Abstract
Insights into the process of HIV-1 neutralization may assist in rational vaccine design. Here, we compared antibody neutralization against the JR-FL primary isolate and trimer binding affinities judged by native PAGE. Monovalent Fabtrimer binding and neutralization showed a direct quantitative relationship, implying that neutralization begins as each trimer is occupied by one antibody. At saturation, three Fab or soluble CD4 molecules engaged each trimer. In contrast, a maximum of one soluble CD4 molecule bound to functional SIV trimers with a truncated a gp41 tail. Remarkably, soluble CD4 was found to trigger dramatic enhancement of this virus. Unlike Fabs, a quantitative correlation between JR-FL trimer binding and neutralization was unclear for certain IgGs, where neutralization was markedly increased, but trimer affinity was largely unchanged. We discuss the implications of these findings in weighing the relative contributions of size, multivalent binding and other possible effects of IgGs to explain their increased potency.
Keywords: HIV-1, SIV, Envelope, glycoprotein, gp120, gp41, antibody, neutralization, stoichiometry, occupancy
INTRODUCTION
Antibodies (Ab) neutralize viruses by binding surface structures, thereby blocking infection (Burton, Saphire, and Parren, 2001; Parren et al., 1998). For human immunodeficiency virus type 1 (HIV-1), neutralization occurs by Ab occupation of functional gp120/gp41 envelope glycoprotein (Env) trimers (Fouts et al., 1997; Parren and Burton, 2000; Poignard et al., 1996). Abs can also recognize non-functional forms of Env present on HIV-1 particle surfaces. In this case, Ab binding does not result in neutralization (Herrera et al., 2003; Moore et al., 2006; Nyambi et al., 1998; Poignard et al., 2003).
The few monoclonal antibodies (mAbs) reported to date capable of broad and potent neutralization of HIV-1 primary isolates include IgG1b12 (directed to an epitope overlapping the CD4 binding site of gp120) (Burton et al., 1994), 2G12 (directed to high mannose carbohydrate epitope of gp120) (Scanlan et al., 2003), 2F5 and 4E10 (directed to adjacent epitopes at the C-terminal ectodomain of gp41) (Muster et al., 1993; Zwick et al., 2001). Neutralization can occur by various mechanisms. IgG1b12 blocks attachment to the primary CD4 receptor on cells, 2G12 blocks coreceptor binding (Binley et al., 2006) and gp41 mAbs neutralize a fusion intermediate complexed with both CD4 and coreceptor (Binley et al., 2003; Crooks et al., 2005; Frey et al., 2008). MAbs directed to the V3 loop and CD4-induced epitopes show intermittent activity, usually at quite high concentrations (Binley et al., 2004; Labrijn et al., 2003; Xiang et al., 2002).
It is thought that 3 molecules of neutralizing Ab (nAb) bind to trimers at saturation. In contrast, non-neutralizing epitopes are buried by Env intersubunit interactions and are therefore only exposed on non-functional components such as gp120 (Bou-Habib et al., 1994; Schonning et al., 1999; Yang et al., 2005a). Interestingly, one report showed that only one molecule of soluble CD4 (sCD4) binds to SIV gp140 trimers, suggesting an exception to 3:1 trimer-ligand stoichiometry (Kim et al., 2001). However, another report showed multimeric binding (Earl, Doms, and Moss, 1992). Moreover, the stoichiometry of CD4 binding to native SIV trimers is unknown.
Many groups have attempted to understand the quantitative aspects of trimer binding and neutralization (Fouts et al., 1997; Klasse and Sattentau, 2002; Kwong et al., 2002; McInerney et al., 1997; Ou et al., 2006; Parren et al., 1998; Salzwedel et al., 2000; Sattentau and Moore, 1995; Schonning et al., 1999; Spenlehauer et al., 2001; Yang et al., 2005a; Yang et al., 2005b, Klasse, 2007 #643; Yang et al., 2006b). Among the questions addressed are; how many subunits need to be occupied to neutralize each trimer?, how many spikes per particle need to be occupied by mAbs to neutralize the virus?, and the effect of mechanism and Ab specificity. Many of these questions remain unresolved.
A variety of techniques have provided insights into the process of neutralization. Some groups have captured inhibitor- and receptor-bound trimers (Frey et al., 2008; Furuta et al., 1998; Mkrtchyan et al., 2005). Others have visualized ligand binding by gel filtration, co-immunoprecipitation, analytical ultracentrifugation and isothermal titration calorimetry (Kim et al., 2001; Kwong et al., 2002; Pancera et al., 2005; Srivastava et al., 2007; Srivastava et al., 2002). Crystallography has revealed the structures of nAbs, alone and in complex with Env proteins and peptides (Calarese et al., 2003; Kwong et al., 1998; Ofek et al., 2004; Saphire et al., 2001; Zhou et al., 2007). However, functional trimers have not been investigated by any of these methods to date, owing to the difficulties in generating sufficient quantities, purification, and instability. Recently, however, native trimer structures (Zanetti et al., 2006; Zhu et al., 2006) and trimer complexes with neutralizing ligands (Bennett et al., 2007) have been visualized by cryoelectron tomography.
The outcome of neutralization assays can be dramatically influenced by the specific conditions of the assay. For example, target cells with a low CCR5 surface density may extend the lifetime of the CD4-liganded trimers, allowing certain Abs to inhibit more effectively (Binley et al., 2004; Choudhry et al., 2006). A post-CD4 binding neutralization format can similarly enhance the effect of some Abs (Crooks et al., 2005; Decker et al., 2005). The size of the ligand may also affect its potency. In some cases, smaller inhibitors can better access cryptic epitopes, as exemplified by mAb X5 that detectably neutralizes as a monovalent Fab, but not as an IgG (Labrijn et al., 2003). On the other hand, IgG1b12 neutralizes much more effectively than its monovalent Fab counterpart (Burton et al., 1994). High valency versions of sCD4 fused with IgG constant domains also neutralize more effectively than sCD4 monomer (Arthos et al., 2002; Bennett et al., 2007; Trkola et al., 1995). A dodecameric CD4 protein was reported to cross-link trimers on separate particles, perhaps also “inactivating” any other trimers that happen to be close to the apposing membranes of the cross-linked particles, as they would not be capable of engaging target cell surfaces (Bennett et al., 2007). Even more surprising was the observation of increased particle "rupturing" by this dodecameric CD4 protein (Bennett et al., 2007). Overall, however, the relative contributions of valency, affinity, avidity, physical bulk or other factors to the increased neutralizing activities of IgGs and other multivalent molecules remain largely unresolved.
