Abstract
Many biological networks can maintain their function against single gene loss. However, the evolutionary mechanisms responsible for such robustness remain unclear. Here, we demonstrate that antagonistic host–parasite interactions can act as a selective pressure driving the emergence of robustness against gene loss. Using a model of host signaling networks and simulating their coevolution with parasites that interfere with network function, we find that networks evolve both redundancy and specific architectures that allow them to maintain their response despite removal of proteins. We show that when the parasite pressure is removed, subsequent evolution can lead to loss of redundancy while architecture-based robustness is retained. Contrary to intuition, increased parasite virulence hampers evolution of robustness by limiting the generation of population level diversity in the host. However, when robustness emerges under high virulence, it tends to be stronger. These findings predict an increased presence of robustness mechanisms in biological networks operating under parasite interference. Conversely, the presence of such mechanisms could indicate current or past parasite interference.
Keywords: coevolution, computational methods, host–parasite interaction, robustness, signaling networks
Introduction
In several organisms, gene deletion studies suggest a large fraction of genes to be dispensable (Giaever et al, 2002; Alonso et al, 2003; Kamath et al, 2003; Wilson et al, 2005). Although part of this observation stems from genes that are required under environments not assayed in the lab, a still appreciable number of genes can be lost with seemingly no phenotypic effect (Hoffmann, 1991; Joyner et al, 1991; Goldstein, 1993; Cadigan et al, 1994). Such robustness can result from duplicate genes that maintain a functional overlap despite molecular divergence (redundancy) (Wagner, 2000a; Gu et al, 2003; Papp et al, 2004) or from system architecture (Edwards and Palsson, 1999; Emmerling et al, 2002). The emergence and maintenance of these two mechanisms pose significant challenges for evolutionary biology (de Visser et al, 2003). Previous work has shown that genetic mutational load could act as a weak selective pressure for redundancy and can result in the emergence and maintenance of robustness under specific conditions (Nowak et al, 1997; Wagner, 2000b). However, no explanation currently exists for how evolution can lead to specific network architectures that are robust against deletion of parts. Furthermore, although fluctuating environments are suggested to drive the evolution of robustness in metabolic networks (Harrison et al, 2007), the relation between ecological factors and robustness is not addressed in detail.
Among the many potential ecological factors that could influence the evolution of networks, parasites stand out for several reasons. Parasite-imposed selective pressure on the host is usually high, as the fitness reductions that result from failure to deal with the parasite are often severe. Conversely, the fitness reductions of parasites that fail to infect, survive and transmit themselves in and among hosts are even more severe (Anderson and May, 1991). This strong antagonistic fitness interaction, combined with the fact that both parasite virulence and host susceptibility are strongly determined by genes, results in a never-ending arms race at molecular level (Schmid-Hempel and Ebert, 2003; de Wit, 2007). This arms race, usually referred to as Red Queen dynamics (Bell, 1982), is a driving force responsible for the selection of many fundamental traits such as sexual reproduction (Jaenike, 1978; Salathé et al, 2008a). At the molecular level, the coevolution gives rise to parasite proteins that promote entry to and growth in the host system, and host proteins that recognize parasite and initiate an immune response against it. It has been suggested that parasites might have lost the race to avoid direct recognition by hosts and have instead shifted their focus to interfering with the host pathways that are mediating defence (Schmid-Hempel, 2005). Indeed, there is ample empirical evidence for parasite interference with host pathways (Sacks and Sher, 2002; Bhavsar et al, 2007; Marques and Carthew, 2007). For example, parasitic protozoa Leishmania and Toxoplasma gondii are shown to inhibit the activity of specific host kinases (Olivier et al, 1992) and block nuclear localization of specific transcription factors (Denkers et al, 2004), respectively. The bacterial pathogens Salmonella enterica and Yersinia have proteins that act as kinases and phosphotases for proteins in the intracellular signaling pathways of their hosts (Terebiznik et al, 2002; Prehna et al, 2006), thereby altering cellular responses. Parasites also seem to have evolved strategies to downregulate expression (Stern-Ginossar et al, 2007) or inhibit proper folding (Tardif and Siddiqui, 2003) of specific host proteins. Ultimately, all these interference strategies will disturb the dynamics and the final response of the host signaling networks, thereby reducing the fitness of the host and benefiting that of the parasite.
Here, we explore the consequences of parasite interference with host signaling networks on the evolution of the latter. In particular, we would like to understand (i) whether parasite interference can lead to evolution of networks with robust responses, (ii) how exactly these networks would achieve robustness and (iii) which parameters would affect the evolution of robustness. Standard population genetic models, which have been successfully applied to the emergence of redundancy in one- or two-loci systems (Nowak et al, 1997; Wagner, 2000b), are not adequate to capture the complexity of such a system. To overcome this limitation, here we use models of host signaling networks and computer simulations that allow us to capture the structure–behavior relation in these networks and their evolution under parasite interference, respectively. We find that robustness against gene loss can evolve in networks, and that it results from either redundancy or specific network structures. We show that robustness based on the latter is maintained over subsequent evolution even in the absence of parasites, whereas redundancy is lost. Furthermore, we find parasite virulence to be a key parameter having a double impact on the evolution of robustness: high virulence limits the emergence of robustness by hampering diversity in the host population, but at the same time it selects for stronger robustness. These findings indicate that antagonistic host–parasite interactions can be a strong ecological factor shaping the global structure of biological networks. Furthermore, they point to an interesting interplay between population level diversity and emergence of complex traits and the influence of parasite virulence on this interplay.
