor. Indeed, one does not have to know HTML to be able to correctly format text on a wiki. This means that anyone who can open a web browser can edit and contribute to wiki-based websites; thus, the average web surfer grows accustomed to publishing data online rather than just reading it.
Where wikis truly excel is in their versatility. A wiki's usage scales as needed. Students have found the wiki to be a better media for note taking than traditional word processors, which indicates that a wiki can prove useful to just one person working alone. At the other end of the spectrum, Wikipedia provides service to millions with more than 75 000 regular contributors. Because biomedical research produces hundreds of thousands of papers annually, the necessity of allowing anyone to be actively involved in editing entries, modifying text, and adding links as new works are published is evident (Giles, 2007). Countless researchers rely on GenBank and EMBL as their primary source for genome annotation, but due to the constantly increasing amount of new sequencing information, a new wiki-based open editing framework may provide a better solution to allow the community to work out the best naming conventions for each gene and discuss alternative annotation-based solutions (Salzberg, 2007). Another helpful schema for wiki use is that of the open notebook. The UsefulChem Project has implemented a wiki to share and broadcast their open source chemistry work. Wiki is an ideal platform for a notebook as it seamlessly tracks changes and annotations made by many different collaborators.
Blogs, short for web logs, have exploded in popularity in recent years (http://cjrarchives.org/issues/2003/5/blog-jensen.asp?printerfriendly=yes). Other than online diaries and journals, which have been around since the dawn of the web, the blog is a phenomenon that has only taken off in the last few years. While a distinction is made between a blog and journal, the line between them is blurred. Technologically there is no difference between an online diary and a blog—the difference lies in the content. A loose definition of a blog is that it is an informal online journal of the author's thoughts. Usually, a blog will focus on a specific topic or issue, although some are a collection of thoughts on everything the blogger finds interesting (McLean et al, 2007). We note in this context that Molecular Systems Biology maintains its own blog, The Seven Stones, which is updated regularly with viewpoints by noted researchers (Box 1). Like most blogs, this content is more up to date than that of full research papers, but left deliberately less balanced and unedited. It is an effective way to keep updated on what is being researched now, rather than waiting for papers to go through a lengthy peer-review and publishing process (Plutchak, 2005) and, equally important, to provide a lively forum for discussions that rarely take place in the literature (Bonetta, 2007). The Technorati search engine is now tracking over 7.8 million blogs and 937 million links and reports, nearly 20 000 of which consider themselves to be science related. The potential use of blogs for cancer patients, scientists, clinical researchers, and practicing oncologists to discuss findings and suggestions has been envisioned in biomedical journals (Hillan, 2003; Kim and Chung, 2007). Many bloggers allow their readers to comment on posts written by the blog owner. Often these comments will take the form of posts on the reader's own blog, using a mechanism called ‘Linkback' or ‘Trackback', which informs web authors of other sites linking and referring to their data. Many communities of people with related interests have sprung into life through this backward-linking and forward-commenting blog based interactivity. Trackback is interesting in its own right as a provenance system in which an author is notified about who is linking to their posted material, although it does require commentators to opt in by including a Trackback link. Linkback takes this beyond blogging and into the desktop. Applications built on the Linkback framework keep track of text copied and pasted between each other. A paragraph of text that was shared between several files would be treated as a single shared data source such that changes in the original document would be automatically mirrored to other documents. Another recent innovation is OpenID, which allows users to identify themselves to other websites by using their own blogging website rather than creating a new account for each blog they potentially visit. It is possible that OpenID and Linkback will become another important part of what is needed to establish chains of provenance and identity for biomedical data across the web by providing identity services.
Applications
Many sites and platforms exist to aid in the collaboration of scientific research, using some or all of the technologies described in the previous sections. Here, we briefly list some examples from the biomedical community:
The Alzheimer Research Forum (AlzForum) has been opening the field of Alzheimer's disease research for scientist and patient alike. The AlzForum team aggregates new developments and papers, and shares them with the community. On top of that, they publish their own material on the site to help those without medical degrees grasp information rather than be bogged down under too many scientific papers to read. AlzForum is so successful that they have a sister site dedicated to search for causes and treatments while understanding the disease schizophrenia. The Schizophrenia Research Forum has been actively tracking papers and research since 2003 when ARF writer Hakon Heimer realized that a site like ARF could be used to help more people than just Alzheimer's patients. Additionally, AlzForum team members have recently been involved in the AlzPharm project, which has been integrating data into RDF (Lam et al, 2007). Also notable is their involvement in the Semantic Web Applications in Neuromedicine (Gao et al, 2006) project, an attempt to create effective specialist knowledge bases and tools for the Alzheimer's disease research community.