In this article, we investigated the quantitative relationship of trimer binding and neutralization. We also examined the effects of ligand specificity, size and valency on trimer binding and neutralization. We also looked at mAb binding to sCD4-trimer complexes, the stoichiometry of sCD4 binding to HIV and SIV trimers and the consequences of 1:1 or 3:1 sCD4:trimer stoichiometry on infection.
RESULTS
Behavior of Trimers from Various Isolates in Native PAGE
In previous reports, we described native PAGE as a method to directly visualize ligand binding to HIV particle-derived functional trimers (Crooks et al., 2007; Crooks et al., 2005; Moore et al., 2006). In addition to trimers, we found various forms of non-functional Env on HIV-1 surfaces, including gp120/gp41 monomer and gp41 stumps. To establish any patterns that might relate to Env phenotype, we initially compared our JR-FL prototype to Envs from various other isolates in BN-PAGE. We chose gp160ΔCT versions of Env analogous to that used for JR-FL (Moore et al., 2006), because expression and processing into gp120/gp41 is more efficient. Previous comparisons of live inactivated virus preparations and pseudoviruses, including those with tail truncations and “SOS” disulfide mutations in native PAGE and virus capture assays suggest that the infectious pseudovirions exactly mirror the behavior of live primary isolates (Binley et al., 2003; Crooks et al., 2005; Moore et al., 2006; Poignard et al., 2003). Furthermore, truncated JR-FL trimers were found to retain comparable function and neutralization resistance to their full-length counterparts (Crooks al., 2007; Crooks et al., 2005; Moore et al., 2006).
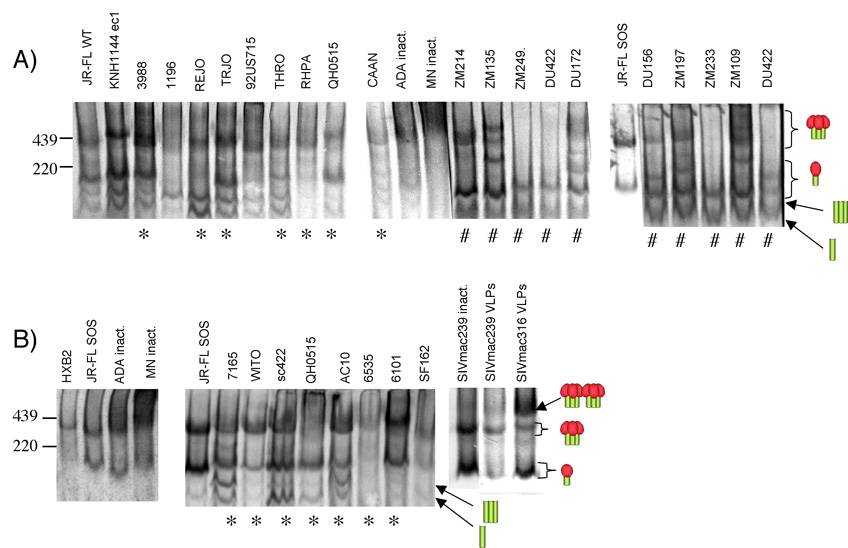
A comparison of trimers from clades A, B and C and SIVmac239 and SIVmac316 pseudoviruses, and inactivated HIV-1 MN and ADA preparations by BN-PAGE is shown in Fig. 1. Many of the clade B and all of the C isolates are derived from published virus panels developed for standardized measurement of neutralization activity (Li et al., 2005; Li et al., 2006). In some cases, trimers formed sharp bands similar to JR-FL (e.g. KNH1144 ec1 (Brown et al., 2005), REJO, TRJO, WITO, 6101, DU156 and ZM214). Three neutralization-sensitive trimers (MN, 1196, and SF162) were rather diffuse, perhaps indicating more "open" trimer structures. On the other hand, HXB2 trimers were somewhat sharper, like JR-FL. Some trimers were efficiently expressed like JR-FLSOS (e.g. KNH1144 ec1, 3988, TRJO, ZM214, and AC10), while others were poorly expressed or absent (e.g. 1196, ZM249, DU422, ZM233, 6535, and SF162). In some cases, the lack of a clear trimer band is mirrored by low infectivity of the pseudoviruses (e.g. isolates ZM249, ZM233, and 6535 each give infectious virus counts of <60,000 relative light units; R.L.U.). However, others (1196, DU422, and SF162) gave rather high infectious counts (>150,000 R.L.U.). This may be because 1) the small quantity of trimers present are highly fusogenic (possibly SF162), 2) the trimers are particularly diffuse and therefore difficult to visualize (1196, DU422), or 3) because trimers may be unusually labile and fall apart in native PAGE (1196, DU422).
Fig. 1. Behavior of native Env from various clade A, B and C HIV-1 viruses and SIVmac239 and SIVmac316 in native PAGE.
Gp160ΔCT Env trimers derived from concentrated pseudovirion stocks were examined in BN-PAGE with reference to our JR-FL gp160ΔCT prototype. JR-FL gp160ΔCT trimers and monomers and gp41 trimers and monomers (Moore et al., 2006) are indicated by cartoons. In part A, and the middle section of part B, blots were probed with a cocktail of HIV+ donor plasmas. In the left and right panels of part B, blots were probed with either the HIV-1 gp120 mAb or SIV mAb cocktails, as described in materials and methods. Asterisks (*) denote Envs derived from a published clade B virus panel (Li et al., 2005) and a sharp sign (#) denotes Envs from a published clade C virus panel (Li et al., 2006).
