Abstract
Gut microbiome–host metabolic interactions affect human health and can be modified by probiotic and prebiotic supplementation. Here, we have assessed the effects of consumption of a combination of probiotics (Lactobacillus paracasei or L. rhamnosus) and two galactosyl-oligosaccharide prebiotics on the symbiotic microbiome–mammalian supersystem using integrative metabolic profiling and modeling of multiple compartments in germ-free mice inoculated with a model of human baby microbiota. We have shown specific impacts of two prebiotics on the microbial populations of HBM mice when co-administered with two probiotics. We observed an increase in the populations of Bifidobacterium longum and B. breve, and a reduction in Clostridium perfringens, which were more marked when combining prebiotics with L. rhamnosus. In turn, these microbial effects were associated with modulation of a range of host metabolic pathways observed via changes in lipid profiles, gluconeogenesis, and amino-acid and methylamine metabolism associated to fermentation of carbohydrates by different bacterial strains. These results provide evidence for the potential use of prebiotics for beneficially modifying the gut microbial balance as well as host energy and lipid homeostasis.
Keywords: galactosyl-oligosaccharides, human baby microbiota, Lactobacillus paracasei, Lactobacillus rhamnosus, metabonomics
Introduction
Adult humans carry ca. 1.5 kg of gut microbial symbiotic and commensal organisms that are in intimate communication with the host metabolic and immune systems (Nicholson et al, 2005; Dethlefsen et al, 2007). This symbiosis is the result of a long period of co-evolution and co-adaptation between the host genotype and the complex and variable microbiome (Gill et al, 2006). Consequently, to be able to understand how the changes in environmental conditions and lifestyle influence human genetics and physiology, one needs to elucidate how these factors determine the distribution, activities and evolution of gut microbes, and subsequently transgenomic metabolic interactions (Xu et al, 2003; Backhed et al, 2005; Nicholson et al, 2005; Tannock, 2005; Sonnenburg et al, 2006; Blaut and Clavel, 2007; Turnbaugh et al, 2007). Thus, the gut microbiota can be regarded as an extra-genomic functional unit providing extra control mechanisms that affect the host's nutritional status and health (Holmes and Nicholson, 2005; Nicholson et al, 2005; Bik et al, 2006; Martin et al, 2006; Eckburg and Relman, 2007). We have recently reported that exogenous gut microbiome components can be transplanted into a host and this results in modulation of the host calorific bioavailability via differential metabolism of bile acids, and we and others have surmised that related metabolic processes might be involved in common metabolic diseases such as obesity or type II diabetes (Dumas et al, 2006; Houten et al, 2006; Watanabe et al, 2006; Martin et al, 2007a).
The effects of consuming live microbial supplements (probiotics) on the microbial ecology and on human health and nutritional status have been investigated extensively over many years (Collins and Gibson, 1999; Rastall, 2005; Sonnenburg et al, 2006; Martin et al, 2007b). It has been reported recently that probiotic consumption can lead to modification of the resident microbiota resulting in modulation of bile acid and lipid metabolism, and alter the recirculation and distribution of fat within the host organisms (Martin et al, 2008). Other reports suggest that the microbiota could be a contributing factor to obesity (Ley et al, 2006; Sonnenburg et al, 2006; Turnbaugh et al, 2006) and can, in addition, regulate host genes controlling lipid transport and deposition (Backhed et al, 2004).
As an alternative, the combined use of prebiotics and probiotics may have beneficial effects on health maintenance through modulating the microbial functional ecology (Collins and Gibson, 1999; Schrezenmeir and de Vrese, 2001). Prebiotics are non-digestible food ingredients, generally oligosaccharides, that modify the balance of the intestinal microbiota by stimulating the activity of beneficial bacteria, such as lactobacilli and bifidobacteria (Gibson and Roberfroid, 1995; Collins and Gibson, 1999). There is now considerable evidence that manipulation of the gut microbiota by prebiotics can beneficially influence the health of the host (Gibson and Roberfroid, 1995; Roberfroid, 1998; Delzenne and Kok, 2001; Sartor, 2004; Lim et al, 2005; Rastall, 2005; Parracho et al, 2007). In particular, many attempts have been made to control serum triacylglycerol concentrations through modification of dietary habits with regard to consumption of pre- and probiotics (Delzenne and Kok, 2001; Pereira and Gibson, 2002). Furthermore, unlike probiotics, prebiotics are not subject to biological viability problems and thus can be incorporated into a wide range of alimentary products (milk, yogurts, biscuits) and they target organisms that are natural residents of the gut microbiota (Gibson and Roberfroid, 1995). For example, oligosaccharides have been suggested to represent the most important prebiotic dietary factor in human milk, promoting the development of a beneficial intestinal microbiota (Kunz et al, 2000; Bode, 2006).
Nowadays, clinical trials support the claims of efficacy of pro- and prebiotic nutritional intervention with regard to various proposed beneficial health effects, and this has raised the requirement for providing additional evidence and for elucidation of the molecular bases of their action. This can be captured effectively only by studying the global system response of an organism to an intervention using top-down systems biology approaches. Metabolic profiling using high-resolution spectroscopic methods with subsequent multivariate statistical analyses is a well-established strategy for differential metabolic pathway profiling (Nicholson et al, 2005; Griffin and Nicholls, 2006; Ellis et al, 2007). Noticeably, the metabolic effects of various dietary modulations of gut microbiota have been successfully characterized using this approach (Wang et al, 2005, 2007; Martin et al, 2006; Stella et al, 2006; Goodacre, 2007; Rezzi et al, 2007). Recently, we have described that germ-free mice re-inoculated with a model of human baby microbiota (HBM mice) offer a simplified microbiome mouse model well adapted to assess the impact of nutritional intervention on the gut microbial functional ecosystem and subsequent effects on host metabolism (Martin et al, 2008). Interestingly, the microbiota model shows a number of similarities with the microbiota found in formula-fed neonates (Mackie et al, 1999). However, we also reported the limitations associated with gut colonization by a non-adapted microbiota and the subsequent alterations of host and microbial metabolism (Martin et al, 2007a).
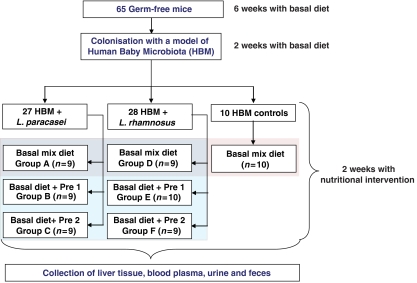
The aim of the present study is to extend our previous investigations evaluating metabolic response to probiotics in HBM mice (Martin et al, 2008). In our previous study, we had shown alterations in carbohydrate and protein fermentation with subsequent effects on host lipid and energy metabolism, which were more marked with Lactobacillus paracasei than L. rhamnosus. In the current study, we compare the effects of consumption of a synthetic galactosyl-oligosaccharide (Pre1) with those due to consumption of an in-house preparation of galactosyl-oligosaccharides (Pre2). We have assessed the impact of prebiotics on the microbial balance and the mammalian metabolism of HBM mice supplemented with a probiotic, L. paracasei or L. rhamnosus (Figure 1). Here, we show a significant association of specific metabotypes obtained from urine, plasma, fecal extracts and intact liver tissue with changes of the gut microbiome induced by the prebiotic supplementation.
Figure 1.
Schematic diagram of the experimental study design.
Results
Effects of pre- and probiotics on microbial composition and animal weight
The effects of prebiotics on the populations of microbiota in the jejunum and the feces are summarized in Table I. Fecal microbiota for the control groups (HBM alone, HBM+L. paracasei and HBM+L. rhamnosus) have previously been published (Martin et al, 2008). In the current and previous studies (Martin et al, 2008), the impact of probiotics with and without prebiotics on gut microbiota was assessed in the upper gut and the feces. The effects of probiotics are indeed expected along the whole gastrointestinal tract due to the great adaptability of lactobacilli to extreme aerobic/anaerobic conditions and low pH conditions (Tannock, 2004). However, most prebiotics are complex carbohydrates that escape digestion in the upper gastrointestinal tract and these are fermented by certain bacteria in the colon (Gibson and Roberfroid, 1995; Collins and Gibson, 1999). Nevertheless, the ability of galactosyl-oligosaccharides to modulate the upper gut microflora remains unclear and was thus also investigated. Our results provide evidence that the populations of microbiota in the fecal and jejunal content were modulated by prebiotic supplementation. In general, prebiotic supplementation slightly reduced the L. paracasei populations in both the fecal and jejunal content and increased fecal populations of Bifidobacterium breve and B. longum. Interestingly, supplementation with Pre2 was correlated with lower fecal populations of Clostridium perfringens in mice regardless of which probiotics they receive. The jejunal population of Bacteroides distasonis was decreased in HBM mice simultaneously supplemented with L. paracasei, whereas the number of fecal Escherichia coli was reduced in HBM mice simultaneously colonized with L. rhamnosus.
Table 1.