The Forum for Collaborative HIV Research is somewhat different. Rather than focusing on broadcasting articles to active researchers, HIV Forum places more of an emphasis on facilitating open discussion on emerging issues in HIV clinical research. Although it is not one of the typical bulletin board style web forums, the HIV forum is all about bringing people together to collaborate in a web-based environment (Miller, 2006). The HIV forum initially focused on creating treatment guidelines for AIDS and HIV, but has recently broadened itself to include global AIDS awareness.
The Cancer Biomedical Informatics Grid (caBIG) is a large-scale National Cancer Institute-sponsored project that aims to maximize the full power of cancer knowledge from experts all over the world. Its underlying architecture is a Grid infrastructure known as caGrid, which is a model-driven and service-oriented architecture. caGrid exposes a number of analytical services and tools as web services. The caBIG community has developed a variety of bioinformatics tools that span the entire continuum of clinical research, including genomics, imaging, and pathology, which greatly facilitate the launch of coordinated cancer studies involving multiple institutions (Bouchie, 2004). caBIG provides perhaps the best example of web services. This is in no small part due to caBIG's community of over 800 contributors. The ultimate vision of caBIG is to provide a full cycle of integrated cancer research, thus defining how the cancer research is conducted in the future (Saltz et al, 2006).
The BioPAX group aims to develop an XML-based standard data exchange format for biological pathway data. BioPAX is presently divided into four levels of semantic markup. The first two levels have already been achieved: small molecule proteins, RNA, DNA, and complexes. Furthermore, the BioPAX group has been successful in collaborating with other markup groups such as SBML and CellML (Stromback and Lambrix, 2005) as well as encouraging its utilization in existing databases including Kegg, Reactome, and BioCyc.
YeastHub is noteworthy as well. It is a small RDF store developed at Yale University and is used to demonstrate RDF as a proof of concept. YeastHub stores and integrates diverse sets of data of a variety of formats. Users are capable of making RDF queries using RQL, SeRQL, and RDQL, or by using a graphical user interface to build up a search query, without requiring any knowledge of query languages (Cheung et al, 2005). Although YeastHub does not have the largest data store, it is significant in that it is a small implementation of a semantic web.
Finally, the Center for the Development of a Virtual Tumor (CViT), part of the National Cancer Institute's Integrative Cancer Biology Program, is building an ever-growing community of researchers around the world dedicated to computational and mathematical cancer modeling. Its online outlet, CViT.org, currently provides participants with all the tools of a community-driven website: wikis, blogs, forums, member profiles, and RSS-based news updates. Where things get really interesting is what lies in the future for CViT: building caBIG-compliant infrastructure tools that help facilitate interaction among its contributing scientists. Currently under development is the core piece of this effort: CViT's digital model repository (Deisboeck et al, 2007). Not only will the repository be a place to store modeling experiments and data, it will do this in an RDF environment that can be queried using the SPARQL language. What this means is that documents will be linked together as an ever-growing Semantic Web. Papers and experiments that a contributing investigator references are then built up into a web-like provenance structure within the repository. What separates CViT from other model repositories such as BioModels is, aside from CViT's semantics, the community of cancer modeling experts involved in CViT. Members of the community come together online on a regular basis to discuss cutting-edge literature on CViT's online forums. Additionally, the social networking aspect of the site will allow teams to collaborate from anywhere around the world in a workflow designed specifically for the cancer modeling community. All this is supplemented by a creative electronic licensing workflow that makes best use of the provenance tracking the system's innovative architecture allows.
Summary
What all these community websites have in common is that they are working to facilitate web-based collaborative biomedical research, education, and outreach. To facilitate this type of global exchange and multidisciplinary interaction, certain challenges must be met head on—machine-readable data representations, data quality, integrity, identity, provenance, and ownership are all lacking in much of the current web. The Semantic Web promises to offer help in connecting and integrating the ever-growing amount of biomedical data, and in combining them with cutting-edge analytical services. However, although the Semantic Web certainly has great potential, it faces a number of hurdles for widespread adoption, not least of which is the difficulty of achieving enough incremental value to fund its development before the network effect provides enormous general value for the Life Sciences and beyond.
The biomedical research community will continue to innovate on whatever current technology is available. AlzForum, caBIG, GO, and others will be central to new developments. Increasingly, scientists will be using semantically enabled applications for the same purpose. New technology efforts centered on communities such as CViT's digital model repository, MyExperiment, and Alzform will help to advance the field in an effort to empower the next generation of community-driven scientific enterprises.
Really Simple Syndication separates content from websites for efficient reading.