Some Envs exhibited more than one oligomeric band, for example, the trimer doublet of 3988 or the multiple bands of ZM135, DU172, ZM197 and ZM109 (Fig. 1A). Further analysis in reducing SDS-PAGE gels indicated the presence of uncleaved Env. Uncleaved Env is known to exist in various multimeric forms, possibly explaining these additional oligomer bands (Moore et al., 2006). A disproportionate fraction of clade C Envs harbored multiple oligomeric bands. The clade B trimers from a virus panel (Li et al., 2005) on the other hand were quite consistent in their expression and mobility, perhaps reflecting a similar degree of neutralization resistance. The additional bands seen below the monomer in Fig. 1 are gp41 trimers and monomers, as identified previously (Moore et al., 2006).
Interestingly, Env from the neutralization-resistant SIVmac239 isolate and its sensitive derivative, SIVmac316 resolved in two distinct oligomer forms (Fig. 1B). The lower band appears to be a trimer, while the upper band is estimated to be ~800 kDa, larger than any band observed with the HIV-1 isolates. We hypothesize that this might represent be a dimers of trimers, analogous to the dimers of gp120-GCN4 trimers reported previously (Pancera et al., 2005). Alternatively, the band might represent trimer complexed with an unknown membrane protein. Reducing SDS-PAGE indicated negligible uncleaved gp120/gp41 precursor (not shown), ruling out this as a possible explanation. The sensitive SIVmac316 bore proportionately more of the larger Env form compared to SIVmac239. The SIVmac316 trimers were also slightly slower moving than the SIVmac239 counterpart (estimated at 420 kDa versus 386 kDa), suggesting that the latter, neutralization-resistant trimers may be more compact. Env derived from live inactivated SIVmac239 particles behaved identically to its VLP counterpart in BN-PAGE, indicating that the patterns observed are not a VLP artifact (Fig. 1B).
Relationship of monovalent Fab-Env trimer binding and neutralization
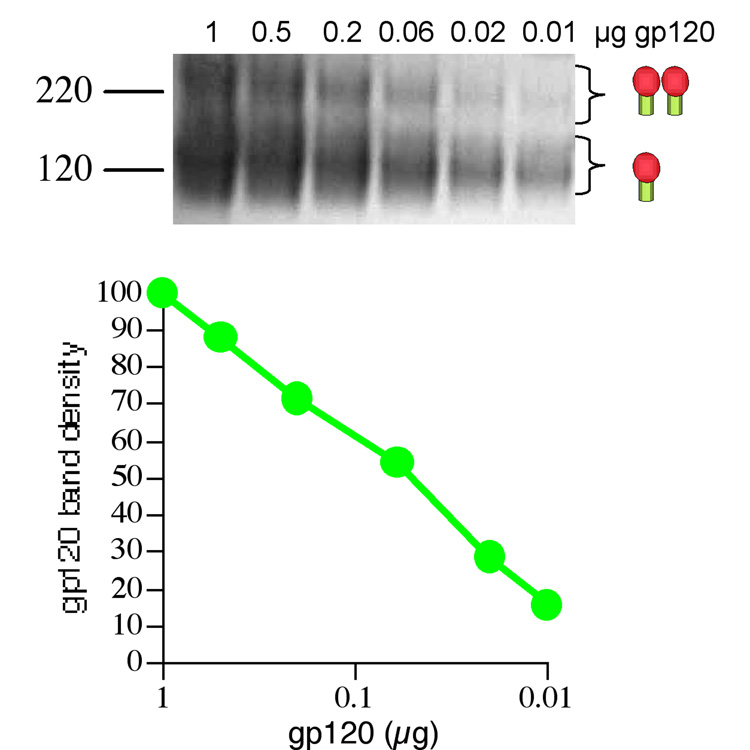
We next investigated the binding of a panel of Fabs to JR-FL trimers over a range of concentrations. As we planned to determine trimer binding affinity by depletion of uncomplexed trimer in the presence of mAb, we first established the relationship of Env loaded in BN-PAGE and its detection in Western blot using densitometry. Gp120 was titrated in BN-PAGE in Fig. 2 (upper panel) and the relationship between densitometry and amount loaded is plotted in the lower panel. Clearly, a direct relationship exits between the amount of gp120 loaded and the band density recorded, suggesting that our intention to measure trimer-Ab affinity by its depletion in BN-PAGE should yield reasonable estimates. Interestingly, the BN-PAGE revealed a fraction of gp120 dimers, as reported by others (Center et al., 2000).
Fig. 2. Relationship of band density and gp120 quantity in BN-PAGE.
Graded concentrations of JR-FL gp120 were loaded in BN-PAGE. Band densities were calculated and plotted against the microgram quantity of gp120.