Microbial species counts in mouse fecal and jejunal contents
| Groups/log10 CFU | HBM (n=10) | HBM+ L. paracasei (n=9) | HBM+ L. paracasei +Pre1 (n=9) | HBM+ L. paracasei+ Pre2 (n=9) | HBM+ L. rhamnosus (n=9) | HBM+ L. rhamnosus +Pre1 (n=10) | HBM+ L. rhamnosus +Pre2 (n=9) |
|---|---|---|---|---|---|---|---|
| Feses | |||||||
| L. paracasei | — | 8.5±0.2 | 8.3±0.3* | 8.1±0.4** | — | — | — |
| L. rhamnosus | — | — | — | — | 7.8±0.2 | 7.6±0.4 | 7.9±0.3 |
| E. coli | 9.2±0.3 | 9.4±0.3 | 9.7±0.3 | 9.3±0.2 | 9.8±0.5 | 9.3±0.2* | 9.3±0.2* |
| B. breve | 9.1±0.2 | 7.78±2.13 | 8.5±1.5 | 8.7±1.5 | 8.7±0.3 | 9.8±0.3** | 10.0±0.4*** |
| B. longum | 8.2±0.6 | 5.6±1.9 | 6.2±1.6 | 6.7±1.8 | 6.3±0.5 | 7.7±1.2** | 9.3±1.04*** |
| S. aureus | 7.4±0.3 | 6.3±0.3 | 6.3±0.5 | 6.1±0.7 | 6.6±0.5 | 6.1±0.4 | 6.4±0.9 |
| S. epidermidis | 4.8±0.4 | 4.9±1.2 | 4.5±0.9 | 3.8±0.4 | 4.0±0.5 | 3.7±0.7 | 6.0±1.5 |
| C. perfringens | 7.2±0.3 | 7.0±0.5 | 6.5±1.0 | 5.9±0.6** | 5.7±1.0 | 6.6±1.1 | <5.0 |
| Bacteroides distasonis | 10.3±0.2 | 10.4±0.2 | 10.1±0.6 | 10.1±0.4 | 10.1±0.4 | 10.2±0.3 | 10.3±0.3 |
| Jejunum | |||||||
| L. paracasei | — | 4.2±1.8 | 2.9±0.8 | 2.6±0.9* | — | — | — |
| L. rhamnosus | — | — | — | — | 3.6±1.3 | 3.1±1.3 | 3.0±0.8 |
| E. coli | 3.3±1.3 | 5.1±1.9 | 4.0±0.8 | 3.7±1.2 | 5.2±1.9 | 4.2±1.3 | 4.4±0.8 |
| B. breve | 2.7±1.4 | 2.5±1.0 | 2.7±1.3 | 3.0±1.4 | 2.4±0.8 | 4.0±1.6** | 4.0±0.9** |
| B. longum | <2.0 | <2.0 | <2.0 | <2.0 | <2.0 | <2.0 | <2.0 |
| S. aureus | 4.1±0.9 | 3.8±1.3 | 4.1±0.9 | 3.7±0.7 | 4.2±0.7 | 3.0±1.3* | 4.0±0.5 |
| S. epidermidis | <3.0 | <3.0 | <3.0 | <3.0 | <3.0 | <3.0 | <3.0 |
| C. perfringens | 3.4±1.1 | 4.5±1.2 | 3.4±0.7 | 2.9±1.1 | 4.7±1.3 | 3.4±1.0* | 3.2±0.5* |
| Bacteroides distasonis | 3.4±1.6 | 4.8±1.6 | 3.2±1.2* | 3.3±0.9* | 3.8±1.7 | 4.0±1.3 | 4.1±1.3 |
Key: Log10 CFU (colony forming unit) given per gram of wet weight of feces or wet weight of jejunal content. Data are presented as mean±s.d. The average values obtained from the HBM+probiotics mice supplemented with prebiotics were compared with corresponding HBM+probiotics control mice, *, ** and *** designate significant difference at 95, 99 and 99.9% confidence level, respectively; —, probiotics not present in the gut microbiota.
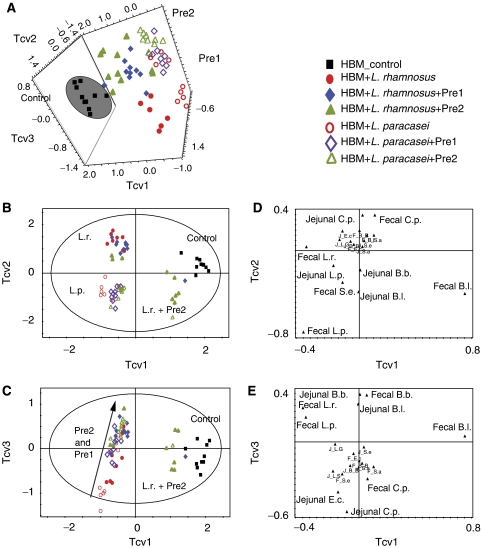
A three-component projection to latent structure discriminant analysis (PLS-DA) model of mean-centered microbial counts in fecal and jejunal contents showed that the HBM control mice samples formed a distinct cluster (Figure 2A, black squares). Two subclusters of samples representing each of the probiotics administered either alone or in combination with prebiotics were observed in the plane described by Tcv1 and Tcv2 (red circles). These groups indicated that each probiotic exerted a systematic and unique effect on the microbiota as described previously (Martin et al, 2008). For clarity, two-dimensional representations of the data scores plots are given in Figure 2B and C along with the corresponding loadings plots (Figure 2D and E). Interestingly, the combination of L. rhamnosus and Pre2 formed a subcluster along the first component Tcv1 closer to the control HBM colonized group than the other nutritional intervention groups, and this was predominantly influenced by higher fecal B. longum and lower C. perfringens populations when compared with other groups. The effects of the two prebiotics (green triangles for Pre1 and blue diamonds for Pre2) superimposed on the probiotic background were further differentiated along component Tcv3. Multivariate data analysis highlighted that prebiotic intervention was correlated with increased B. longum and B. breve, and lower numbers of E. coli and C. perfringens.
Figure 2.
PLS-DA scores plots (A–C) and loading plots for the three predictive components (D, E) derived from PLS-DA model of log10 CFU (colony forming unit) for the different bacterial species measured for fecal and jejunal samples from HBM control (black square), HBM+L. rhamnosus (red dot), HBM+L. rhamnosus+Pre1 (blue diamond), HBM+L. rhamnosus+Pre2 (green triangle), HBM+L. rhamnosus (red circle), HBM+L. rhamnosus+Pre1 (purple open diamond) and HBM+L. rhamnosus+Pre2 (green open triangle). Loadings represent the bacterial populations, beginning with J or F for jejunal or fecal counts, respectively. The model has been calculated with four predictive components and mean-centered data, RX2=76.5%, QY2=51.3%. Key: B.a., Bacteroides distasonis; B.b., Bifidobacterium breve; B.l., Bifidobacterium longum; C.p., Clostridium perfringens; E.c., Escherichia coli; L.p., Lactobacillus paracasei; L.r., Lactobacillus rhamnosus; S.a., Staphylococcus aureus; S.e., Staphylococcus epidermidis.
No effect of prebiotic supplementation on animal body weight was observed (Supplementary Table 1).
Quantification of short-chain fatty acids in the cecum
Several short-chain fatty acids (SCFAs), namely acetate, propionate, isobutyrate, n-butyrate and isovalerate, were identified and quantified from the cecal content using gas chromatography (GC) with flame ionization detection. The results, presented in Table II, are given in μmol per gram of dry cecal material and as mean±s.d. for each group of mice. The data for the control groups (HBM colonized mice without further intervention and HBM colonized mice after administration of a probiotic) have previously been published, but are included here for comparative purposes (Martin et al, 2008). The effect of prebiotic treatment on the production of SCFAs was limited to a reduction in the production of propionate and butyrate in HBM mice receiving L. rhamnosus combined with Pre2 and a reduction in isobutyrate in HBM mice receiving L. paracasei combined with Pre2 (Table II). Although cecal L- and D-lactate were not actually measured in the present study, only minor changes in lactate were observed in similar experiments with a slight reduction in cecal lactate when feeding L. rhamnosus+Pre2 (unpublished data).
Table 2.
SCFA content in the cecum from the different groups
| SCFA concentration | Acetate | Propionate | Isobutyrate | Butyrate | Isovalerate |
|---|---|---|---|---|---|
| HBM+L. paracasei (n=7) | 59.7±11.4 (65.4±4.4) | 25.3±6.6 (27.7±3.9) | 1.4±0.5 (1.6±0.4) | 1.7±0.5 (1.9±0.3) | 3.1±0.5 (3.5±0.7) |
| HBM+L. paracasei+Pre1 (n=9) | 73.4±25.0 (70.1±4.9)* | 23.9±5.3 (23.6±3.6) | 1.1±0.2 (1.1±0.2)** | 2.3±0.5 (2.3±0.6) | 2.8±0.4 (2.9±0.7) |
| HBM+L. paracasei+Pre2 (n=9) | 80.4±42.2 (72.3±5.7)* | 22±3.4 (21.8±4) | 1.1±0.1* (1.1±0.4)* | 1.6±0.9 (1.6±0.7) | 2.9±0.2 (3.1±1.2) |
| HBM+L. rhamnosus (n=9) | 40.6±8.0 (61.3±4.1) | 20.3±2.8 (31±3.7) | 0.8±0.2 (1.3±0.3) | 2.1±0.4 (3.3±0.5) | 2.1±0.5 (3.2±0.5) |
| HBM+L. rhamnosus+Pre1 (n=10) | 54.7±22.7 (68.2±5.2)** | 19±4.1 (24.9±4.1)** | 0.9±0.2 (1.2±0.3) | 1.9±0.4 (2.5±0.6)*** | 2.3±0.4 (3.2±0.9) |
| HBM+L. rhamnosus+Pre2 (n=9) | 45.3±19.5 (69.8±6)* | 14.7±4.4* (24.1±4.9)* | 0.8±0.3 (1.3±0.4) | 0.9±0.3* (1.5±0.4)*** | 2.0±0.8 (3.3±1.1) |
Keys: Data are presented as μmol per gram of dry cecal content and as means±s.d. The relative composition of SCFAs in the total content is given in percentage in parentheses. The average values obtained from the HBM+probiotics mice supplemented with prebiotics were compared with corresponding HBM+probiotics alone mice; *, ** and *** designate significant difference at 95, 99 and 99.9% confidence level, respectively.
1H NMR metabolic profiles of plasma, liver, fecal extracts and urine
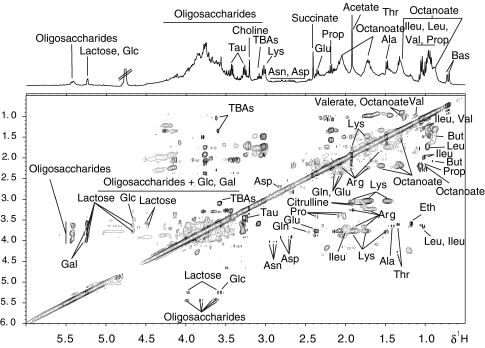
A comprehensive 1H nuclear magnetic resonance (NMR) spectral comparison was carried out using orthogonal projection to latent structure discriminant analysis (O-PLS-DA) to characterize metabolic changes associated with prebiotic supplementation and the metabolic changes were verified by two-dimensional NMR experiments as shown in Figure 3. Several amino acids, glucosides and SCFAs were readily assigned, as exemplified in Table III. Some 1H-1H cross-peaks at δ5.42:3.98, 5.52:3.79, 5.40:3.61 and 5.40:3.80 show values typical of short-chain oligosaccharides, which are as yet unidentified (Figure 3).
Figure 3.
1H-1H TOCSY NMR spectrum of a fecal extract acquired at 400 MHz. Key: Ala, alanine; Arg, arginine; Asn, asparagine; Asp, aspartate; BAs, bile acids; But, butyrate; Eth, ethanol; Gal, galactose; Glc, glucose; Gln, glutamine; Glu, glutamate; Ileu, isoleucine; Leu, leucine; Lys, lysine; Phe, phenylalanine; Pro, proline; Prop, propionate; Tau, taurine; TBAs, tauro-conjugated bile acids; Trp, tryptophan; Val, valine.
Table 3.