Results
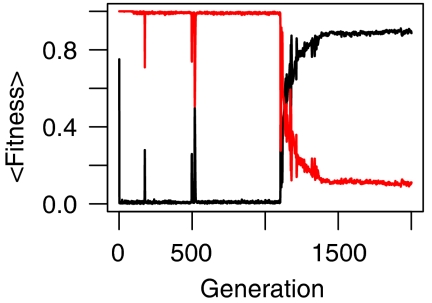
We used a mathematical model of signaling networks to study the consequences of antagonistic host–parasite interaction at the molecular level. Briefly, this model assumes that a signaling network consists of proteins that exist in two forms, active and inactive (Soyer and Bonhoeffer, 2006; Soyer et al, 2006). Each protein, when active, can influence the state of other proteins in the network with which it interacts. Two arbitrary proteins are taken as receptor and effector, allowing the network behavior to be quantified as changes in the active effector concentration in response to incoming signals received at the receptor (see the section Network model). We ran evolutionary simulations starting with a homogenous host population, where each organism contained the same founder network of four proteins, and a heterogeneous parasite population, where each organism contained a single protein interfering with one specific protein in the host network. Host networks were free to evolve through biologically plausible mutations, whereas parasites could evolve only the specificity of their interference (i.e. the identity of the protein they target in the host; see the section Evolutionary simulations). The fitness of a host was defined as the ability of its network to produce the same dynamical response to an incoming signal as that of the founder, in the presence of parasite interference (see Equation (4)). To account for the fact that the fitness effects of parasite interference on network dynamics might be of different strength, we adjust the effects of the parasite by a global parameter, which we refer to as the virulence of the parasite. By doing so, we follow the common notation of virulence as parasite-induced fitness reduction in the host (Anderson and May, 1991; Bull, 1994). Parasite fitness relates directly to the effect it has on network dynamics of its host (see Equation (5)). The resulting antagonistic fitness relation quickly led to the emergence of highly fit parasites that interfered with a key protein in the host network (see Figure 1). As evolution proceeded, hosts slowly improved their fitness, eventually escaping the parasite interference. The resulting host networks were able to maintain a high fitness upon random removal of single proteins, that is, they are robust to gene loss (see Figure 2). For the sample simulation shown, the most frequent network in the final population could maintain its response to an incoming signal similar to that of the founder when any one of its proteins was removed (see Supplementary Figure 1).
Figure 1.
Host and parasite fitness (black and red lines, respectively) for a sample simulation with virulence equal to 1. For robustness of networks from the final host population resulting from this simulation, see Figure 2. For the dynamics of the most frequent network in the final population, see Supplementary Figure 1.
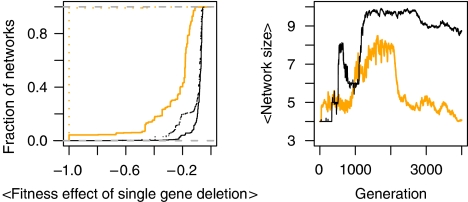
Figure 2.
Left: The average fitness effect of single gene deletion on the networks of the population. Straight lines show the data for the population at the end of evolution with parasites (generation 2000 on the right panel). Dashed lines show the data for the population at the end of subsequent evolution without parasites (generation 4000 on the right panel). Orange and black indicate simulations starting from the same founder but with virulence values 0.1, and 1.0, respectively. The straight black line corresponds to the simulation shown in Figure 1. Data are shown as empirical cumulative distributions; each vertical line represents the fraction of networks in the population for which average fitness effect of a gene deletion is below, or equal to, the value shown on the x-axis. Right: Network size (i.e. number of proteins in the network) averaged over the host population during the course of evolution with (first 2000 generations) and without (last 2000 generations) parasites.
Achieving such complete robustness requires redundancy at each protein or a specific system architecture that could tolerate the loss of each of the proteins. Although we find both mechanisms evolving in the host networks, only the latter seems to be maintained over evolution. Subsequent evolution of sample host populations—taken from simulations where coevolution led to high robustness—in the absence of parasites but under stabilizing selection (see Materials and methods) showed that robustness was either lost almost completely or barely affected. In the former case, evolution in absence of parasites resulted in a significant reduction in the average network size, strongly indicating that loss in robustness was due to loss of redundant proteins (see Figure 2). In contrast, host populations that maintained high robustness also maintained network size, suggesting that in these cases, robustness was due to network architecture. These indications were further supported by the analysis of most frequent network architectures in the host population at the end of evolution with and without parasites (see Figure 3). These results suggest that architecture-based robustness is more stable than pure redundancy and can be maintained independent of the generative selective pressures. Once a specific network architecture arises that is robust against parasite interference, it could not be changed over subsequent evolution even when parasites are absent because the network has to maintain specific dynamics (that of the founder). On the other hand, purely redundant proteins can be easily lost without any dynamical constraints. Fitness data from single protein deletions in the absence of parasites indicate that networks exhibiting redundancy-based robustness consist of more proteins whose removal has exceedingly small fitness effects than networks exhibiting architecture-based robustness (data not shown). Thus, despite showing high robustness, networks with redundancy-based robustness can become smaller at a faster rate than networks with architecture-based robustness, because in the former, a higher proportion of deletions are effectively neutral (i.e. the selection coefficient is smaller than one over population size).