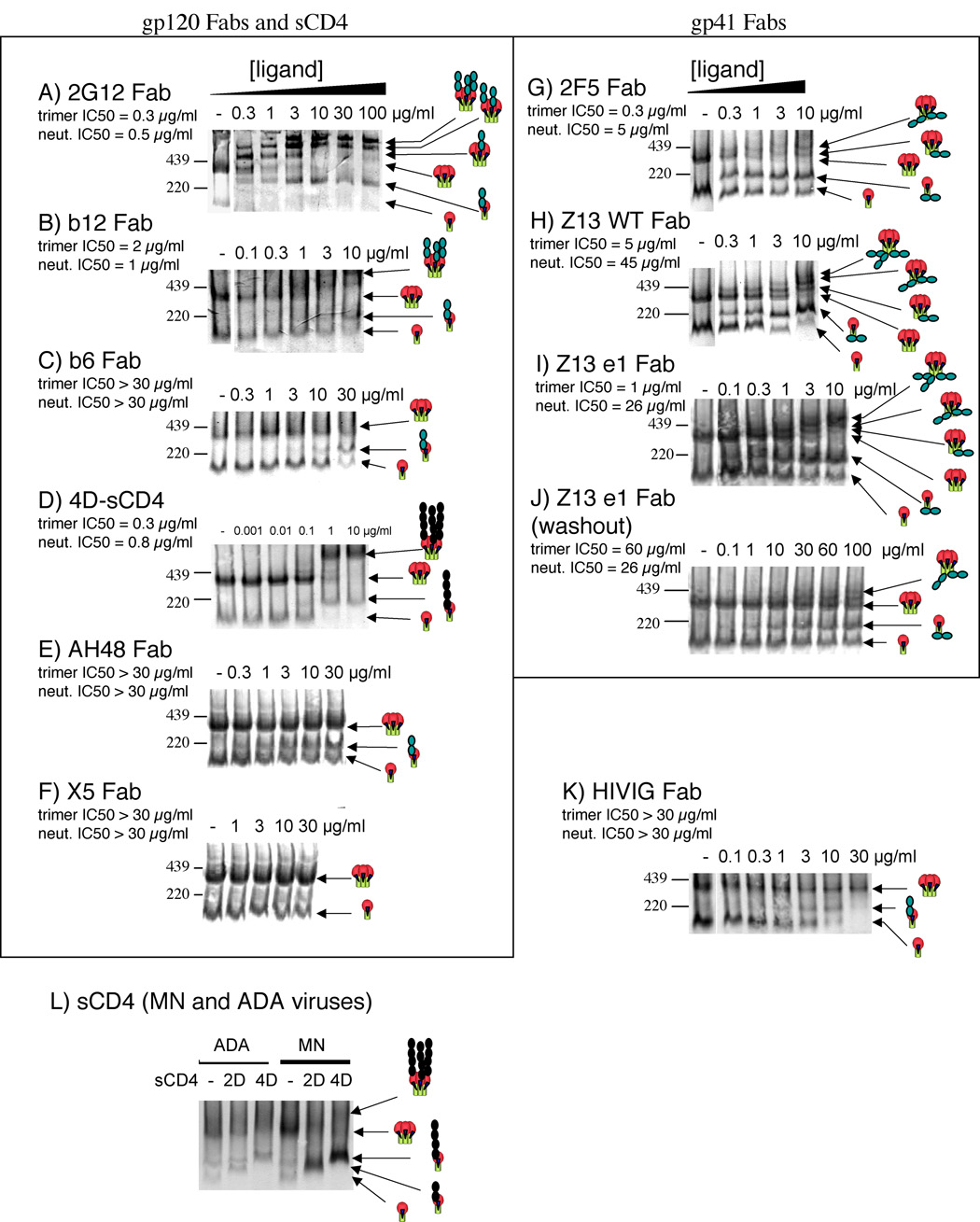
In BN-PAGE, neutralizing Fabs (2G12, b12, 2F5, Z13 WT, and Z13 e1) and sCD4 were able to shift JR-FL trimers (Fig. 3). Specificity was confirmed using trimers with point mutations at residues 295 and 368 that selectively eliminate b12 or 2G12 binding, respectively (E. Crooks and J. Binley, in preparation). We determined affinities by measuring depletion of unliganded trimer and did not factor liganded complexes into the estimates since the detection Ab cocktail may react differently with trimers in complex with test mAbs. The affinities of each neutralizing Fab for trimers and monomers were similar, indicating little or no energy barrier for trimer compared to monomer binding. In contrast, most non-neutralizing Fabs bound to monomer, but their epitopes were conformationally occluded on trimers (b6, AH48, HIVIG, T2, T3, P7 in Fig. 3 and Table 1), confirming the exclusive relationship of trimer binding and neutralization. Fab X5 did not bind effectively to gp120/gp41 monomers and may therefore recognize other forms of Env, such as monomeric gp120 or receptor-bound forms of Env.
Fig. 3. Quantitative Ligand Binding to JR-FL trimers.
Graded concentrations of Fabs or 4D-sCD4 were incubated with JR-FL SOS-VLPs Env trimers were then resolved by BN-PAGE. In one experiment (panel J), VLPs were washed to remove unbound Fab before native PAGE. Neutralizing IC50s and estimated trimer IC50s are indicated. Cartoons depict gp120/gp41 trimers and monomers and bound ligands. Black circles depict 4D-sCD4 and shaded circles depict Fab. The molecular weights of trimers and monomers were previously estimated as 420 kDa and 140 kDa (Moore et al., 2006). The masses of trimers complexed with saturating concentrations of neutralizing Fabs were estimated as follows: 2G12 (780 kDa), b12 (614 kDa), 4D-sCD4 (620 kDa), 2F5 (493 kDa), Z13 WT (551 kDa) and Z13e1 (533 kDa). In the case of the gp41 Fabs, the complexes with three Fabs were unclear, so the molecular weights given are estimated of trimers with two Fabs bound. Panel L shows the effects of saturating (20µg/ml) 2D-and 4D-sCD4 on live inactivated MN and ADA preparations.
Table 1. Summary of the neutralizing titers of various ligands and their affinities for JR-FL Env trimers and monomers.
IC50s are given in µg/ml and the corresponding titers in nM are given in parentheses, assuming molecular weights of ligands as follows: Fabs 50 kDa, IgG 150 kDa, CD4-IgG2 170 kDa, T-20 peptide 4 kDa, C34-Ig dimer (60 kDa). In some cases, titers are given in the presence of a fixed concentration of sCD4 (5 µg/ml). Post-CD4 neutralization by mAbs AH48, 2F5 and X5 was measured on CCR5 only-expressing cell lines (Crooks et al., 2005). IC50s are representative of several repeat assays. An asterisk (*) denotes experiments in which trimer binding was measured in a format where unbound ligand was washed away prior to detergent treatment of VLPs.