Summary of influential metabolites for discriminating 1H NMR spectra of liver, fecal extracts and urine
| Metabolite | Chemical shift and multiplicity | HBM+L.p. | HBM+L.p. +Pre1 | HBM+L.p.+Pre2 | HBM+L.r. | HBM+L.r. +Pre1 | HBM+L.r.+Pre2 |
|---|---|---|---|---|---|---|---|
| Plasma | QY2<0 | QY2<0 | QY2<0 | QY2<0 | |||
| Liver | QY2=52%, RX2=49% | QY2=57%, RX2=57% | QY2=38%, RX2=39% | QY2=49%, RX2=38% | |||
| Glu | 2.34(m) | 0.4±0.1 | 0.6±0.2* | 0.5±0.1 | 0.5±0.2 | 0.9±0.2** | 0.6±0.1 |
| Gln | 2.44(m) | 1.3±0.4 | 1.8±0.4* | 2.0±0.6* | 1.6±0.6 | 2.0±0.6* | 1.8±0.4 |
| TGL | 1.27(s) | 60.9±13.5 | 42.7±13.9* | 43.4±9.5** | 44.8±16.2 | 31.9±11.9 | 42.6±15.8 |
| PUFAs | 5.26(m) | 0.2±0.1 | 0.4±0.1* | 0.4±0.1*** | 0.4±0.1 | 0.5±0.1* | 0.4±0.1 |
| Ileu | 0.94(t) | 1.9±0.3 | 2.4±0.5* | 2.4±0.3* | 2.3±0.5 | 2.9±0.6a | 2.4±0.3 |
| Glycogen | 5.45 (m) | 1.1±0.4 | 1.8±1.0a | 1.5±0.4a | 2.5±1.2 | 2.1±0.1 | 1.3±0.5a |
| TMA | 2.91 (s) | 0.1±0.04 | 0.2±0.1** | 0.2±0.04*** | 0.2±0.1 | 0.2±0.04 | 0.2±0.1 |
| TMAO | 3.27(s) | 13.1±3.7 | 14.3±4.8 | 20.6±4.8** | 20.7±5.1 | 18.2±6.9 | 26.3±9.8a |
| Gly | 3.56(s) | 3.4±0.5 | 4.1±1.1 | 4.7±1.0* | 5.2±1.7 | 5.0±1.7 | 5.0±1.5 |
| Ala | 1.46(d) | 6.4±1.9 | 5.2±1.9 | 6.7±1.8 | 7.2±2.3 | 8.0±1.0 | 9.6±3.5 |
| PC | 3.20(s) | 17.6±3.5 | 16.1±2.1 | 20.8±3.9 | 19.2±4.3 | 22.5±4.9 | 24.5±5.4a |
| Feces | QY2=87%, RX2=41% | QY2=87%, RX2=40% | QY2=85%, RX2=49% | QY2=91%, RX2=43% | |||
| Oligosaccharides O3 | 3.67(m) | 5.6±0.2 | 6.1±0.2*** | 6.8±0.2*** | 5.9±0.1 | 6.1±0.1 | 7.1±0.3*** |
| Oligosaccharides O2 | 5.43(m) | 1.2±0.1 | 1.2±0.1 | 0.9±0.1*** | 1.6±0.1 | 1.2±0.2*** | 0.9±0.1*** |
| Oligosaccharides O1 | 3.94(m) | 4.7±0.2 | 5.3±0.4** | 5.7±0.2*** | 4.8±0.4 | 4.9±0.4 | 6.1±0.6*** |
| Arg | 1.66(m) | 2.0±0.1 | 2.7±0.1*** | 2.1±0.2 | 2.1±0.1 | 2.8±0.2*** | 1.9±0.2 |
| Citrulline | 3.15(t) | 1.4±0.1 | 2.1±0.1*** | 1.4±0.1 | 1.4±0.1 | 1.9±0.2*** | 1.4±0.1 |
| Octanoate | 1.27(m) | 4.2±0.7 | 2.4±0.2*** | 3.8±0.7 | 2.0±0.2 | 2.5 ±0.4*** | 3.8±0.6*** |
| Lys | 3.02(m) | 5.1±0.3 | 5.4±0.9 | 3.5±0.4*** | 4.9±0.1 | 4.4±0.9 | 3.9±0.6* |
| Butyrate | 2.16(t) | 4.4±0.3 | 4.3±0.3 | 3.8±0.2*** | 4.6±0.4 | 4.2±0.6 | 3.7±0.4** |
| Isovalerate | 2.03(t) | 4.3±0.4 | 4.0±0.3 | 3.5±0.3*** | 4.1±0.4 | 3.8±0.2 | 3.5±0.2** |
| Propionate | 1.06(t) | 4.1±0.6 | 3.5±0.7 | 2.8±0.8** | 2.9±0.7 | 2.1±0.5 | 2.2±0.6 |
| Glu | 2.34(m) | 3.0±0.3 | 3.3±0.3 | 3.1±0.1 | 3.3±0.2 | 2.8±0.2*** | 2.9±0.4** |
| Lactose | 5.23(d) | 1.9±0.2 | 1.5±0.2** | 1.6±0.3* | 2.0±0.1 | 1.3±0.1*** | 1.7±0.3* |
| Glucose | 5.23(d) | 1.3±0.1 | 1.3±0.1 | 1.1±0.1* | 1.6±0.1 | 1.3±0.1*** | 1.2±0.1*** |
| Urine | QY2=66%, RX2=50% | QY2=72%, RX2=47% | QY2=63%, RX2=45% | QY2=81%, RX2=50% | |||
| Creatinine | 4.05(s) | 15.0±3.9 | 14.3±2.9 | 10.8±5.5 | 15.9±2.4 | 11.9±7.0 | 8.4±1.9*** |
| PAG | 7.37(m) | 1.5±0.2 | 1.1±0.3* | 1.2±0.5 | 1.2±0.4 | 1.1±0.3 | 1.1±0.2 |
| Tryptamine | 7.70(d) | 0.4±0.1 | 0.2±0.1* | 0.2±0.1 | 0.3±0.2 | 0.2±0.1 | 0.2±0.1 |
| ULp | 1.27(m) | 2.9±0.5 | 1.9±0.2** | 2.2±0.3* | 1.8±0.2 | 1.8±0.3 | 2.8±0.7*** |
| Nac | 2.06(s) | 0.3±0.1 | 0.2±0.1* | 0.2±0.1 | 0.4±0.2 | 0.2±0.1 | 0.2±0.1 |
| 1-NMN | 4.48(s) | 0.7±0.1 | 1.4±0.3*** | 1.6±0.3** | 1.1±0.5 | 1.3±0.7 | 1.1±0.1 |
| Creatine | 3.92(s) | 4.5±1.5 | 4.8±1.3 | 12.2±6.0** | 3.5±0.3 | 9.9±8.0** | 8.2±3.4*** |
| Glycerate | 4.04 (m) | 5.1±0.3 | 5.3±0.3 | 7.2±1.3* | 5.2±0.5 | 5.0±0.3 | 5.4±0.4 |
| Citrate | 2.55(d) | 5.1±2.1 | 10.4±5.1* | 6.6±4.4 | 11.9±8.5 | 7.4±4.0 | 7.7±4.2 |
| TMA | 2.91(s) | 1.1±0.1 | 1.4±0.5 | 1.9±0.6** | 1.0±0.2 | 1.0±0.5 | 1.6±0.3*** |
| α-Keto-isovalerate | 1.13(d) | 4.4±1.8 | 4.0±2.1 | 2.1±0.8a | 3.7±1.7 | 2.4±1.3 | 1.8±0.4* |
| α-Keto-glutarate | 3.19(t) | 4.1±2.2 | 3.5±2.5 | 1.8±0.1 | 2.7±1.9 | 3.1±2.7 | 2.3±0.1 |
| Arg | 1.66(m) | 3.7±0.5 | 3.4±0.2 | 2.5±0.3** | 3.4±0.2 | 2.7±0.2*** | 2.3±0.1*** |
| Citrulline | 1.88(m) | 3.1±0.5 | 2.9±0.2 | 2.3±0.4* | 3.1±0.6 | 2.4±0.2* | 2.1±0.2*** |
| Taurine | 3.44(t) | 10.7±8.6 | 10.5±5.0 | 9.8±5.3 | 9.0±7.2 | 31.8±12.6** | 17.8±10.4* |
| U1 | 4.30(t) | 1.6±0.3 | 2.0±0.3* | 1.9±0.2a | 1.7±0.2 | 2.7±0.9** | 2.2±0.5* |
| U2 | 4.21(s) | 1.5±0.2 | 1.5±0.2 | 1.9±0.4a | 1.2±0.1 | 1.6±0.6 | 2.2±1.0*** |
Data are presented as area-normalized intensities (101 a.u.) of representative metabolite signals expressed as means±s.d. The values for the HBM mice supplemented with probiotics in combination with prebiotics were compared with HBM control mice fed with the probiotics alone. a, *, ** and *** designate significant difference at 90, 95, 99 and 99.9% confidence level, respectively.
Key: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets. Ala, alanine; Arg, arginine; Gln, glutamine; Glu, glutamate; Gly, glycine; Ileu, isoleucine; Leu, leucine; Lys, lysine; PUFAs, polyunsaturated fatty acids; PC, phosphocholine; TGL, triglycerides; TMA, trimethylamine; TMAO, trimethylamine-N-oxide. For abbreviations of other metabolites, refer to keys in Figures 3 and 4.
Goodness of fit (RX2) and predictability (QY2) values for O-PLS-DA models calculated separately for NMR spectroscopic data for plasma, urine, fecal extracts and intact liver tissues are presented in Table III. All the models were calculated using one predictive component and two orthogonal components using the NMR data as the X matrix and the type of prebiotics treatment (none, Pre1 or Pre2) as a dummy Y variable. No impact of prebiotic supplementation on the metabolic profiles of plasma was observed as indicated by the negative values of QY2 (Table III). However, prebiotic supplementation showed significant effects on murine metabolic profiles of urine, fecal extracts and liver, as reflected by the high values of QY2 for each model (Table III). The O-PLS-DA coefficients plots for models based on NMR spectra of fecal extracts, liver tissue and urine are presented in Figures 4 and Supplementary Figures 1 and 2 together with metabolites with the highest coefficient values responsible for the discriminatory variation listed as mean±s.d. in Table III.