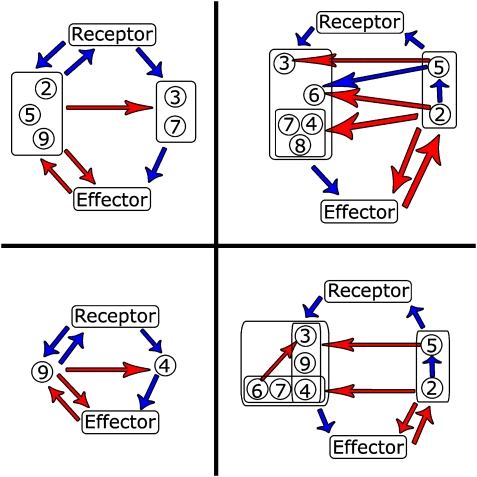
Figure 3.
Cartoon representation of most frequent networks for the simulations shown in Figure 2. Top: Most frequent network in the final population resulting from the simulation with parasite virulence equal to 1.0 (right) and 0.1 (left). Bottom: Most frequent networks in the corresponding final populations resulting from subsequent evolution in the absence of parasites. Red and blue arrows represent activating and inhibiting interactions, respectively. Arrows pointing from a single protein to a box with many proteins inside indicate that the former interacts with all of the proteins inside the box. Note that proteins with no outgoing interactions are not shown in these cartoons for clarity, as these proteins do not affect network behavior (see Supplementary information).
Repeating the evolutionary simulations with more than 60 different founder networks and with different virulence values (see Materials and methods), we find that the evolution of neither mechanism is trivial; in 40% of the simulations we ran, hosts were not able to escape parasite pressure, and robustness against gene loss did not evolve. What are the determining factors behind this observation? One possibility is that founder network properties and/or the stochastic nature of the evolutionary dynamics dictate the outcome of these simulations. As network evolution is akin to a random move in the topology space, both the starting location and the path taken might matter (Wagner, 2005). Although we see some effect of stochasticity in the emergence of robustness (see Supplementary Figure 6), we could not find any clear relation between evolution of robustness and the founder network properties. An alternative explanation is that the emergence of robustness relates to the global virulence parameter. In the presented model, virulence controls the effect of parasite interference on the host fitness and effectively tunes the strength of the selective pressure for robustness. Hence, we would expect that increased virulence, by means of increasing selective pressure, should favor evolution of robustness. Surprisingly, we found the opposite, that is, evolution of robustness was hampered under high virulence. This paradox was resolved once we analyzed the diversity in host populations.
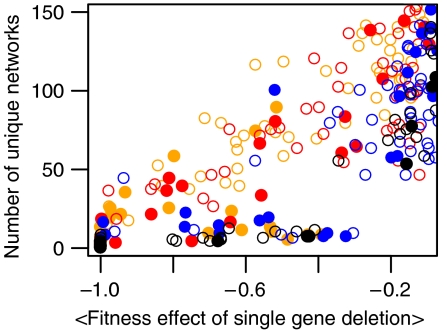
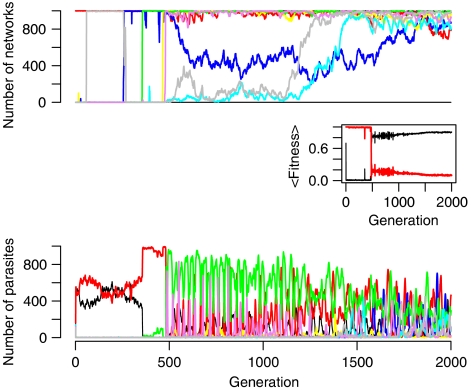
As shown in Figure 4, we found a high correlation between structural diversity in the population (number of networks with a unique structure) and emergence of robust networks (average correlation coefficient (R2) is 0.83). It was very typical in these evolutionary simulations that emergence of high host fitness (i.e. robustness) was always preceded by an increase in diversity. This is demonstrated in Figure 5 for a sample simulation, where we show the number of networks utilizing a given protein and the number of parasites interfering with that protein over the course of evolution along with the fitness of the two populations. Early on, the parasite population is dominated by two genotypes that are interfering with the two intermediary proteins in the founder network. As it is highly likely that any mutant network arising in the host population will also utilize these key proteins, it is difficult for such mutants to arise in frequency and generate some diversity in the host population. However, even when a small amount of diversity is generated in the host population, it allows other parasite genotypes to coexist with the dominant ones. This decreases the selective pressure on the host population, further enhancing generation of diversity. The resulting feedback eventually leads to a Red Queen dynamics where several host and parasite genotypes enter an oscillatory cycle of negative frequency-dependent selection (see Figure 5). The side effect of the resulting diversity in the host population is a more efficient search of the aforementioned topology space, eventually leading to robust networks and an increase in fitness. As high virulence makes initial emergence and establishment of mutant networks more difficult, it hinders the emergence of diversity and consequently robustness. However, if hosts manage to generate some diversity under high virulence and break parasite pressure early, they tend to evolve higher robustness than under low virulence (see Supplementary Figure 2). Hence, increased parasite virulence acts as a two-sided sword, both limiting the emergence of precursors of robust networks by hampering population diversity and promoting evolution toward higher robustness by providing increased selective pressure for it.
Figure 4.