| IC50 concentration In µg/ml (nM) | ||||
|---|---|---|---|---|
| Epitope | Ligand | Monomer shift | Trimer Shift | Neutralization |
| CD4bs | b12 Fab | 2(40) | 2(40) | 1(20) |
| b12 IgG | >30 (200) | 2(13) | 0.05 (0.3) | |
| b6 Fab | 15(300) | >50 (>1,000) | >50 (>1,000) | |
| mannose cluster | 2G12 Fab | 0.2(2) | 0.3(3) | 0.5(5) |
| 2G12 IgG | 1(7) | 1(7) | 0.21 (1.4) | |
| 2G12 IgG+ 4D-sCD4 | N.D. | N.D. | 0.08(0.5) | |
| V3loop | AH48 Fab | 3(60) | >50 (>1,000) | >50 (>1,000) |
| AH48 Fab + 4D-sCD4 | 0.03(1.6) | 0.3(6) | 0.001(0.02) | |
| cluster I g p41 | T2 Fab | >50(>1,000) | >50 (>1,000) | >50 (>1,000) |
| cluster II tgp41 | T3 Fab | >50 (>1,000) | >50 (>1,000) | >50 (>1,000) |
| CD4 | 2D-sCD4 | 0.8(32) | 0.8(32) | 0.5(20) |
| 4D-sCD4 | 0.3(6) | 0.3(6) | 0.8(16) | |
| CD4-IgG2 | >30(180) | 0.5(3) | 0.015(0.09) | |
| gp41 MPER | 2F5 Fab | 0.3(6) | 0.3(6) | 5(100) |
| 2F5 Fab* | 15(300)* | 15(300)* | 5(100) | |
| 2F5 Fab + 4D-sCD4* | 7(140)* | 7(140)* | N.D. | |
| 2F5 IgG | 10(66) | 10(66) | 0.2(1.4) | |
| 2FS IgG + 4D-sCD4 | N.D. | N.D. | 0.2(0.14) | |
| 4E10 Ig G | 5(33) | 5(33) | 2(14) | |
| Z13 WT Fab | 2(40) | 5(100) | 45(900) | |
| Z13 e1 Fab | 0.3(6) | 1(20) | 26(520) | |
| Z13 e1 Fab* | 60(1,200)* | 60(1,200)* | 26(520) | |
| Z13e1 IgG | 10(66) | 30(200) | N.D. | |
| CD41 | X5 Fab | >50 (>1,000) | >50 (>1,000) | >50 (>1,000) |
| X5 Fab +4D-sCD4 | 0.06(l.2) | 0.3(6) | 0.0009(0.018) | |
| gp41 prehirpin | T-20 | N.D. | N.D. | 0.1(25) |
| T-20 + 4D-sCD4 | N.D. | N.D. | 0.04(10) | |
| C1 conformational | P7 Fab | 2(40) | > 50 (> 1,000) | >50(> 1,000) |
Incremental occupation of trimers by 1, 2 and 3 molecules of Fab 2G12 was observed over a broad concentration range (Fig. 3A). Binding was detectable at <0.3 µg/ml, but did not become fully saturated even at 100 µg/ml (Fig. 3A). The intensity of the bands became brighter with increasing concentration, presumably because the 2G12 remains bound to trimers after Western blotting, leading to increased detection sensitivity. Similar progressive binding of 1, 2 and 3 gp41 subunits of trimers was observed for gp41 Fabs. The pattern for Z13 WT was particularly clear (Fig. 3H). In contrast, 4-domain sCD4 (4D-sCD4) saturated at a precise concentration. This may indicate triggering of a structural change at a threshold concentration, exposing all 3 CD4 binding sites (Fig. 3D). Similar results were obtained with 2-domain sCD4 (2D-sCD4) (Table 1). For comparative purposes, we examined the effect of sCD4 on ADA and MN trimers (Fig 3L). A ladder of partially and fully liganded trimers was not evident for Fab b12 as it was for the other neutralizing Fabs (Fig. 3B). This may be due to Fab b12's enzymatic production from IgG1b12 by papain digestion, yielding a somewhat heterogeneous mixture of Fab products of slightly differing sizes, contrasting the more consistent expression of Fabs by phagemid vector, as for the other Fabs. It follows that binding of this Fab b12 mixture to trimers results in the rather diffuse complexes observed. At saturation, the estimated molecular weight of the largest trimer-Fab complexes suggests that 3 Fab molecules bind in each case. Saturating concentrations of 2G12 shifted trimers to an estimated molecular weight of ~780 kDa, dramatically larger than with Fab b12 and 4D-sCD4, where saturated complexes were ~620 kDa (compare Fig. 3A, B and D). This is consistent with the inherent dimeric nature of 2G12 Fabs, resulting from a unique domain exchange conformation (Calarese et al., 2003), making it approximately twice the size of the other Fabs and 4D-sCD4.
Comparing affinity with neutralization potency revealed a generally in good agreement for gp120 ligands, suggesting a simple quantitative relationship (Fig. 3 and Table 1). In contrast, gp41 Fabs 2F5, Z13 WT and Z13 e1 bound to trimers more potently than they neutralized (Fig. 3G–I, Table 1). A much closer correlation of neutralization and trimer binding IC50s existed, however, when we measured Z13 e1 trimer shifts in a format where unbound Fab was washed away prior to detergent treatment of VLPs in preparation for BN-PAGE (compare Fig. 3I and J, Table 1). Similar results were obtained with other gp41 membrane-proximal ectodomain region (MPER)-specific mAbs 2F5 and Z13 WT (Table 1 and not shown). In contrast, gp120 ligands bound equivalently, regardless of an additional wash step. This suggests that there may be constraints on MPER exposure on native spikes that may be released after trimers are liberated by detergent.