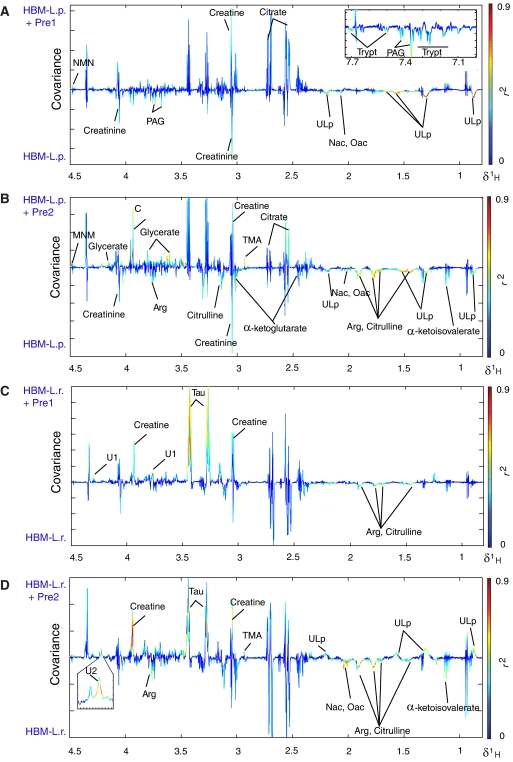
Figure 4.
O-PLS-DA coefficients for a model derived from 1H NMR spectra of urine based on the discrimination between HBM mice fed with probiotics only (negative) and HBM mice fed with probiotics and prebiotics (positive): L. paracasei supplementation with and without prebiotics Pre1 and Pre2 is shown in (A) and (B), whereas L. rhamnosus supplementation with and without prebiotics Pre1 and Pre2 is shown in (C) and (D). Key: Arg, arginine; GPC, glycerophosphorylcholine; Nac, N-acetylated glycoproteins; MNM, 1-methylnicotinamide; PAG, phenylacetylglycine; Tau, taurine; TMA, trimethylamine; Trypt, tryptamine; ULp, unidentified lipids.
Fecal metabolic profiles
Pre1 and Pre2 caused marked effects on the metabolic profiles of fecal extracts of mice colonized with HBM and L. paracasei, and these effects included a marked increase in the concentrations of some as yet unassigned oligosaccharide resonances (O1, O3), which were associated with decreased levels of resonances derived from other oligosaccharides (O2) for Pre2 (Table III and Supplementary Figure 1). In HBM mice supplemented with L. rhamnosus, Pre1 and Pre2 induced some degree of reduction in the levels of unassigned oligosaccharides (O2). Pre2 treatment was also correlated with increases in the other unassigned oligosaccharides (O1 and O3).
Further O-PLS-DA models of a pairwise comparison between Pre1 and Pre2 showed clear differences in the content of oligosaccharides O1 and O3 between the groups of HBM mice supplemented with either L. paracasei or L. rhamnosus (Supplementary Figure 3A and B). The changes in the fecal content of oligosaccharides O1 and O3 may thus result from differences in the content of products obtained from the digestion of prebiotics by the gut microbiota.
In HBM mice supplemented with L. paracasei, unique effects of Pre1 included elevated levels of arginine and citrulline, and reduced octanoic acid (caprylate) in the fecal composition, whereas unique features of Pre2 ingestion included a decrease in the levels of glucose, lysine, butyrate, isovalerate and propionate. When combined with L. paracasei supplementation, both prebiotic treatments were associated with a reduction in the content of lactose. In addition, in the feces of HBM mice supplemented with L. rhamnosus, Pre1 induced higher levels of arginine and citrulline, whereas Pre2 caused a decrease in lysine, butyrate and isovalerate. In HBM mice supplemented with L. rhamnosus, both prebiotic treatments were associated with a reduction in the content of lactose, glucose, glutamate and octanoate.
Liver metabolite profiles
The liver of mice colonized with L. paracasei and receiving either of the prebiotics was metabolically differentiated from those fed with probiotics alone, as indicated by the increased levels of glycogen, trimethylamine (TMA), polyunsaturated fatty acids (PUFAs) and a range of amino acids (i.e. leucine, isoleucine, glutamine, glutamate, glycine) and a decreased concentration of triglycerides (Supplementary Figure 2A and B and Table III). Moreover, Pre2 induced specific increases in the levels of trimethylamine-N-oxide (TMAO) in L. paracasei colonized animals.
In addition, L. rhamnosus colonized HBM mice supplemented with Pre1 were characterized by increased levels of amino acids and PUFAs. Supplementation with Pre2 was specifically associated with a reduction in the level of glycogen and an increase in TMAO and phosphatidylcholine.
Urinary metabolite profiles
Prebiotic administration also affected the urinary metabolic profiles of mice colonized with L. paracasei. These changes were mainly manifested in decreased concentrations of a putative mixture of lipids (unidentified lipids (ULp), chemical shifts: 0.89(m), 1.27(m), 1.56(m), 2.25(m)) and an increase in 1-methylnicotinamide in mice fed with Pre1 or Pre2. In addition, consumption of prebiotics was correlated with higher levels of an unknown compound U1 (1H NMR chemical shifts: 3.80(m), 4.30(t) as given by statistical total correlation spectroscopy (STOCSY) analysis; Cloarec et al, 2005a) in urine. Pre1 also caused decreased concentrations of phenylacetylglycine, N-acetyl- and O-acetyl-glycoproteins, and tryptamine and increased levels of citrate. Animals supplemented with Pre2 showed elevation in the levels of glycerate, creatine and TMA, which was associated with a reduction of α-keto-isovalerate, arginine and citrulline. In addition, consumption of Pre2 was correlated with higher levels of U1 and another unknown compound U2 (1H NMR chemical shift: 4.21(s)).
In contrast, L. rhamnosus colonized mice treated with both prebiotics showed higher urinary excretion of creatine, taurine and U1, and a reduction in urinary levels of arginine and citrulline. Feeding HBM mice with L. rhamnosus and Pre2 led to increased urinary concentrations of ULp, TMA and U2, and decreased levels of α-keto-isovalerate and creatinine.
Correlation analysis of inter-compartment metabolite functional relationships
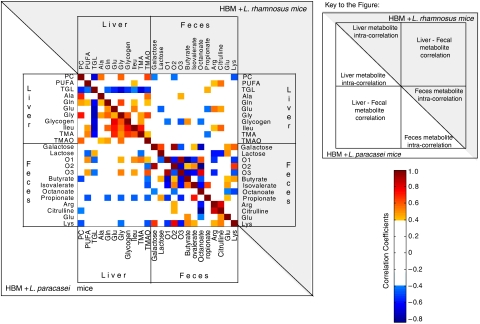
As the major changes following intervention with combined use of pre- and probiotics occurred in the fecal and liver matrices, a correlation analysis was conducted to identify any latent metabolic links between these two biological compartments (Figure 5). Such analyses have been carried out on groups of animals that received the same probiotic combined with prebiotics or not. Pixel maps obtained from the two groups of animals showed different intra- and inter-compartment correlation patterns, which highlighted the metabolic differences previously described. In HBM mice receiving L. paracasei supplementation, hepatic PUFAs and isoleucine showed positive associations with the oligosaccharide resonances O1 and O3, whereas hepatic triglycerides were negatively correlated to O1 and O3. These data suggested a direct relationship between carbohydrate digestion and liver lipid metabolism. These metabolic links were not observed in the matrix of correlations obtained from HBM+L. rhamnosus. Interestingly, negative correlations between O1 and O3 with fecal SCFAs illustrate bacterial fermentation of dietary carbohydrates in both groups. The pixel map also highlighted the positive correlations between glucogenic amino acids, glycogen and PUFAs in the liver, suggesting functional relationships between glucogenesis and gluconeogenesis.
Figure 5.
Integration of inter-compartment metabolic correlations. The pixel maps were derived from correlations between liver and fecal metabolites found to be significantly different with nutritional intervention in each group of mice colonized with one type of probiotic. The intra- and inter-compartmental metabolite correlations are displayed for HBM mice supplemented with L. paracasei probiotics down the diagonal from top-left to bottom-right, and with L. rhamnosus probiotics up the diagonal from top-left to bottom-right. The cutoff value of 0.4 was applied to the absolute value of the coefficient ∣r∣ for displaying the correlations between metabolites. Correlation values are displayed as a color-coded pixel map according to correlation value (gradient of red colors for positive values and gradient of blue colors for negative values). Key: Ala, alanine; Arg, arginine; Gln, glutamine; Glu, glutamate; Gly, glycine; Ileu, isoleucine; Lys, lysine; PC, phosphocholine; PUFA, polyunsaturated fatty acid; TGL, triglycerides; TMA, trimethylamine; TMAO, trimethylamine-N-oxide.
Correlation analysis of microbiotal variation and SCFAs
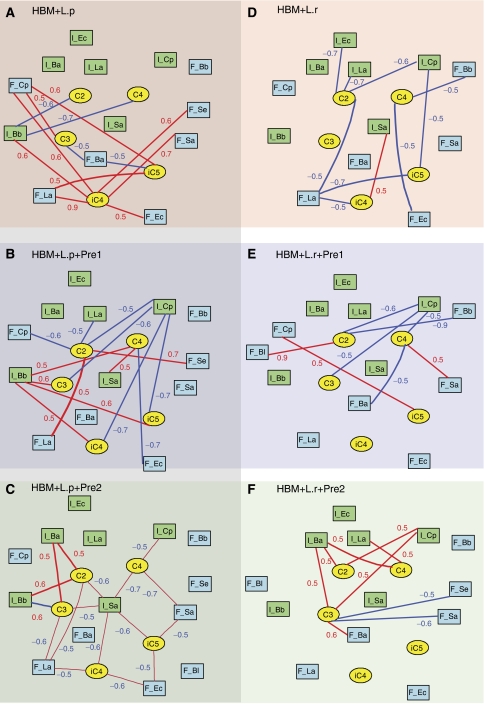
A correlation analysis was applied to investigate the connections between levels of fecal and jejunal microbiota and the cecal SCFAs using bipartite graphical modeling (Figure 6). Positive and negative correlations between nodes show the multicolinearity between SCFAs and gut bacteria, whose concentrations are interdependent such as in the case of substrate–product biochemical reactions. Correlation analysis derived from SCFAs and fecal/jejunal microbiota profiles offered a unique approach to describe intra-group sources of variability and subtle alterations in SCFAs in relation to gut bacterial changes. By comparing the networks obtained with different treatments, we can highlight significant differential patterns, suggesting different functional ecology in relation to different microbial populations and activities. HBM mice supplemented with different probiotic/prebiotic combinations show remarkably different SCFA/microbial correlation networks (Figure 6), indicating that probiotic and prebiotic modulation of the microbiome can result in specific functional ecological changes. In particular, we observed that microbial changes in the upper gut and fecal pellet showed a functional relationship with the intestinal content of SCFAs. Such data can help to generate testable hypotheses on differential bacterial metabolism in response to a stressor.
Figure 6.