Number of unique network topologies in the final host population versus the average fitness effect of a single gene loss averaged over the final host population. Points with different colors represent data from simulations with different virulence; for orange, red, blue and black, virulence equals 0.1, 0.2, 0.5 and 1.0, respectively. Open (filled) circles represent simulations where the average fitness of the final host population is above (below) that of the parasite population (i.e. indicating hosts escaping parasite pressure).
Figure 5.
Diversity in host and parasite population shown as the number of networks in the host population that utilizes a given protein (top) and the number of parasites interfering with a given protein in the host network (bottom). Each colored line represents a protein identity with the color coding in two panels being the same; black, red, blue, yellow, green, cyan, violet and gray represent proteins 2, 3, 4, 5, 6, 7, 8 and 9. Note that receptor and effector in the host network are excluded from this plot, as they are not involved in parasite targeting (see Supplementary information). The inset shows the mean fitness for host (black) and parasite (red) population. Data are from a sample simulation with virulence equal to 1.
What is the significance of parasite interference for the evolution of robustness against gene loss in host networks? As parasites provide an ever-changing and coadapting environment for their hosts, it is possible that such robustness could also result simply from evolution under changing or multiple environments. A similar argument is made in the case of metabolic networks, namely that the evolution toward metabolizing multiple sources (i.e. adaptation to a new environment) might result in robustness against gene loss as a by-product (Harrison et al, 2007). Alternatively, robustness against gene loss could result as a response to genetic mutations or developmental errors. To test such alternative scenarios and understand the specific effect of parasites on the evolution of robustness against gene loss, we ran additional simulations. As summarized in Table I and Supplementary Figure 3, we do not find any significant amount of robustness against gene loss when networks evolve under stabilizing selection or under environmental fluctuations, modeled as changing fitness requirements (see Materials and methods). The former result is interesting, as another kind of robustness (that against fluctuations in kinetic rates) can evolve under stabilizing selection (Wagner, 1996; Siegal and Bergman, 2002). To have a more direct comparison to the parasite scenario, we create an alternative environment model where a given environment corresponds to knockout of a specific gene in the network (see Materials and methods). Biologically, this scenario corresponds to the environment containing a toxin or drug that interferes with a specific protein in the network. The environment fluctuates over time as the specificity of the drug changes. When such changes are modeled to occur as frequent as every generation, this scenario would correspond to developmental errors (Nowak et al, 1997). We find that networks evolving under this scenario become highly robust to gene loss, when fluctuations occur as frequent as every 200 generations or faster (see Table I and Supplementary Figure 3). However, parasite interference tends to result in stronger robustness more often (see Supplementary Figure 4). Among the 10 simulations considered, the average fitness effect of deletions (e.g. robustness) over the population reached −0.3 or higher seven times for parasite interference but only three times for environmental fluctuations (for k=50). Furthermore, we find that environmental robustness is more readily lost upon subsequent evolution compared with that evolved under parasite interference. Assuming that simulations where evolution under interference resulted in a mean robustness of −0.5 or higher and where further evolution did not reduce robustness to levels below this threshold correspond to evolutionarily stable robustness, we found 37.5% stability in robustness evolving under environmental fluctuations with k=50 (33% for k=200). In the 10 simulations starting from the same founders, parasite-driven robustness had a stability of 57%.
Table 1.
Average robustness calculated from simulations corresponding to different evolutionary scenarios (see Materials and methods)
| Parasite interference | Environmental fluctuation A |
Environmental fluctuation B |
Mutational load |
||||
|---|---|---|---|---|---|---|---|
| k=200 | k=50 | k=1 | k=200 | k=50 | μ=0.005 | μ=0.05 | |
| −0.35 | −0.45 | −0.40 | −0.38 | −0.80 | −0.85 | −0.99 | −0.97 |
Each robustness value corresponds to the fitness effect of single gene deletion averaged over the networks found in the final populations from 10 simulations. For each evolutionary scenario, the considered simulations start from the same 10 founder networks. Environmental fluctuations A and B correspond to interference and changes in fitness requirements, respectively, whereas the combination of the former with k=1 corresponds to developmental errors (see main text).
Taken together, these findings show that interference with network dynamics can lead to evolution of robustness against gene loss and that such robustness might be maintained even after removal of interference (i.e. be evolutionarily stable). Following the empirical evidence that parasites interfere with host networks, we show how and under which conditions host–parasite coevolution leads to evolution of robustness and predict an increased presence of robustness mechanisms in biological networks operating under parasite interference.
Discussion
Here we have shown that antagonistic interaction between parasites and their host at molecular level may lead to host networks evolving both architecture- and redundancy-based robustness toward gene loss. The emergence and the extent of these robustness strategies will depend on the virulence of the parasites and the ability of the host to generate diversity at molecular level. The key prediction of this study is that signaling networks (and other biological systems) operating under parasite interference would show increased redundancy or specific architectures that can tolerate removal of parts. Conversely, presence of such properties in a network could be taken as a sign of current or past parasite interference. In particular, we point out that in at least one specific plant–viroid system, parasite success seems to relate to the redundancy of host proteins (Kalantidis et al, 2007). Specific knockout experiments in this and similar systems can verify the relation between redundancy in the host and the fitness of the parasite.
The main assumptions of the model are that the parasite exerts a specific and strong interference with a single protein in the host network and benefits directly from this. Hence, our findings would apply to any host–parasite system in which these conditions are met (see also Materials and methods). Although we do not include protein sequences in the model, we expect that generation of sequence diversity would be another strategy for the host to avoid parasite interference. Interestingly, signal transducers are observed to show gene number expansion and increased sequence diversity in disease-vector mosquitoes (Waterhouse et al, 2007).