To keep track of ever-increasing amounts of content on the web, scientists are taking advantage of RSS (for Really Simple Syndication or Rich Site Summary). RSS is yet another way of publishing data online. When a new post is made on an RSS-enabled site, that entry appears on the web page as well as in the RSS feed. This feed is composed entirely of the content of that post plus some metadata for tracking purposes. It can be read in a feed reader or aggregator, such as RSS Owl, Google Reader, or NewsFox (see illustration). The advantage of this is that news and updates from many different sites are collected in one place for easy navigation and filtration. Articles and entries appear in a system that resembles a familiar email inbox. Letting the computer automatically collect reading material from a set of favorite sites is far more efficient than tracking all those sites manually (Scarsbrook, 2007).
Acknowledgments
This work has been supported in part by NIH grant CA 113004 and by the Harvard-MIT (HST) Athinoula A Martinos Center for Biomedical Imaging and the Department of Radiology at Massachusetts General Hospital.
References
- Aranguren ME, Bechhofer S, Lord P, Sattler U, Stevens R (2007) Understanding and using the meaning of statements in a bio-ontology: recasting the Gene Ontology in OWL. BMC Bioinformatics 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauheim CC, Wymore F, Nitzberg M, Zachariah ZK, Jin H, Skene JP, Ball CA, Sherlock G (2007) OntologyWidget—a reusable, embeddable widget for easily locating ontology terms. BMC Bioinformatics 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SI, Iyengar R, Ma'ayan A (2007) AVIS: AJAX viewer of interactive signaling networks. Bioinformatics (Oxford, England) 23: 2803–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berners-Lee T, Hendler J (2001) Publishing on the semantic web. Nature 410: 1023–1024 [DOI] [PubMed] [Google Scholar]
- Bonetta L (2007) Scientists enter the blogosphere. Cell 129: 443–445 [DOI] [PubMed] [Google Scholar]
- Bouchie A (2004) Coming soon: a global grid for cancer research. Nat Biotechnol 22: 1071–1073 [DOI] [PubMed] [Google Scholar]
- Boulos MN, Maramba I, Wheeler S (2006) Wikis, blogs and podcasts: a new generation of Web-based tools for virtual collaborative clinical practice and education. BMC Med Educ 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrere S, Gouzy J (2006) REMORA: a pilot in the ocean of BioMoby web-services. Bioinformatics (Oxford, England) 22: 900–901 [DOI] [PubMed] [Google Scholar]
- Cheung KH, Yip KY, Smith A, Deknikker R, Masiar A, Gerstein M (2005) YeastHub: a semantic web use case for integrating data in the life sciences domain. Bioinformatics 21 (Suppl 1): i85–i96 [DOI] [PubMed] [Google Scholar]
- Clark T, Martin S, Liefeld T (2004) Globally distributed object identification for biological knowledgebases. Brief Bioinformatics 5: 59–70 [DOI] [PubMed] [Google Scholar]
- Cote RG, Jones P, Apweiler R, Hermjakob H (2006) The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisboeck TS, Zhang L, Martin S (2007) Advancing cancer systems biology: Introducing the Center for the Development of a Virtual Tumor, CViT. Cancer Informatics 5: 1–8 [PMC free article] [PubMed] [Google Scholar]
- Digiampietri LA, Medeiros CB, Setubal JC (2005) A framework based on Web service orchestration for bioinformatics workflow management. Genet Mol Res 4: 535–542 [PubMed] [Google Scholar]
- Feigenbaum L, Martin S, Roy MN, Szekely B, Yung WC (2007) Boca: an open-source RDF store for building Semantic Web applications. Brief Bioinformatics 8: 195–200 [DOI] [PubMed] [Google Scholar]
- Gao Y, Kinoshita J, Wu E, Miller E, Lee R, Seaborne A, Cayzer S, Clark T (2006) Web semantics: science, services, and agents on the World Wide Web. J Web Semantics 4: 222–228 [Google Scholar]
- Garcia Castro A, Thoraval S, Garcia LJ, Ragan MA (2005) Workflows in bioinformatics: meta-analysis and prototype implementation of a workflow generator. BMC Bioinformatics 6: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles J (2007) Key biology databases go wiki. Nature 445: 691. [DOI] [PubMed] [Google Scholar]
- Hendler J (2003) Communication. Science and the semantic web. Science (New York, NY) 299: 520–521 [DOI] [PubMed] [Google Scholar]
- Hillan J (2003) Physician use of patient-centered weblogs and online journals. Clin Med Res 1: 333–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel Boulos MN, Wheeler S (2007) The emerging Web 2.0 social software: an enabling suite of sociable technologies in health and health care education. Health Info Libr J 24: 2–23 [DOI] [PubMed] [Google Scholar]