Integration of SCFAs and microbiota correlations. The bipartite graphs were derived from correlations between microbiota (fecal and jejunal) and SCFAs in each of the six groups of mice. HBM mice were supplemented with L. paracasei without prebiotics (A), with Pre1 (B) and with Pre2 (C), and HBM mice were supplemented with L. rhamnosus (D), with Pre1 (E) and with Pre2 (F). The cutoff value of 0.5 was applied to the absolute value of the coefficient ∣r∣ for displaying the correlations between microbiota and SCFAs. SCFAs, and intestinal and fecal bacteria correspond to yellow ellipse nodes, and blue and green rectangle nodes, respectively. Edges are coded according to correlation value: positive and negative correlations are displayed in blue and red, respectively. Key: Ba., Bacteroides distasonis; Bb., Bifidobacterium breve; Bl, Bifidobacterium longum; C2, acetate; C3, propionate; C4, butyrate; iC4, isobutyrate; C5, valerate; iC5, isovalerate; Cp, Clostridium perfringens; Ec, Escherichia coli; La, Lactobacilli probiotics; Sa, Staphylococcus aureus; Se, Staphylococcus epidermidis. Bacterial names starting with ‘F_' and ‘I_' correspond to fecal and intestinal bacteria, respectively.
In particular, network analysis for HBM mice supplemented with L. paracasei revealed that dietary oligosaccharide supplementation induced significant changes in the functional linkage between the acetate and propionate levels, and lactobacilli, bifidobacteria, Bacteroides distasonis and C. perfringens. Interestingly, fecal bacterial changes showed strong correlations with the cecal composition of SCFAs in mice not supplemented with prebiotics (Figure 6A). Animals receiving prebiotics showed a greater number of statistically significant correlations between the jejunal microbiota changes and the SCFAs. Moreover, Pre1 induced negative and positive correlations between SCFAs and C. perfringens and B. breve, respectively (Figure 6B). Pre2 supplementation was associated with positive correlations between SCFAs (acetate and propionate) and both Bacteroides distasonis and B. breve (Figure 6C). In contrast, negative correlations were observed for S. aureus with most of the SCFAs (Figure 6C).
When HBM mice received L. rhamnosus with or without prebiotics, the microbiome/SCFAs network showed a simpler structure. Such observations suggest different bacterial interactions in both HBM+probiotic models. Available microbial data support this idea as seen, for instance, with the specific inhibition of B. breve growth with L. paracasei but not with L. rhamnosus. In contrast with animals supplemented with L. paracasei alone, variation in the intestinal bacterial populations from mice that received L. rhamnosus was strongly correlated with the cecal composition in SCFAs (Figure 6D). Moreover, Pre1 induced correlations between the cecal content in acetate and the balance between B. breve, B. longum and C. perfringens. Interestingly, Pre2 appeared to initiate functional relationships between the main SCFAs (acetate, propionate and butyrate) and Bacteroides distasonis, C. perfringens and lactobacilli.
Discussion
We have shown specific effects of two prebiotics on the microbial populations of HBM mice when co-administered with two probiotics. These microbial changes were associated with specific host metabolic phenotypes, for example, variations in the fecal carbohydrate content, reduction in the levels of hepatic triglyceride content and increased hepatic concentrations of PUFAs and hepatic glucogenic amino acids. These data provide further evidence for the critical involvement of prebiotics in host metabolism through modulation of the gut microbiome.
Effects of prebiotics on gut functional ecology
Previous work on humans has described the fragile equilibrium between the host and beneficial gut bacteria (lactobacilli, bifidobacteria) and potentially detrimental species (Clostridium spp, Staphylococcus spp and the members of the Enterobacteria and Bacteroides groups), and this ultimately determines the health and nutritional status of the host (Collins and Gibson, 1999; Pereira and Gibson, 2002; Gopal et al, 2003; Martin et al, 2006; Sonnenburg et al, 2006). Our data provide additional evidence that the populations of beneficial bacteria in the gastrointestinal tract can, to some extent, be controlled with dietary interventions, here based on supplementation with galactosyl-oligosaccharides. Here, increases in the fecal populations of beneficial bacteria, namely B. longum and B. breve (Table I), were specifically associated with supplementation of prebiotics, Pre2 offering a greater ability to modulate the gut microbiota in HBM mice compared with Pre1. Galacto-oligosaccharides are the preferred growth substrates for bifidobacteria and lactobacilli that have an extraordinary ability to acquire and degrade oligosaccharides (Kikuchi et al, 1992; Ito et al, 1993; Rycroft et al, 2001; Tzortzis et al, 2005; Macfarlane et al, 2006). Pure bacterial cultures indicated that L. rhamnosus and L. paracasei were not able to grow on culture media containing Pre1, whereas B. breve and B. longum did not show any difference in growth when cultured on Pre1, glucose or lactose media (unpublished data). In addition, the ability of B. longum (Hopkins et al, 1998; Macfarlane et al, 2008) and B. breve (Djouzi et al, 1995) to utilize extensively galacto-oligosaccharides for growth was previously reported in vivo, whereas only a few Lactobacillus species could use this substrate efficiently, unlike L. rhamnosus (Gopal et al, 2001). This bacterial capacity is strongly dependent on the pattern of glycosidic linkages present in the galacto-oligosaccharides and thus on the existence of specific β-galactosidases, as evidenced for B. longum by genome analysis (Schell et al, 2002).
It has been previously reported that bacterial fermentation of carbohydrate may result in inhibition of the growth of pathogens by acidification of the environment through the production of large quantities of carboxylic acids (Kikuchi et al, 1992; Ito et al, 1993; Rowland, 1993; Gibson and Roberfroid, 1995; Djouzi and Andrieux, 1997). In the current study, prebiotic supplementation resulted in a reduction in E. coli and C. perfringens bacterial counts in the feces (Table I), which is in agreement with previous reports (Rycroft et al, 2001; Tzortzis et al, 2005). Our results suggest that the association of Pre2 with L. rhamnosus enables a more significant reduction in pathogenic C. perfringens and an increase in health-promoting B. longum populations in feces and jejunal content. However, measures of cecal SCFAs did not reveal significant alterations in total concentrations and composition with prebiotic treatment (Table II), whereas changes were more marked in the stool (Table III). Therefore, it remains unclear if the gut microbial changes result directly from the fermentation of galactosyl-oligosaccharides and acidification of the luminal environment. Moreover, the significant increase in fecal bifidobacteria suggests that a different bacterial fermentation may occur in the colon, and measures of cecal pH and of colonic content of SCFAs will help in the interpretation in future studies.
However, our results show that the type of prebiotic (Pre1 or Pre2) entering the large intestine has differential effects on bacterial metabolism and is in agreement with the different abilities of bacteria such as lactobacilli, bifidobacteria and Bacteroides spp to hydrolyze carbohydrates, as reported previously (Hidaka and Hirayama, 1991; Djouzi and Andrieux, 1997). Application of network analysis to display the relationships between cecal SCFAs and microbial profiles revealed different intra-group patterns (Figure 6). These observations suggest that prebiotics induced a specific functional ecology in relation to different microbial populations and activities. For instance, the negative correlations between SCFAs and C. perfringens and E. coli, and positive correlations between SCFAs and bifidobacteria, lactobacilli and Bacteroides distasonis consistently indicate a link between SCFA production and certain changes in bacterial populations, such as reduction in pathogens. Moreover, C. perfringens being a primary butyrate producer, the specific anti-correlation with butyrate may also indicate that in response to stressors, C. perfringens activities may be shifted and stimulated. In particular, measures of bacterial activities in similar mouse models showed that the reduction of C. perfringens counts was associated with an increased activity in response to Pre2 supplementation (unpublished data).
In HBM mice supplemented with L. paracasei, populations of lactobacilli were slightly reduced with both prebiotic supplementations, and bifidobacteria showed only upward trends, which suggested a competition for the prebiotics between bifidobacteria and L. paracasei. Moreover, the observation of higher fecal content of oligosaccharides O1 and O3 specific to HBM mice fed with prebiotics and L. paracasei indicated that the microbiota may use these substrates poorly when compared with groups with L. rhamnosus. This information suggests a higher efficiency of bacterial hydrolysis and intestinal absorption of dietary oligosaccharides in animals fed with prebiotics and L. rhamnosus (Table III and Supplementary Figures 1 and 3).
Furthermore, ingestion of galacto-oligosaccharides or fructo-oligosaccharides is known to specifically induce bacterial hydrolysis of the substrate (Djouzi and Andrieux, 1997), as well as to modulate some bacterial activities, including glycolytic properties, hydrolysis of oligosaccharides and glucuronides, reduction of nitro-compounds, and formation of phenols and indoles (Mitsuoka et al, 1987; Ito et al, 1993; Rowland, 1993). In the current study, decreased levels of lysine in feces (Table III), isobutyrate in cecum (Table II) and N-acetyl-glycoproteins in the urine (Table III) suggest that prebiotic treatment decreased overall bacterial proteolysis in animals also receiving L. paracasei (Macfarlane et al, 1992; Hallson et al, 1997; Metges, 2000). Investigation of the metabolite changes in urinary excretion showed a significant decrease in the concentrations of microbial co-metabolites PAG and tryptamine (Goodwin et al, 1994; Smith and Macfarlane, 1996), which also supports decreased bacterial proteolytic activities. Altogether, our data suggest that prebiotics intervention may reduce proteolytic activities previously ascribed to the basal metabolism of L. paracasei on casein medium (Martin et al, 2008), which is in agreement with the reduced number of these bacteria observed in this study.
In addition, methylamines, as a class of compounds, are another well-documented example of metabolites derived from host–microbial interactions produced within the large intestine (Smith et al, 1994). A significant fraction of ingested choline is converted by microbial enzymes to TMA (Zeisel et al, 1983), which is either oxidized to TMAO in the liver or excreted into the urine (Smith et al, 1994). Increases in the levels of TMA and TMAO in the liver and TMA in the urine indicate that changes in methylamine metabolism were induced by prebiotics, the changes being more marked with Pre2 supplementation (Table III).