We find that another possible evolutionary force that could generate such robustness against gene loss in host networks is fluctuations in environmental factors that interfere with network dynamics (i.e. cause knockout of a specific protein). To evolve robustness that is similar in strength and stability to that observed under host–parasite coevolution, such fluctuations would have to occur as frequent as every 50 generations. Interestingly, we do not find any robustness evolving under mutational load (even when mutation rate is very high) or under fluctuating fitness requirements. The latter would correspond to changes in dynamic requirements from the network (e.g. the need to lengthen response), which could result from environmental or internal fluctuations.
To conclude, this study clearly demonstrates the need to consider evolutionary dynamics and ecological factors, such as host–parasite interactions and environmental constraints, for achieving a better understanding of system level properties in molecular biology.
Materials and methods
Network model
To capture the dynamics and structure of host networks, we use a generic mathematical model that has been described in detail before (Soyer and Bonhoeffer, 2006). Briefly, the model describes a network as a collection of n interacting proteins, each of which can be in an active (Pi*) or inactive state (Pi). When active, each protein can activate or deactivate any of the other proteins, depending on an interaction coefficient. Biologically, such an effect can be due to phosphorylation, methylation or any other type of chemical or structural interaction. The concentration of the active and inactive forms of each protein adds up to 1 (i.e. [Pi*]=1−[Pi]). Two proteins (the first and the last) in the network are arbitrarily defined as receptor, where the network interacts with an external signal, and effector, where a resulting network response is measured. The biochemical dynamics of the active protein concentrations in the network are governed by the set of differential equations of the form
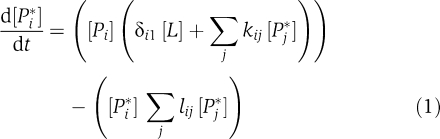 |
where the interaction coefficients kij and lij denote the strength of activation and deactivation, respectively, of protein j on protein i, [L] is the signal concentration and δ is the Kronecker delta (i.e. δi1=1 when i=1 and δi1=0 otherwise). It is assumed that the effect of protein j on protein i is either activating or deactivating, but not both (i.e. kijlij=0), and that proteins cannot have an effect on themselves (i.e. kij=lij=0 when i=j). Finally, the maximum value that an interaction coefficient can attain is set to 1.
To quantify network behavior (or dynamics) D, we use three measures: (i) the steady-state activity before a signal, (ii) the response in the presence of a signal and (iii) the steady-state activity after the signal. Biologically, this means that to be deemed functional a network should have stable dynamics and the ability to produce a detectable response to incoming signal (defined here as at least 10% of the maximum possible). To obtain D for a given network, we first set [Pi*]=[Pi]=0.5 for all proteins in the network and [L]=0. We equilibrate the system by integrating the set of differential equations resulting from (1) for 1000 iterations or until steady state is reached (determined by an eigenvalue analysis) and record the active effector concentration at the end of this period as the pre-signal steady state of the system, [Peff*]pre. We then introduce a signal by setting [L]=1 and integrate for 100 iterations, after which the signal disappears (i.e. [L]=0). During these 100 iterations, we deduce the response of the network as the sum of absolute differences between the current active effector concentration and [Peff*]pre, that is, the dynamic response of a network to an incoming signal, r, is given by
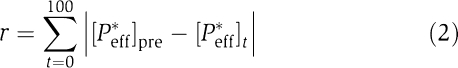 |
After the signal has passed, we let the system equilibrate again for 1000 iterations or until steady state is reached. We record the active effector concentration at the end of this period as the post-signal steady state of the system [Peff*]post. With these measurements, we can write network dynamics as
In summary, the presented network model captures the basic biochemistry of biological networks and allows us to quantify their function using the dynamic response to incoming signals. Although this model is inspired by biological signaling networks, at an abstract level it can be used to capture the dynamics of any biological system of interacting units such as gene networks. The basic assumptions of the model are that interacting molecules are found in two states (e.g. on and off) and that each molecule could interact with any other. The latter assumption allows generality in the model without imposing limitations; every network structure that could be constructed in the presented model could be constructed in nature (possibly requiring a larger system).
Evolutionary simulations
To study the coevolution of parasite and host at molecular level, we run evolutionary simulations, starting from initial populations of the host and parasite. The initial host population is homogeneous and contains N replicates of a founder individual (reported results are for N=1000). A host organism is modeled simply as a biochemical network that has the form described above. The network of the founder individual is a random, functional network that consists of four proteins including receptor and effector. This random network is generated by creating each of the possible connections among the four proteins with probability 0.5. If an interaction is generated, an interaction coefficient (between −1 and 1) is chosen randomly to represent type and strength of the interaction (positive values correspond to activation, negative values correspond to deactivation). No direct interaction is allowed between effector and receptor. The dynamics of such founder network is considered the wild-type dynamics Dwt for the host population.