Effects of sCD4 binding on trimer epitope exposure
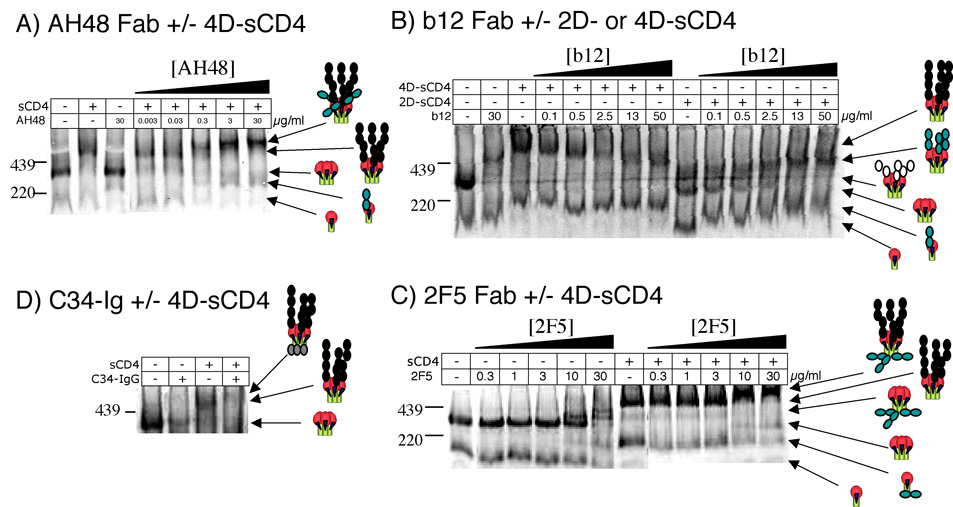
Entry inhibitors may be categorized as CD4-inhibiting, CD4-induced, or CD4 impartial. We attempted to visualize these scenarios in BN-PAGE. Soluble CD4 complexing with trimers greatly enhanced the binding of V3 Fab AH48 (Fig. 4A), consistent with its drastically increased neutralizing activity in a post-CD4 format (Table 1). Similar results were observed with CD4i mAbs (Table 1 and ref. (Crooks et al., 2005)). The molecular weight of the trimer fully complexed with 4D-sCD4 and AH48 (Fig. 4A) was estimated as ~780 kDa. With reference to the molecular weight of 4D-sCD4 liganded trimers of ~620 kDa, this suggests that 3 copies of Fab AH48 bind per sCD4-saturated trimer. Interestingly, the stoichiometry of X5 binding to CD4-trimer complexes differed from AH48 (BN-PAGE shown in Fig. 3B of ref. (Crooks et al., 2005)). The molecular weight of Fab X5 complexes with 4D-sCD4 and trimer were estimated at ~653 kDa, suggesting a 1:1 stoichiometry of X5: sCD4/trimer binding. This result was further supported using different sized ligands: 2D-sCD4 gave a shift to ~515 kDa (suggesting 3:1 CD4:trimer stoichiometry), to which the addition of a single chain Fv X5 fragment only increased the complex molecular weight to 548 kDa (Crooks et al., 2005). This suggest that there may be steric constraints on CD4i mAb-trimer binding. Given the similarity of the CD4i epitope to the CCR5 binding site, (Trkola et al., 1996), it will be of interest to investigate whether trimer/sCD4:CCR5 binding is also limited to a 1:1 stoichiometry.
Fig. 4. Ligand binding to JR-FL in the presence of sCD4.
The binding of various ligands to JR-FL SOS-VLP trimers was determined in the presence or absence of 2D- or 4D-sCD4 (5 µg/ml). A) Fab AH48. B) Fab b12. C) Fab 2F5. D) C34-Ig. In the case of 2F5, unbound Fab was washed away prior to sample preparation, but in all other cases, ligand was left in. A final concentration of 40 µg/ml (666 nM) C34-Ig was added to VLPs. Cartoons indicate liganded and unliganded trimers. Black circles depict 4D-sCD4, white circles depict 2D-sCD4, shaded circles depict Fab and C34-Ig. In part A, the size of the 4D-sCD4+AH48 saturated trimer (far right lane) was estimated as 780 kDa. In part C, the size of trimer+sCD4+2F5 complexes at near saturation (right lane) was estimated as 795 kDa. In part D, the sizes of complexed trimers were estimated as: lane 2 (trimer+C34-Ig) 447 kDa; lane 3 (trimer+4D-sCD4) 620 kDa; lane 4 (trimer+4D-sCD4+C34-Ig) 672 kDa.
In contrast to Fab AH48, Fab b12 competed with sCD4 for trimer binding (Fig. 4B). Addition of increasing concentrations of b12 in the presence of a fixed 4D-sCD4 concentration caused a decrease in the size of the complexed trimer, as b12 replaced sCD4’s positions on each gp120 component. The mobility of 4D-sCD4 complexed trimers was somewhat slower than b12-trimer complexes, despite the similar molecular weights of these two ligands (~50 kDa) (Fig. 3 compare parts B and D). This may be because 4D-sCD4 is an extended polyprotein of four IgG subdomains, whereas the Fab is a more compact heterodimer of two IgG subdomains. In a similar experiment, titration of Fab b12 in the presence of a fixed 2D-sCD4 concentration resulted in liganded trimers of increasing mobility, as b12 replaced the smaller 2D-sCD4’s positions on each gp120 subunit (Fig. 4B).
4D-sCD4 binding resulted in a slight enhancement Fab 2F5 affinity (Fig. 4C, Table 1), consistent with a mild increase in neutralization potency in the post-CD4 format (Crooks et al., 2005). The molecular weight of trimer+sCD4+2F5 at close to saturation was estimated as 795 kDa, consistent with a 3:1 stoichiometry of 2F5 binding to the trimer:sCD4 complexes. A similar slight increase in affinity was also observed for 2G12 (not shown) (Binley et al., 2006).
The low mass peptides based on the C-terminal helix of gp41 would make their binding impossible to visualize in BN-PAGE. However, C34-Ig, a molecule consisting of a C34 peptide fused to a human Fc fragment (Si et al., 2004) was able to perceptibly bind to trimers, especially in the presence of 4D-sCD4 (Fig. 4D). Consistent with this observation, the activity of a related peptide, T-20, was also slightly induced in the presence of sCD4 (Table 1) (Binley et al., 2003; Yuan et al., 2004). Insufficient material precluded an estimate of C34-Ig's neutralizing IC50, but it was clearly greater than 5 µg/ml (>83 nM). Thus, C34-Ig appears to be somewhat less potent than the T-20 peptide. The magnitude of shifts in the absence (~27 kDa) or presence of sCD4 (~52 kDa) was consistent with binding of one molecule of C43-Ig dimer (~60 kDa) or monomer (~30 kDa), rather than 3 molecules per trimer. It is unclear whether this is due to a weaker affinity or to the limited access of this bulkier molecule to its target.