Impact of prebiotics on host energy and lipid homeostasis
In parallel to gut microbial changes, relative reduction of hepatic triglycerides and increased concentrations of PUFAs were observed in mice supplemented with prebiotics. Non-digestible but fermentable carbohydrates were reported to decrease triglycerides in both serum and liver via modulation of the activity and gene expression of the lipogenic enzymes (Delzenne and Kok, 1998, 2001; Roberfroid and Delzenne, 1998; Pereira and Gibson, 2002). For instance, fatty acid synthase is sensitive to nutrients and hormones (Girard et al, 1997), whereas insulin and glucose are essential factors regulating fatty acid and triglyceride synthesis (Katsurada et al, 1990; Girard et al, 1997). Moreover, fructan-type prebiotic feeding may reduce the ability of isolated hepatocytes to synthesize and secrete triglycerides by 54% (Kok et al, 1996), as well as their ability to esterify fatty acids into triacylglycerols (Fiordaliso et al, 1995). A similar mechanism might exist with galactosyl-oligosaccharides, which may explain the relative increase in the NMR signals of PUFA-containing phospholipids in the current study. PUFAs can act by directing fatty acids away from triglyceride storage and toward oxidation, and can also enhance glucose flux to glycogen (Kliewer et al, 1997). These processes are supported by the higher content of hepatic glutamine and branched-chain amino acids observed here, which would have a lower contribution to the citric acid cycle. No significant effects of prebiotics on the plasma metabolic profiles were observed here (Table III). Previously, we described that single probiotic supplementation reduced the levels of plasma lipoproteins in HBM mice by modulating the absorption of the dietary long-chain PUFAs (Martin et al, 2008). Here, we report that similar blood plasma metabolic profiles, associated with reduction of the triglyceride content in the liver, can be obtained when combining the probiotic with prebiotic supplements (Table III).
Notably, animals fed with L. paracasei in combination with prebiotics showed the most significant hepatic reduction in triglycerides, which was associated with a high fecal content of oligosaccharides. Previous studies showed that some prebiotics induce changes in lipogenic enzyme activities by reducing postprandial insulinemia and glycemia (Kok et al, 1998; Delzenne and Kok, 2001) through stimulation of the intestinal release of hormonal mediators (Morgan, 1996), or through modification of the intestinal absorption of carbohydrates (Stanley and Newsholme, 1985) and shortening small intestinal transit time (Roberfroid and Delzenne, 1998). Our results suggest that a similar mechanism may be involved in mice supplemented with prebiotics co-administered with L. paracasei, as the higher concentrations of fecal oligosaccharides may reflect poorer digestion and absorption resulting in lower energy generation from carbohydrates, with a consequent switch to fat metabolism. However, further work is needed to understand the functional link between the residual fecal carbohydrate and the digestion of prebiotics by the gut microbiota, for instance by assessing experimentally the metabolic abilities of the bacterial species to utilize the prebiotics. Moreover, recent studies showed that the microbial processing of dietary oligosaccharides modifies monosaccharide uptake from the gut by regulating the activity of host monosaccharide transporters, which can result in various changes in hepatic metabolism, including modulation of synthesis and deposition of triglycerides in adipocytes and increased glycogenesis (Backhed et al, 2004). Here, prebiotic supplementation was associated with relatively increased levels of glutamate, glutamine, branched-chain amino acids and alanine in the liver, as well as hepatic glycogen accumulation when mice were specifically fed with L. paracasei, which suggests stimulated gluconeogenesis and glycogenesis (Table III).
The animals that received L. rhamnosus in combination with either prebiotics showed elevated levels of urinary taurine and creatine, which is likely to be related to higher muscular activity due to supplementation of the feed with new sources of carbohydrates (Cuisinier et al, 2001). Moreover, it has been shown that adipocyte-derived hormones, whose expression correlates with adipocyte lipid content, can increase energy expenditure in mice (Backhed et al, 2004). These changes suggest that HBM supplemented with L. rhamnosus supplies the host metabolism with new sources of carbohydrates more efficiently, which leads to changes in energy expenditure.
In conclusion, integrative systemic metabolic and microbiome profiling demonstrated the importance of nutritional intervention based on prebiotics and probiotic combinations in determining the host metabolic status and the levels of a diverse range of compounds in multiple pathways. Our data highlight that prebiotic nutritional intervention is a key factor in determining the resulting host metabolic phenotypes. The perspective of inducing unique changes in the host metabolism triggered by unique combinations of prebiotics and probiotics establishes an important step forward in the efforts to develop tailored nutritional solutions at an individual level.
Materials and methods
Animal handling procedure and supplementation of probiotics and prebiotics
All animal studies were carried out under appropriate national guidelines at the Nestlé Research Center (Lausanne, Switzerland). The model of HBM consists of seven bacterial strains that were isolated, using a previously described method (Guigoz et al, 2002), from the stool of a 20-day-old female baby who was given birth by normal delivery and breast-fed. A total of seven bacterial species were isolated, namely E. coli, B. breve and B. longum, Staphylococcus epidermidis and S. aureus, C. perfringens and Bacteroides distasonis, and they were mixed in equal amounts (approximately 1010 cells/ml for each strain) for gavage. Bacterial cell mixtures were kept in frozen aliquots until use. L. paracasei NCC2461 and L. rhamnosus NCC4007 probiotics were obtained from the Nestlé Culture Collection (Lausanne, Switzerland).
A total of 65 C3H female germ-free mice, aged 6 weeks, received a single dose of HBM bacterial mixture and were fed with a standard semisynthetic germ-free rodent diet for 2 weeks, as described previously, to allow establishment of the HBM (Martin et al, 2008). The full trial design is given in Figure 1. A control group of HBM mice (n=10) received a saline drink containing Man, Rogosa and Sharpe (MRS) culture medium and were fed with a basal diet containing 2.5% of a glucose–lactose mix (1.25% each) for 2 additional weeks. Two groups of HBM mice were given daily a probiotic supplement, either L. paracasei (group A, n=9) or L. rhamnosus (group D, n=9), containing 108 probiotic bacteria in MRS per day mixed with saline solution and were also fed with the basal diet. Two groups of HBM mice were fed with a diet containing 3 g per 100 g diet of commercially available galactosyl-oligosaccharide prebiotics (Vivinal-GOS, Borculo Domo Ingredients, The Netherlands), called Pre1 here. Pre1 comprises 75% dry matter in syrup and on a dry matter basis is composed of 23% lactose, 22% glucose, 0.8% galactose and 57% galactosyl-oligosaccharides with a degree of polymerization (DP) ranging between 3 and 9, and primarily composed of β-1,4 linkages. Amaretti et al (2007) previously reported the relative sugar composition of Vivinal GOS. DP 3 oligomer formed a major part of the oligosaccharides used and accounted for 37% of total carbohydrates, whereas the concentration of other oligomers decreased with the increase in DP (Amaretti et al, 2007). Two additional groups of HBM mice were fed with a diet containing 3 g per 100 g diet of an in-house preparation of galacto-oligosaccharides, called Pre2 here. Pre2 is composed of 80% of Pre1 and 20% of a mixture containing additional galactosyl-oligosaccharide structures, the latter being primarily composed of DP 3 oligomers with β-1,3 and β-1,6 linkages. The control diet was supplemented with lactose and glucose to control for the lactose and glucose that were added to the experimental diets by means of galactosyl-oligosaccharide preparations. Additionally, all groups received daily a fresh probiotic, either L. paracasei (group B, n=9; group C, n=9) or L. rhamnosus (group E, n=10; group F, n=9). With an average consumption of 5 ml of drinking water per mouse per day, each animal received 108 CFU probiotics per day. Fecal pellets and morning spot urine samples were collected and frozen for NMR spectroscopy at the end of the 2 weeks of nutritional intervention. An additional fecal pellet was also collected in sterile condition for microbial profiling. Urine samples were not obtained for every animal, as some mice had an empty bladder at the time of termination (i.e. total urine samples per group was A, n=6; B, n=8; C, n=6; D, n=8; E, n=6; F, n=7). Animals were weighed and then euthanized. Blood (400 μl) was collected into Li-heparin tubes and the plasma was obtained after centrifugation and then frozen at −80°C. For jejunal microbiota analysis, the first 8 cm of the jejunum was collected into sterile tubes containing Ringer solution (Oxoid, UK), homogenized and kept on ice before microbial profiling. The cecal content was collected in Eppendorf™ tubes and a central section of the median lobe of the liver was dissected. Samples were snap-frozen immediately and kept at −80°C before analysis.
Microbial profiling of fecal and jejunal contents
Briefly, for each mouse, a fecal pellet was homogenized in 0.5 ml Ringer solution supplemented with 0.05% (w/v) L-cysteine (HCl). For fecal and jejunal samples, solutions at different dilutions were plated on selective and semiselective culture media to assess the bacterial populations, B. breve and B. longum on Eugon Tomato medium (Chemie Brunschwig, Switzerland), L. paracasei and L. rhamnosus on MRS medium (Chemie Brunschwig) with antibiotics (phosphomycin, sulfamethoxazole and trimethoprim) medium (Sigma, Switzerland), C. perfringens on NN-agar medium (Chemie Brunschwig), E. coli on Drigalski medium (Bio-Rad, Switzerland), Bacteroides distasonis on Shaedler Neo Vanco medium (BioMérieux, Switzerland) and S. aureus and S. epidermidis on Chapman medium (BioMérieux). The bacterial cultures of E. coli, S. aureus and S. epidermidis were incubated at 37°C under aerobic conditions for 24 h and those of B. longum, B. breve, L. rhamnosus, L. paracasei, Bacteroides distasonis and C. perfringens under anaerobic conditions for 48 h.
Gas chromatographic analysis of cecal content
Cecal extracts were obtained from an aliquot from the cecum with 4 ml buffer (0.1% (w/v) HgCl2 and 1% (v/v) H3PO4) containing 0.045 mg/ml 2,2-dimethylbutyric acid (as an internal standard) per gram fresh weight. The resulting slurry was centrifuged for 30 min at 5000 g at 4°C and the supernatant containing SCFAs was analyzed using a gas chromatograph (HP 6890) equipped with flame ionization detector and a DB-FFAP column (J&W Scientific, MSP Friedli & Co., Switzerland) of 30 m length, 530 μm diameter and 1 μm film thickness. The system was run with helium gas at an inlet constant pressure of 10 psi at 180°C. A cleaning injection of 1.2% formic acid was used before each analysis. Samples were run at an initial temperature of 80°C for 1.2 min followed by heating to 145°C in 6.5 min, heating to 200°C in 0.55 min and an additional 0.5 min at 200°C. SCFAs were identified and quantified using the internal standard as well as external standards consisting of acetate, propionate, isobutyrate, n-butyrate, isovalerate and n-valerate.
1H NMR spectroscopic analysis of biofluids and extracts
Plasma samples (100 μl) were introduced into a 5 mm NMR tube with 450 μl of saline solution containing 10% D2O as the locking substance. Urine samples were prepared by mixing 20 μl of samples with 30 μl of a phosphate buffer solution containing 90% D2O and 0.25 mM 3-trimethylsilyl-1-[2,2,3,3-2H4] propionate (TSP), which was used as a chemical shift reference, into 1.7 mm NMR tubes. Fecal pellets were homogenized in 650 μl of a phosphate buffer solution containing 90% D2O and 0.25 mM TSP. The homogenates were sonicated at ambient temperature (298 K) for 30 min to destroy bacterial cells and then centrifuged at 6000 g for 20 min. The supernatants were removed and centrifuged again at 6000 g for 10 min. Aliquots of 550 μl were then pipetted into 5 mm NMR tubes. Intact liver samples were bathed in an ice-cold saline D2O solution. A portion of the tissue (∼15 mg) was packed into a zirconium oxide 4 mm outer-diameter rotor.