The initial parasite population is heterogeneous (reflecting its faster evolution) and contains N randomly generated individuals. Parasites are modeled as organisms with a single gene that specifies which protein it interferes with in the host network (the target protein x of the parasite). Biologically, parasite interference can have different forms such as suppressing gene expression (Stern-Ginossar et al, 2007), inhibiting protein folding (Tardif and Siddiqui, 2003) or competitive binding at protein active site (or at its substrate) (Terebiznik et al, 2002; Denkers et al, 2004; Prehna et al, 2006). All these different types of interferences at protein level will influence network dynamics, as the interplay of the affected protein with other proteins will be halted or reduced. Here we model an extreme case of interference, where we set all interaction coefficients in the host network that involve the target protein to 0. In other words, parasite interference blocks all activity of the targeted protein. Modeling interference in such a way is motivated by the fact that when a parasite trait is introduced in the coevolutionary simulations that codes for the strength of interference, it quickly leads to evolution of parasites with maximum interference ability (data not shown). Furthermore, we assume that parasite interference at one target protein will also affect any duplicates of it that may emerge during evolution (see below) equally (i.e. interference is carried on to the duplicates of x). The parasite is never allowed to interfere with the effector or the receptor of a network, as the host has no means of escaping such interference in this model (see below). In summary, this model allows us to capture the complexity of the parasite–host interactions envisaged here. The often used population genetics approach, where host and parasite interact at least at two loci with at least two alleles, and a specified interaction matrix defines whether an infection occurs or not (see for example, Frank, 1994; Agrawal and Lively, 2002; Kouyos et al, 2007; Salathé et al, 2008b), is not adequate for the analysis presented here.
After the host and parasite populations are initialized, an evolutionary simulation is run for 2000 generations. At each generation, every host in the population is infected with a parasite that is drawn randomly from the parasite population and the resulting interaction is used to determine the fitness of both host and parasite. The host fitness is defined as the ability of its network to adhere to wild-type dynamics after parasite interference. It is calculated as
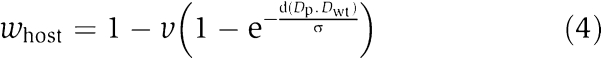 |
where v refers to parasite virulence and d is a function to capture the difference between the current dynamics obtained after parasite interference (Dp) and that of the wild type (Dwt). The latter returns the sum of the absolute difference between current and wild-type steady-state values and the response. The parameters σ and virulence define the strength of selection imposed by network dynamics and parasites, respectively. Here, we report results from simulations where σ was set to 1. In general, increasing values of σ would result in decreasing selection toward attaining the wild-type dynamics and decreasing fitness effects of the parasite. Hence, we find that increasing σ (weak selection on network dynamics) helps the emergence of robustness whereas decreasing σ impairs it (data not shown). Hosts that harbor networks with wild-type dynamics will have maximum fitness irrespective of virulence and σ values (i.e. d(Dp, Dwt)=0 and w=1). Hosts with a non-functional network, as defined above, are assigned a minimal fitness (of 10−10). Fitness of a parasite is simply given by its ability to disturb the network dynamics of its host and is calculated as
For parasites that were involved in multiple interactions (i.e. infections), the final fitness value is taken as the average over all interactions.
At the end of each generation, new host and parasite populations are produced from the current one using random drawing with replacement. A random individual is picked up from the population and is cloned into the new population if a randomly generated number (from the interval [0,1]) is below its fitness. Then it is put back into the current population and a new draw is made. The process is continued until the new population contains N individuals. During replication of individuals, mutations can occur. For parasites, mutations occur with a probability of 0.01 and result in a random change in their target, x, in the host network. For asexually reproducing hosts, mutations occur with a probability of 0.005 per protein in their network. These modeling choices result in parasites evolving twice as fast as their host, and reflect the general biological observation that parasites evolve faster than their hosts. In reality, the exact difference in the rate of evolution will be determined by several factors, including number of parasite generations per host generation, susceptibility of the host and infectivity of the parasite. We find that evolution of robustness is impaired with increasing relevant rate of parasite evolution, but that hosts can still evolve robustness when parasites are evolving as high as 20-fold faster (see Supplementary Figure 5).
When a mutation occurs in the host, it will cause one of the following outcomes at network level (with the corresponding probabilities in parentheses): deletion (P=0.4), formation (P=0.1) or adjustment (P=0.2) of an interaction, and deletion (P=0.2) or duplication (P=0.1) of a protein. These probabilities are arbitrarily chosen to represent the generally accepted view that in general deleterious mutations are more common. The observation that mutations leading to deletion of interactions and proteins are more deleterious than insertions has been shown before for similar models (Soyer, 2007).
Mutations involving an interaction are modeled by choosing a random protein A (that is already part of the network) and then choosing a random protein B (that is either part of the network or not). If the mutation to be introduced involves interaction formation, it is made sure that an interaction between A and B does not yet exist, and that A and B are not duplicates. We then create this interaction with a random interaction coefficient from the interval [−1,1]. For mutations resulting in interaction deletion or adjustment, A and B are selected such that there exists at least one interaction between them. In the first case, the new interaction coefficient is set to 0, whereas in the latter case, the new interaction coefficient results from adding a Gaussian noise (mean 0, s.d. 0.1) to the original value. In both cases, the changes made are carried over to all duplicates of A and B accordingly. Biologically, this corresponds to the situation where a mutation in protein A (B) changes the way it interacts with protein B (A)—clearly, all duplicates of B (A) are affected equally.
Deleting a protein is modeled by setting the coefficient of all incoming and outgoing interactions of that protein to 0. Duplicating a protein is implemented by picking a random protein in the current network and duplicating it with all its existing interactions. A new protein generated in this way will be considered a duplicate of the original one as long as the two have the same interactions and the total difference in interaction coefficients is less than a set threshold (reported results are with a threshold of 0.1). As duplicate relations also have an effect on parasite–host interaction (see above), this threshold could influence the evolutionary dynamics in these simulations. However, sample simulations with a threshold of 0.5 indicated that reported results are qualitatively robust to the value of this parameter.