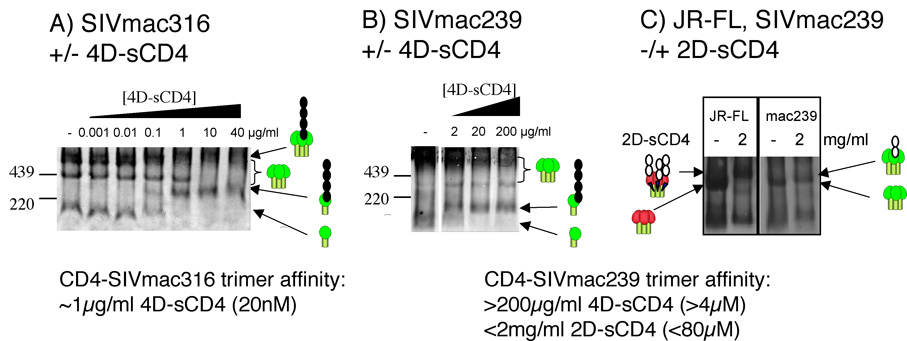
Affinity and stoichiometry of CD4 binding to SIV trimers
Previous studies have variously described of 3:1 or 1:1 sCD4-trimer binding stoichiometry for soluble gp140 versions of SIV trimers (Earl, Doms, and Moss, 1992; Kim et al., 2001). We reinvestigated this question here using functional SIV trimers. Neutralization-sensitive SIVmac316 trimers bound potently to 4D-sCD4, with an IC50 of ~1 µg/ml (~20 nM) (Fig. 5A). Although 4D-sCD4-trimer complexes did not resolve clearly, complexes with the putative trimer dimer were observed (right 3 lanes of Fig. 5A). The high mass and diffuse nature of these complexes made it difficult to accurately assign a molecular weight and therefore to determine how many CD4 molecules were bound. Soluble CD4 bound to SIVmac316 monomers and trimers with similar affinity, suggesting that trimers do not impose constraints on sCD4 binding, as observed with JR-FL (compare Fig. 3D and Fig. 5A). Soluble CD4 bound to monomers from the neutralization-resistant SIVmac239 isolate with an affinity similar to that for SIVmac316 (Fig. 5B). However, 4D-sCD4 did not bind SIVmac239 trimers at concentrations up to 200 µg/ml (4 µM) (Fig. 5B). This suggests that inter-subunit associations within SIVmac239 trimers conformationally disfavor sCD4 binding. Further investigation using a very high concentration of 2D-sCD4 (2 mg/ml; 80 µM) revealed a modest SIVmac239 trimer shift, compared to the marked shift induced in JR-FL trimers (Fig. 5C). The estimated molecular weights of trimer-CD4 complexes were ~443 kDa and ~510 kDa, respectively (assuming unliganded trimers are each ~420 kDa), consistent with a binding of one ~25 kDa sCD4 molecule per SIV trimer and 3 per JR-FL trimer.
Fig. 5. Soluble CD4 affinity and stoichiometry for SIV trimers.
Graded concentrations of 4D-sCD4 were mixed with A) SIVmac316 or B) SIVmac239 Env trimers in BN-PAGE. C) Saturating concentrations (2 mg/ml; 80 µM) of 2D-sCD4 were mixed with JR-FL and SIVmac239 Env trimers in BN-PAGE. Cartoons indicate bound and unbound forms of Env.
The relationship between IgG-trimer binding and neutralization
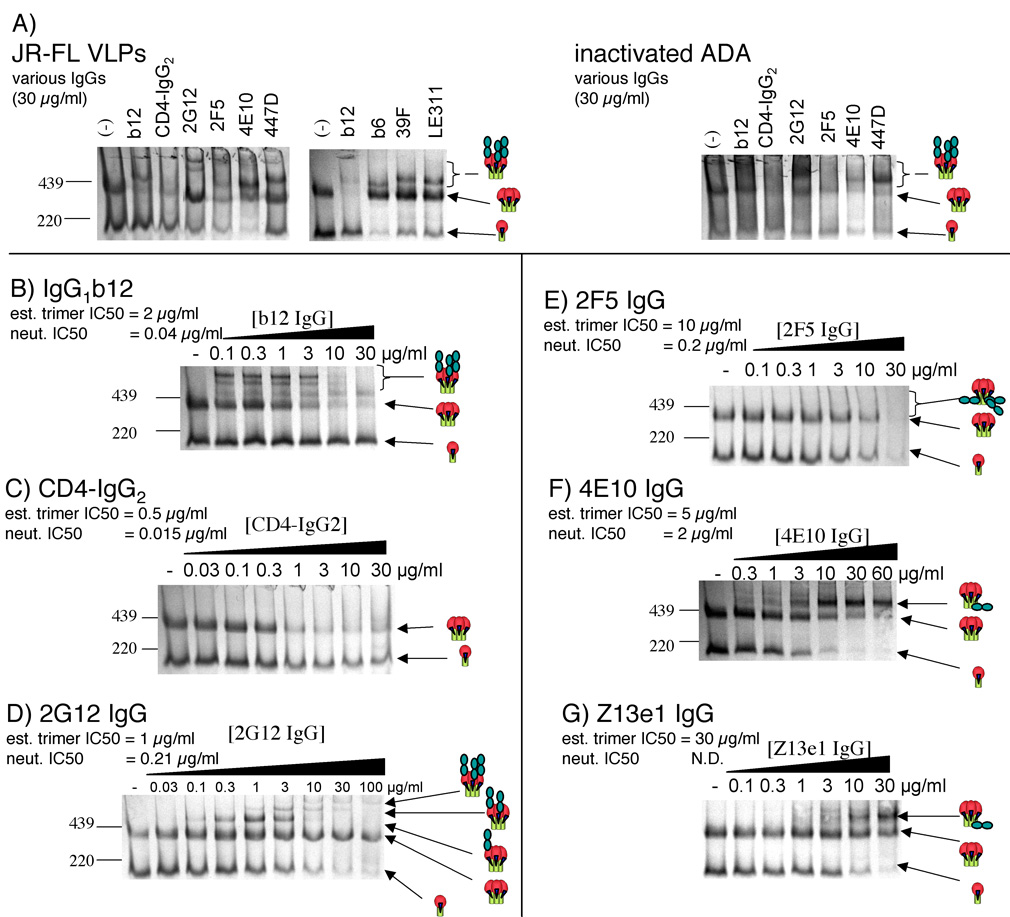
We next investigated the relationship between neutralization titer and trimer affinity using IgG versions of mAbs in a similar manner to the Fab analysis in Fig. 3. One challenge in using whole IgGs for BN-PAGE is in Western blot detection. Anti-Fc conjugate can cross-react with any residual free IgG that is loaded in the gel. Preliminary studies revealed that uncomplexed IgG resolves as a smear, and reduces the quality of Western blots. To eliminate this problem, we included a wash step before detergent treatment of VLPs and BN-PAGE, to remove any unbound IgG.