All 1H NMR spectra were recorded on a Bruker DRX 600 NMR spectrometer (Bruker Biospin, Rheinstetten, Germany) operating at 600.11 MHz for 1H observation. 1H NMR spectra of plasma, urine and fecal extracts were acquired with a Bruker 5 mm TXI triple-resonance probe at 298 K. 1H NMR spectra of intact liver tissues were acquired using a standard Bruker high-resolution MAS probe under magic-angle-spinning conditions at a spin rate of 5000 Hz (Waters et al, 2000). Tissue samples were regulated at 283 K using cold N2 gas to minimize any time-dependent biochemical degradation.
1H NMR spectra of urine and fecal extracts were acquired using a standard one-dimensional pulse sequence (D1-90°-t1-90°-tm-90°-free induction decay (FID)). NMR spectra of plasma and tissues were acquired using the Carr-Purcell-Meiboom-Gill (CPMG, D1-90°-(τ-180°-τ-)n-FID) spin-echo pulse sequence with water suppression. Standard spectra were acquired with a relaxation delay D1 of 2 s during which the water resonance was selectively irradiated, and a fixed interval t1 of 3 μs. The water resonance was irradiated for a second time during the mixing time tm of 100 ms. CPMG spin-echo spectra were registered using a spin-echo loop time (2nτ) of 160 ms for plasma and 200 ms for tissue (Meiboom and Gill, 1958) and a relaxation delay of 2.5 s. A total of 128 and 256 transients were collected into 32K data points for standard and CPMG spectra respectively, with a spectral width of 20 ppm.
The FIDs were multiplied by an exponential weighting function corresponding to a line broadening of 0.3 Hz. The acquired NMR spectra were manually phase- and baseline-corrected using the software package XwinNMR 3.5 (Bruker Biospin), and referenced to the chemical shift of the methyl resonance of alanine at δ 1.466 for plasma and tissue spectra and that of TSP at δ 0.00 for urine and fecal extract samples.
For assignment purposes, 2D COrrelation SpectroscopY (COSY) (Nagayama, 1980) and TOtal Correlation SpectroscopY (TOCSY) (Bax and Davis, 1985) NMR spectra were acquired on selected samples using a Bruker AV 400 spectrometer operating at 400.13 MHz for 1H observation equipped with a Bruker 5 mm SEI (1H-13C) inverse probe with a z-axis field gradient coil at 298 K. Further assignment of the metabolite peaks was also accomplished with the use of STOCSY on 1D spectra.
Multivariate statistical analysis and visualization
Statistical analysis of the changes in animal weights, bacterial populations and in the cecal composition of SCFAs obtained by GC was carried out using a two-tailed Mann–Whitney test.
The 1H NMR spectra were converted into 22K data points over the range of δ 0.2–10.0 using an in-house-developed MATLAB routine. The regions containing the water resonance (δ 4.5–5.19) and, for urine spectra only, the urea resonance (δ 4.5–6.2) were removed. The spectra were normalized to a constant total sum before chemometric analyses. The multivariate pattern recognition techniques used in this study were based on the O-PLS-DA approach with unit-variance scaling (Trygg and Wold, 2003). The O-PLS-DA loadings plots were processed according to the method described by Cloarec et al (2005b). Here, the test for the significance of Pearson product-moment correlation coefficient was used to calculate the cutoff value of the correlation coefficients at the level of P<0.05. To test the validity of the model against overfitting, the cross-validation parameter Q2 was computed and the standard seven-fold cross-validation method was used (Cloarec et al, 2005b). Additional validation of the statistical modeling on urine, liver, feces and bacterial counts was performed using permutation testing based on cross-model validation methods recently published by Westerhuis et al (2008). The means of the distributions of the Q2 parameters obtained using random permutations are significantly different and lower than the experimental Q2 parameters at 95% confidence interval using a one-tailed t-test. These data provide compelling evidence of the statistical validity of the models generated, and the results are provided as Supplementary information (Supplementary Figures 4 and 5).
Pixel map representation of the inter-compartment metabolic correlation
A statistical correlation analysis was applied to normalize the intensities of spectral peaks found to be significantly different with nutritional intervention to establish possible association between metabolites across different biological compartments. Pearson's correlation coefficients were computed between influential metabolite relative intensities derived from liver and fecal metabolic profiles from the same mice within each group of mice colonized with one type of probiotic. Pixel maps were used to display the correlation matrices, and a cutoff value of 0.4 was applied to the absolute value of the coefficient ∣r∣ so that the map represents only those correlations between two metabolites that are above the cutoff. The value and the sign of the correlation were then color-coded (gradient of red colors for positive values, gradient of blue colors for negative values). The presence of colored pixels between specific metabolites reveals a correlation (above the cutoff) between these molecules that may reflect a functional association.
Bipartite graph representation of connectivities between SCFAs and microbial profiles
The bipartite graph (Rgraphsviz) package from R (Free Software Foundation General Public License, USA, Version 2.5.1) was used to display the correlation matrix derived from cecal SCFAs and microbial profiles (jejunal and fecal) to assess the prebiotic-induced changes in the microbial metabolism. Pearson's correlation coefficients were computed between cecal SCFA variables and microbiota variables from the same mice and a cutoff value of 0.5 was applied to the absolute value of the coefficient ∣r∣ so that the bipartite graph represents only those correlations between the two types of nodes (microbiota and SCFAs) that are above the cutoff (Martin et al, 2007a). The sign of the initial correlation was then color-coded (red positive, blue negative) and the correlation value displayed on the bipartite graph. In that context, presence of edges between two specific nodes (one of each type) reveals a correlation (above the cutoff) between these entities that may reflect a functional association.
Supplementary Material
Supplementary Information
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Supplementary Data 4
Supplementary Data 5
Acknowledgments
We acknowledge the help and input of Ivan Montoliu Roura, Olivier Cloarec and Marc-Emmanuel Dumas for statistical analysis; Isabelle Rochat, Catherine Murset and Gloria Reuteler for microbial analysis; and Rodrigo Bibiloni and Enea Rezzonico for helpful discussions on gut bacterial metabolism. We thank John Newell, Monique Julita, Massimo Marchesini, Catherine Schwartz and Christophe Maubert for provision of the animal facilities and expertise. This work received financial support from Nestlé (to F-PJM, YW) and from the International Study of Macro/micronutrient and Blood Pressure grant 1-R01-HL084228-01A1 (to IKSY).
References
- Amaretti A, Bernardi T, Tamburini E, Zanoni S, Lomma M, Matteuzzi D, Rossi M (2007) Kinetics and metabolism of Bifidobacterium adolescentis MB 239 growing on glucose, galactose, lactose, and galactooligosaccharides. Appl Environ Microbiol 73: 3637–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley R, Sonnenburg J, Peterson D, Gordon J (2005) Host-bacterial mutualism in the human intestine. Science 307: 1915–1920 [DOI] [PubMed] [Google Scholar]
- Bax A, Davis D (1985) MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson 65: 355–360 [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA (2006) Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA 103: 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaut M, Clavel T (2007) Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137: 751S–755S [DOI] [PubMed] [Google Scholar]
- Bode L (2006) Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr 136: 2127–2130 [DOI] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson J (2005a) Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem 77: 1282–1289 [DOI] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Trygg J, Craig A, Barton RH, Lindon JC, Nicholson JK, Holmes E (2005b) Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem 77: 517–526 [DOI] [PubMed] [Google Scholar]
- Collins MD, Gibson GR (1999) Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am J Clin Nutr 69: 1052S–1057S [DOI] [PubMed] [Google Scholar]
- Cuisinier C, Ward RJ, Francaux M, Sturbois X, de Witte P (2001) Changes in plasma and urinary taurine and amino acids in runners immediately and 24 h after a marathon. Amino Acids 20: 13–23 [DOI] [PubMed] [Google Scholar]
- Delzenne NM, Kok N (1998) Effect of non-digestible fermentable carbohydrates on hepatic fatty acid metabolism. Biochem Soc Trans 26: 228–230 [DOI] [PubMed] [Google Scholar]
- Delzenne NM, Kok N (2001) Effects of fructans-type prebiotics on lipid metabolism. Am J Clin Nutr 73: 456S–458S [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Fall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouzi Z, Andrieux C (1997) Compared effects of three oligosaccharides on metabolism of intestinal microflora in rats inoculated with a human faecal flora. Br J Nutr 78: 313–324 [DOI] [PubMed] [Google Scholar]
- Djouzi Z, Andrieux C, Pelenc V, Somarriba S, Popot F, Paul F, Monsan P, Szylit O (1995) Degradation and fermentation of alpha-gluco-oligosaccharides by bacterial strains from human colon: in vitro and in vivo studies in gnotobiotic rats. J Appl Microbiol 79: 117–127 [DOI] [PubMed] [Google Scholar]
- Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK (2006) Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA 103: 12511–12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Relman DA (2007) The role of microbes in Crohn's disease. Clin Infect Dis 44: 256–262 [DOI] [PubMed] [Google Scholar]
- Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R (2007) Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 8: 1243–1266 [DOI] [PubMed] [Google Scholar]
- Fiordaliso M, Kok N, Desager JP, Goethals F, Deboyser D, Roberfroid M, Delzenne N (1995) Dietary oligofructose lowers triglycerides, phospholipids and cholesterol in serum and very low density lipoproteins of rats. Lipids 30: 163–167 [DOI] [PubMed] [Google Scholar]
- Gibson GR, Roberfroid MB (1995) Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125: 1401–1412 [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J, Ferre P, Foufelle F (1997) Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu Rev Nutr 17: 325–352 [DOI] [PubMed] [Google Scholar]
- Goodacre R (2007) Metabolomics of a superorganism. J Nutr 137: 259S–266S [DOI] [PubMed] [Google Scholar]
- Goodwin BL, Ruthven CR, Sandler M (1994) Gut flora and the origin of some urinary aromatic phenolic compounds. Biochem Pharmacol 47: 2294–2297 [DOI] [PubMed] [Google Scholar]