Throughout mutations at network level, duplication of effector and receptor, and formation of a direct interaction between the two are not allowed. The maximum number of proteins in any host network is limited to 10. We have run evolutionary simulations under four different parasite virulence values (0.1, 0.2, 0.5 and 1.0) for each of the 63, randomly generated, founder hosts. Furthermore, we have repeated evolutionary simulations five times for two selected founders, resulting in 284 simulations. The latter runs indicate that the qualitative conclusions drawn here are robust to the stochastic nature of these evolutionary simulations (see Supplementary Figure 6).
Alternative evolutionary scenarios
To further analyze evolution of robustness against gene loss, we tested several alternative scenarios. First, we simulated network evolution under stabilizing selection. In these simulations, host fitness was calculated as
where d is the difference between the current and the wild-type dynamics (Dwt) as before. In this case, current network dynamics simply reflect the status of the network as it evolves in the absence of any external influence (i.e. in the absence of parasites or environmental effects).
Second, we simulated network evolution under the effect of fluctuating environments. In particular, we modeled the environment (i) as the fitness requirement under which networks evolve (in other words, the dynamics of the wild type (Dwt)) or (ii) as deletion of a specific protein in the network. The latter scenario would correspond to the environment having a toxin or drug that would interfere with the function of a given protein. Under this scenario, changes in the environment would correspond to changes in the specificity of the toxin. We implemented this scenario as a homogenous parasite population that does not evolve. The first scenario is biologically easier to imagine and simply suggests that the environment determines the functional selective pressures under which the network evolves. In this scenario, changes in the environment would simply correspond to changes in the dynamics required from the network.
We run simulations under each scenario for 10 arbitrarily chosen founders (see Supplementary Figure 3). We implemented environmental fluctuations by changing the environment every k generations. Under the first scenario, this meant adding a Gaussian noise to the three dynamical properties determining network dynamics (namely, steady-state levels of active effector and network response) so that a new wild-type dynamics (Dwt) is generated. Under the second scenario, change in the environment involved changing the identity of the protein being targeted. At every kth generation, we picked the most frequent gene in the population at that moment as the new target (we pick the second most frequent protein if most frequent is the same as the current target). We run simulations for k=200 and 50. Finally, we used the second scenario with k=1 to model developmental errors.
Robustness analyses
To evaluate their robustness toward loss of genes, we subjected networks to single gene deletions. For each network, we first reduced the network to its core by removing all proteins that did not have any outgoing interactions, as these do not affect network dynamics (i.e. networks would be naturally robust to their removal). We then deleted every intermediary gene that participated in this core network, excluding the receptor and effector, one at a time. The final robustness of the network was calculated using
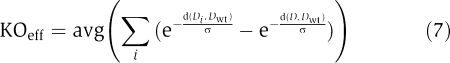 |
where Di corresponds to the network dynamics resulting from deletion of protein i and D corresponds to the network dynamics without any deletion. Giving the average fitness effect of a single deletion, values of KOeff that are close to 0 indicate higher robustness of the network dynamics against gene loss. Note that in the robustness calculation, we used the same value for σ as in the corresponding evolutionary simulations.
It is also possible to analyze the dynamical effects of single protein deletions by plotting the response (i.e. active effector concentration) of the resulting disturbed networks. Supplementary Figure 1 demonstrates such an analysis for the most frequent network found in the final host population, resulting from a sample evolutionary simulation. Considering only those simulations where hosts were able to escape the parasite pressure (i.e. mean fitness of the final host population was higher than that of parasites), we find that the mean KOeff for the host population increases with increasing virulence (see Supplementary Figure 2).
Evolutionary stability of robustness
To test whether evolved robustness in host networks is evolutionarily stable, we evolved final host populations resulting from the coevolutionary simulations for another 2000 generations in the absence of any parasite but under stabilizing selection (see above).
Evolving final populations from 34 sample coevolutionary simulations further, we find that in 11 cases all the networks in the population completely lost their robustness against gene loss. Such loss was always accompanied by a reduction in the average network size. Eleven cases resulted in minimal or no reduction in robustness and network size remaining steady over evolution. The remaining cases gave intermediate results where reductions in both robustness and network size were observed.
Code and data availability
All simulations are written in Java. Source code and results from sample evolutionary simulations are available as Supplementary informaton.
Supplementary Material
Supplementary Figure 1–6
Supplementary Information
Acknowledgments
We are grateful to Csaba Pàl for critical comments on the manuscript and Michela Denti for pointing out the viroid literature. MS acknowledges the support of the Swiss National Science Foundation. OSS acknowledges the support of Italian Ministry of University and Research.