- Kawas E, Senger M, Wilkinson MD (2006) BioMoby extensions to the Taverna workflow management and enactment software. BMC Bioinformatics 7: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chung DS (2007) Characteristics of cancer blog users. J Med Libr Assoc 95: 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HY, Marenco L, Clark T, Gao Y, Kinoshita J, Shepherd G, Miller P, Wu E, Wong GT, Liu N, Crasto C, Morse T, Stephens S, Cheung KH (2007) AlzPharm: integration of neurodegeneration data using RDF. BMC Bioinformatics 8 (Suppl 3): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HY, Marenco L, Shepherd GM, Miller PL, Cheung KH (2006) Using web ontology language to integrate heterogeneous databases in the neurosciences. AMIA Annu Symp Proc AMIA Symp 2006: 464–468 [PMC free article] [PubMed] [Google Scholar]
- McLean R, Richards BH, Wardman JI (2007) The effect of Web 2.0 on the future of medical practice and education: Darwikinian evolution or folksonomic revolution? Med J Aust 187: 174–177 [DOI] [PubMed] [Google Scholar]
- Miller V (2006) Buprenorphine and HIV primary care: report of a forum for collaborative HIV research workshop. Clin Infect Dis 43 (Suppl 4): S254–S257 [DOI] [PubMed] [Google Scholar]
- Muin M, Fontelo P (2006) Technical development of PubMed interact: an improved interface for MEDLINE/PubMed searches. BMC Med Informatics Decision Making 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjea S (2005) Information retrieval and knowledge discovery utilising a biomedical Semantic Web. Brief Bioinformatics 6: 252–262 [DOI] [PubMed] [Google Scholar]
- Neumann E (2005) A life science Semantic Web: are we there yet? Sci STKE 2005: pe22. [DOI] [PubMed] [Google Scholar]
- Neumann EK, Quan D (2006) BioDash: a Semantic Web dashboard for drug development. Pac Symp Biocomput 11: 176–187 [PubMed] [Google Scholar]
- Noy NF, Crubezy M, Fergerson RW, Knublauch H, Tu SW, Vendetti J, Musen MA (2003) Protege-2000: an open-source ontology-development and knowledge-acquisition environment. AMIA Annu Symp Proc 2003: 953. [PMC free article] [PubMed] [Google Scholar]
- Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, Li P (2004) Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics (Oxford, England) 20: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Piwowar HA, Day RS, Fridsma DB (2007) Sharing detailed research data is associated with increased citation rate. PLoS ONE 2: e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutchak TS (2005) I see blog people. J Med Libr Assoc 93: 305–307 [PMC free article] [PubMed] [Google Scholar]
- Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, Doherty D, Forsberg K, Gao Y, Kashyap V, Kinoshita J, Luciano J, Marshall MS, Ogbuji C, Rees J, Stephens S, Wong GT, Wu E, Zaccagnini D, Hongsermeier T et al. (2007) Advancing translational research with the Semantic Web. BMC Bioinformatics 8 (Suppl 3): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P (2006) caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics (Oxford, England) 22: 1910–1916 [DOI] [PubMed] [Google Scholar]
- Salzberg SL (2007) Genome re-annotation: a wiki solution? Genome Biol 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsbrook AF (2007) Open-source software for radiologists: a primer. Clin Radiol 62: 120–130 [DOI] [PubMed] [Google Scholar]
- Seringhaus MR, Gerstein MB (2007) Publishing perishing? Towards tomorrow's information architecture. BMC Bioinformatics 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Cheung KH, Yip KY, Schultz M, Gerstein MK (2007) LinkHub: a Semantic Web system that facilitates cross-database queries and information retrieval in proteomics. BMC Bioinformatics 8 (Suppl 3): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromback L, Lambrix P (2005) Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics (Oxford, England) 21: 4401–4407 [DOI] [PubMed] [Google Scholar]
- Turinsky AL, Ah-Seng AC, Gordon PM, Stromer JN, Taschuk ML, Xu EW, Sensen CW (2005) Bioinformatics visualization and integration with open standards: the Bluejay genomic browser. In Silico Biol 5: 187–198 [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P (2007) STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35: D358–D362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gorlitsky R, Almeida JS (2005) From XML to RDF: how semantic web technologies will change the design of ‘omic' standards. Nat Biotechnol 23: 1099–1103 [DOI] [PubMed] [Google Scholar]
- Wilkinson M, Schoof H, Ernst R, Haase D (2005) BioMOBY successfully integrates distributed heterogeneous bioinformatics Web Services. The PlaNet exemplar case. Plant Physiol 138: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S, Jones BF, Uzzi B (2007) The increasing dominance of teams in production of knowledge. Science (New York, NY) 316: 1036–1039 [DOI] [PubMed] [Google Scholar]
- Zhao J, Goble C, Stevens R, Turi D (2007) Mining Taverna's Semantic Web of Provenance. Concurrency Comput Pract Exp 20: 463–472 [Google Scholar]