- Gopal PK, Prasad J, Gill HS (2003) Effects of the consumption of Bifidobacterium lactis HN019 (DR10 (TM)) and galacto-oligosaccharides on the microflora of the gastrointestinal tract in human subjects. Nutr Res 23: 1313–1328 [Google Scholar]
- Gopal PK, Sullivan PA, Smart JB (2001) Utilisation of galacto-oligosaccharides as selective substrates for growth by lactic acid bacteria including Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20. Int Dairy J 11: 19–25 [Google Scholar]
- Griffin JL, Nicholls AW (2006) Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics 7: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Guigoz Y, Rochat F, Perruisseau-Carrier G, Rochat I, Schiffrin E (2002) Effects of oligosaccharide on the faecal flora and non-specific immune system in the elderly people. Nutr Res 22: 13–25 [Google Scholar]
- Hallson PC, Choong SK, Kasidas GP, Samuell CT (1997) Effects of Tamm-Horsfall protein with normal and reduced sialic acid content upon the crystallization of calcium phosphate and calcium oxalate in human urine. Br J Urol 80: 533–538 [DOI] [PubMed] [Google Scholar]
- Hidaka H, Hirayama M (1991) Useful characteristics and commercial applications of fructo-oligosaccharides. Biochem Soc Trans 19: 561–565 [DOI] [PubMed] [Google Scholar]
- Holmes E, Nicholson J (2005) Variation in gut microbiota strongly influences individual rodent phenotypes. Toxicol Sci 87: 1–2 [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Cummings JH, McFarlane GT (1998) Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosaccharides and other simple carbohydrate sources. J Appl Microbiol 85: 381–386 [Google Scholar]
- Houten SM, Watanabe M, Auwerx J (2006) Endocrine functions of bile acids. EMBO J 25: 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Deguchi Y, Matsumoto K, Kimura M, Onodera N, Yajima T (1993) Influence of galactooligosaccharides on the human fecal microflora. J Nutr Sci Vitaminol (Tokyo) 39: 635–640 [DOI] [PubMed] [Google Scholar]
- Katsurada A, Iritani N, Fukuda H, Matsumura Y, Nishimoto N, Noguchi T, Tanaka T (1990) Effects of nutrients and hormones on transcriptional and post-transcriptional regulation of fatty acid synthase in rat liver. Eur J Biochem 190: 427–433 [DOI] [PubMed] [Google Scholar]
- Kikuchi H, Andrieux C, Szylit O (1992) Effects of galacto-oligosaccharides on bacterial enzymatic activities and metabolite production in rats associated with a human flora. Proc Nutr Soc 51: 7A1508932 [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM (1997) Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA 94: 4318–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok N, Roberfroid M, Robert A, Delzenne N (1996) Involvement of lipogenesis in the lower VLDL secretion induced by oligofructose in rats. Br J Nutr 76: 881–890 [DOI] [PubMed] [Google Scholar]
- Kok NN, Taper HS, Delzenne NM (1998) Oligofructose modulates lipid metabolism alterations induced by a fat-rich diet in rats. J Appl Toxicol 18: 47–53 [DOI] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S (2000) Oligosaccharides in human milk: structural, functional and metabolic aspects. Annu Rev Nutr 20: 699–722 [DOI] [PubMed] [Google Scholar]
- Ley R, Turnbaugh P, Klein S, Gordon J (2006) Human gut microbes associated with obesity. Nature 444: 1023–1024 [DOI] [PubMed] [Google Scholar]
- Lim CC, Ferguson LR, Tannock GW (2005) Dietary fibres as ‘prebiotics': implications for colorectal cancer. Mol Nutr Food Res 49: 609–619 [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR, Beatty E, Cummings JH (1992) Estimation of short-chain fatty-acid production from protein by human intestinal bacteria based on branched-chain fatty-acid measurements. FEMS Microbiol Ecol 101: 81–88 [Google Scholar]
- Macfarlane GT, Steed H, Macfarlane S (2008) Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol 104: 305–344 [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT, Cummings JH (2006) Review article: prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther 24: 701–714 [DOI] [PubMed] [Google Scholar]
- Mackie RI, Sghir A, Gaskins HR (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69: 1035S–1045S [DOI] [PubMed] [Google Scholar]
- Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, Sprenger N, Fay LB, Kochhar S, van Bladeren P, Holmes E, Nicholson JK (2007a) A top-down systems biology view of microbiome–mammalian metabolic interactions in a mouse model. Mol Syst Biol 3: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Verdu EF, Wang Y, Dumas ME, Yap IK, Cloarec O, Bergonzelli GE, Corthesy-Theulaz I, Kochhar S, Holmes E, Lindon JC, Collins SM, Nicholson JK (2006) Transgenomic metabolic interactions in a mouse disease model: interactions of Trichinella spiralis infection with dietary Lactobacillus paracasei supplementation. J Proteome Res 5: 2185–2193 [DOI] [PubMed] [Google Scholar]
- Martin FP, Wang Y, Sprenger N, Holmes E, Lindon JC, Kochhar S, Nicholson JK (2007b) Effects of probiotic Lactobacillus paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res 6: 1471–1481 [DOI] [PubMed] [Google Scholar]
- Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, Kochhar S, Lindon JC, Holmes E, Nicholson JK (2008) Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol 4: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiboom S, Gill D (1958) Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum 29: 688–691 [Google Scholar]
- Metges CC (2000) Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr 130: 1857S–1864S [DOI] [PubMed] [Google Scholar]
- Mitsuoka T, Hidaka H, Eida T (1987) Effect of fructo-oligosaccharides on intestinal microflora. Nahrung 31: 427–436 [DOI] [PubMed] [Google Scholar]
- Morgan LM (1996) The metabolic role of GIP: physiology and pathology. Biochem Soc Trans 24: 585–591 [DOI] [PubMed] [Google Scholar]
- Nagayama K (1980) Experimental techniques of two-dimensional correlated spectroscopy. J Magn Reson 40: 321–334 [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID (2005) Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 3: 431–438 [DOI] [PubMed] [Google Scholar]
- Parracho H, McCartney AL, Gibson GR (2007) Probiotics and prebiotics in infant nutrition. Proc Nutr Soc 66: 405–411 [DOI] [PubMed] [Google Scholar]
- Pereira DI, Gibson GR (2002) Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit Rev Biochem Mol Biol 37: 259–281 [DOI] [PubMed] [Google Scholar]
- Rastall RA (2005) Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol Ecol 52: 145–152 [DOI] [PubMed] [Google Scholar]
- Rezzi S, Ramadan Z, Martin FP, Fay LB, van Bladeren P, Lindon JC, Nicholson JK, Kochhar S (2007) Human metabolic phenotypes link directly to specific dietary preferences in healthy individuals. J Proteome Res 6: 4469–4477 [DOI] [PubMed] [Google Scholar]
- Roberfroid MB (1998) Prebiotics and synbiotics: concepts and nutritional properties. Br J Nutr 80: S197–S202 [PubMed] [Google Scholar]
- Roberfroid MB, Delzenne NM (1998) Dietary fructans. Annu Rev Nutr 18: 117–143 [DOI] [PubMed] [Google Scholar]
- Rowland I (1993) The effects of transgalactosylated oligosaccharides on gut flora metabolism in rats associated with a human fecal microflora. J Appl Bacteriol 74: 667–674 [DOI] [PubMed] [Google Scholar]
- Rycroft CE, Jones MR, Gibson GR, Rastall RA (2001) A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J Appl Microbiol 91: 878–887 [DOI] [PubMed] [Google Scholar]
- Sartor RB (2004) Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology 126: 1620–1633 [DOI] [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F (2002) The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA 99: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeir J, de Vrese M (2001) Probiotics, prebiotics, and synbiotics—approaching a definition. Am J Clin Nutr 73: 361S–364S [DOI] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT (1996) Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol 81: 288–302 [DOI] [PubMed] [Google Scholar]
- Smith JL, Wishnok JS, Deen WM (1994) Metabolism and excretion of methylamines in rats. Toxicol Appl Pharmacol 125: 296–308 [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI (2006) Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol 4: e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley JC, Newsholme EA (1985) The effect of dietary guar gum on the activities of some key enzymes of carbohydrate and lipid metabolism in mouse liver. Br J Nutr 53: 215–222 [DOI] [PubMed] [Google Scholar]
- Stella C, Beckwith-Hall B, Cloarec O, Holmes E, Lindon JC, Powell J, van der Ouderaa F, Bingham S, Cross AJ, Nicholson JK (2006) Susceptibility of human metabolic phenotypes to dietary modulation. J Proteome Res 5: 2780–2788 [DOI] [PubMed] [Google Scholar]
- Tannock GW (2004) A special fondness for lactobacilli. Appl Environ Microbiol 70: 3189–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW (2005) Commentary: remembrance of microbes past. Int J Epidemiol 34: 13–15 [DOI] [PubMed] [Google Scholar]
- Trygg J, Wold S (2003) O2-PLS, a two-block (X–Y) latent variable regression (LVR) method with an integrated OSC filter. J Chemom 17: 53–64 [Google Scholar]
- Turnbaugh P, Ley R, Mahowald M, Magrini V, Mardis E, Gordon J (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449: 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzortzis G, Goulas AK, Gee JM, Gibson GR (2005) A novel galactooligosaccharide mixture increases the bifidobacterial population numbers in a continuous in vitro fermentation system and in the proximal colonic contents of pigs in vivo. J Nutr 135: 1726–1731 [DOI] [PubMed] [Google Scholar]
- Wang Y, Lawler D, Larson B, Ramadan Z, Kochhar S, Holmes E, Nicholson JK (2007) Metabonomic Investigations of aging and caloric restriction in a life-long dog study. J Proteome Res 6: 1846–1854 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Nicholson JK, Hylands PJ, Sampson J, Holmes E (2005) A metabonomic strategy for the detection of the metabolic effects of chamomile (Matricaria recutita L.) ingestion. J Agric Food Chem 53: 191–196 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439: 484–489 [DOI] [PubMed] [Google Scholar]
- Waters NJ, Garrod S, Farrant RD, Haselden JN, Connor SC, Connelly J, Lindon JC, Holmes E, Nicholson JK (2000) High-resolution magic angle spinning 1H NMR spectroscopy of intact liver and kidney: optimisation of sample preparation procedures and biochemical stability of tissue during spectral acquisition. Anal Biochem 282: 16–23 [DOI] [PubMed] [Google Scholar]
- Westerhuis JA, Hoefsloot HC, Smit S, Vis DJ, Smilde AK, van Velzen EJ, van Duijnhoven JP, van Dorsten FA (2008) Assessment of PLSDA cross validation. Metabolomics 4: 81–89 [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chaing HC, Hooper LV, Gordon JI (2003) A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299: 2074–2076 [DOI] [PubMed] [Google Scholar]
- Zeisel SH, Wishnok JS, Blusztajn JK (1983) Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 225: 320–324 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Supplementary Data 4
Supplementary Data 5