References
- Agrawal A, Lively CM (2002) Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res 4: 79–90 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, NY) 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM (1991) Infectious Diseases of Humans. Oxford: Oxford University Press [Google Scholar]
- Bell G (1982) The Masterpiece of Nature: The Evolution and Genetics of Sexuality. Berkeley: University of California Press [Google Scholar]
- Bhavsar AP, Guttman JA, Finlay BB (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449: 827–834 [DOI] [PubMed] [Google Scholar]
- Bull JJ (1994) Perspective—virulence. Evolution 48: 1423–1437 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ (1994) Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proc Natl Acad Sci USA 91: 6324–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, Gibson G, Hansen TF, Krakauer D, Lewontin RC, Ofria C, Rice SH, von Dassow G, Wagner A, Whitlock MC (2003) Perspective: evolution and detection of genetic robustness. Evol Int J Org Evol 57: 1959–1972 [DOI] [PubMed] [Google Scholar]
- de Wit PJ (2007) How plants recognize pathogens and defend themselves. Cell Mol Life Sci 64: 2726–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkers EY, Butcher BA, Del Rio L, Kim L (2004) Manipulation of mitogen-activated protein kinase/nuclear factor-kappaB-signaling cascades during intracellular Toxoplasma gondii infection. Immunol Rev 201: 191–205 [DOI] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (1999) Systems properties of the Haemophilus influenzae Rd metabolic genotype. The Journal of Biological Chemistry 274: 17410–17416 [DOI] [PubMed] [Google Scholar]
- Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wuthrich K, Bailey JE, Sauer U (2002) Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol 184: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA (1994) Recognition and polymorphism in host–parasite genetics. Philos Trans R Soc London 346: 283–293 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goldstein LS (1993) Functional redundancy in mitotic force generation. J Cell Biol 120: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Harrison R, Papp B, Pal C, Oliver SG, Delneri D (2007) Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci USA 104: 2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FM (1991) Drosophila abl and genetic redundancy in signal transduction. Trends Genet 7: 351–355 [DOI] [PubMed] [Google Scholar]
- Jaenike J (1978) An hypothesis to account for the maintanence of sex within populations. Evol Theory 3: 191–194 [Google Scholar]
- Joyner AL, Herrup K, Auerbach BA, Davis CA, Rossant J (1991) Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science (New York, NY) 251: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Kalantidis K, Denti MA, Tzortzakaki S, Marinou E, Tabler M, Tsagris M (2007) Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J Virol 81: 12872–12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kouyos RD, Salathé M, Bonhoeffer S (2007) The Red Queen and the persistence of linkage-disequilibrium oscillations in finite and infinite populations. BMC Evol Biol 7: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Carthew RW (2007) A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet 23: 359–364 [DOI] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388: 167–171 [DOI] [PubMed] [Google Scholar]
- Olivier M, Brownsey RW, Reiner NE (1992) Defective stimulus–response coupling in human monocytes infected with Leishmania donovani is associated with altered activation and translocation of protein kinase C. Proc Natl Acad Sci USA 89: 7481–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2004) Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429: 661–664 [DOI] [PubMed] [Google Scholar]
- Prehna G, Ivanov MI, Bliska JB, Stebbins CE (2006) Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell 126: 869–880 [DOI] [PubMed] [Google Scholar]
- Sacks D, Sher A (2002) Evasion of innate immunity by parasitic protozoa. Nat Immunol 3: 1041–1047 [DOI] [PubMed] [Google Scholar]
- Salathé M, Kouyos RD, Bonhoeffer S (2008a) The state of affairs in the kingdom of the Red Queen. Trends Ecol Evol (in press) [DOI] [PubMed] [Google Scholar]
- Salathé M, Kouyos RD, Regoes RR, Bonhoeffer S (2008b) Rapid parasite adaptation drives selection for high recombination rates. Evol Int J Org Evol 62: 295–300 [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P (2005) Natural insect host–parasite systems show immune priming and specificity: puzzles to be solved. BioEssays 27: 1026–1034 [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P, Ebert D (2003) On the evolutionary ecology of specific immune defence. Trends Ecol Evol 18: 27–32 [Google Scholar]
- Siegal ML, Bergman A (2002) Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci USA 99: 10528–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer OS (2007) Emergence and maintenance of functional modules in signaling pathways. BMC Evol Biol 7: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer OS, Bonhoeffer S (2006) Evolution of complexity in signaling pathways. Proc Natl Acad Sci USA 103: 16337–16342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer OS, Pfeiffer T, Bonhoeffer S (2006) Simulating the evolution of signal transduction pathways. J Theor Biol 241: 223–232 [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O (2007) Host immune system gene targeting by a viral miRNA. Science (New York, NY) 317: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif KD, Siddiqui A (2003) Cell surface expression of major histocompatibility complex class I molecules is reduced in hepatitis C virus subgenomic replicon-expressing cells. J Virol 77: 11644–11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terebiznik MR, Vieira OV, Marcus SL, Slade A, Yip CM, Trimble WS, Meyer T, Finlay BB, Grinstein S (2002) Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat Cell Biol 4: 766–773 [DOI] [PubMed] [Google Scholar]
- Wagner A (1996) Does evolutionary plasticity evolve? Evolution 50: 1008–1023 [DOI] [PubMed] [Google Scholar]
- Wagner A (2000a) Robustness against mutations in genetic networks of yeast. Nat Genet 24: 355–361 [DOI] [PubMed] [Google Scholar]
- Wagner A (2000b) The role of population size, pleiotropy and fitness effects of mutations in the evolution of overlapping gene functions. Genetics 154: 1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A (2005) Circuit topology and the evolution of robustness in two-gene circadian oscillators. Proc Natl Acad Sci USA 102: 11775–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A et al. (2007) Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science (New York, NY) 316: 1738–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Ching YH, Farias M, Hartford SA, Howell G, Shao H, Bucan M, Schimenti JC (2005) Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res 15: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1–6
Supplementary Information