We first compared IgG binding of JR-FL and ADA trimers using fixed high concentrations of IgGs. The neutralizing IgGs bound to both trimers, as evidenced by a depletion of unliganded trimer, coupled in most cases with the appearance of high molecular weight complexes or smears (Fig. 6A). IgG binding exhibited several idiosyncrasies compared to the more straightforward Fab binding (compare Fig. 3 and Fig. 6). First, IgG-trimer complexes with CD4-IgG2, IgG1b12, and 2F5 largely formed unresolved smears, in contrast to the clear bands observed for the equivalent Fab-trimer complexes. Second, a band of near identical size to unliganded trimer was observed in the presence of 2G12 IgG. This is not surprising, considering that Fab 2G12-monomer complexes moved slightly ahead of unliganded trimers in Fig. 3A and factoring the somewhat increased size of the IgG version. Bands of similar size to trimers were also observed for IgG1b12 and 2G12 binding to inactivated ADA particles (Fig 6A, right panel). Third, unliganded monomer was observed with JR-FL VLPs incubated with high concentrations of certain IgGs, in particular CD4-IgG2 and IgG1b12 (Fig 6A, and compare right lanes of Fig. 6B and C to those of Fig. 6D–G). The poor resolution of ADA made it difficult to determine whether this phenomenon also applied. We noted however, that non or weakly neutralizing IgGs b6, 39F and LE311 effectively bound and depleted monomer, suggesting that this may be a phenomenon associated only with certain neutralizing IgGs (Fig. 6A, middle panel).
Fig. 6. Binding of whole IgG mAbs to JR-FL trimers.
A) Various IgGs (at 30 µg/ml) were incubated with JR-FL SOS-VLPs or ADA inactivated particles and resolved by BN-PAGE. B)-G) Increasing concentrations of IgG1b12, CD4-IgG2, 2G12, 2F5, 4E10, or Z13e1 were mixed with JR-FL SOS-VLPs. Unbound IgG was removed by a wash with PBS. Cartoons indicate liganded and unliganded trimers. Though the ligands are in the form of IgGs, for convenience, they are depicted as two shaded circles. N.D. = not done. Neutralizing IC50s and estimated trimer IC50s are indicated, where available. The trimer complexes with one 2G12 IgG molecule (part D, lane 5) were estimated at 696 kDa, those of 4E10 IgG and Z13e1 IgG were estimated as 626 kDa and 720 kDa, respectively.
To further investigate, we titrated the IgGs in BN-PAGE. Despite neutralizing HIV-1 JR-FL approximately 100-fold more potently (on a nM scale, in Table 1), IgG1b12 and CD4-IgG2 bound trimers the increases in affinity compared to their monovalent Fab and 4D-sCD4 counterparts were relatively small (2–10 fold; compare Fig. 3B, D and Fig. 6B, C; Table 1). Similarly, 2F5 IgG neutralized more than 70-fold more potently but its trimer affinity was only 5 fold higher than its Fab counterpart (Table 1). In contrast, 2G12 and 4E10 IgG trimer affinities matched their neutralizing IC50s (Fig. 6D, F). The activity of 2G12 IgG was in fact similar to that of the equivalent Fab (Table 1). Trimer affinity estimation was particularly complicated for 2G12 because liganded monomer almost comigrated with unliganded trimer in BN-PAGE. In estimating the 2G12 trimer affinity, we therefore took into account the density of this monomer+2G12 band at saturation, as well as the appearance of trimer complexes. Our affinity estimate closely matches that of 2G12 Fab (Fig. 3A, Table 1). This was expected, considering that 2G12 IgG is monovalent, so the Fab and IgG differ by mass but not valency (Calarese et al., 2003). Z13e1 IgG bound trimers only slightly more strongly than the Fab (Table 1). Although we did not perform neutralization assays in house with this IgG, studies in another lab (MBZ), show that it has a similar potency to the Fab version. Therefore Z13e1 IgG most likely exhibits matching trimer and neutralizing IC50s, as with 2G12 and 4E10.
Interestingly, 4E10 and Z13e1 both gave a visible IgG-trimer band (Fig. 6F, G), similar to the multiple bands observed for 2G12, but distinct from the rather more diffuse or completely absent trimer complexes with IgG1b12, CD4-IgG2 and 2F5 IgG. The estimated molecular weights of these bands were estimated as 626 kDa and 720 kDa, respectively. This appears to suggest trimer complexes with a one or two IgGs respectively. However, the calculation of molecular weights using IgGs may be a slight overestimate in contrast to the simpler Fab ligands. This is exemplified by the fact that the prominent band observed in lane 5 of Fig. 6D (2G12 at 1µg/ml) gave a molecular weight of 696 kDa. The banding pattern in this figure, with two further higher molecular weight 2G12-trimer complexes appearing at higher concentrations (Fig. 6D, right lanes), makes it clear that despite the large estimated size (~276kDa larger than uncomplexed trimer), this is in fact trimer complexed with a single 2G12 IgG. By similar reasoning, we suggest that the IgG-trimer bands of 4E10 IgG and Z13e1 IgG also reflect a single IgG molecule bound to a trimer.
One possible explanation for the disagreement in trimer binding affinities and neutralization potencies for CD4-IgG2, IgG1b12, and 2F5 IgG may be that we are underestimating trimer affinities due to a technical difficulty similar to that for 2G12, i.e. as IgG-complexed gp120/gp41 monomer co-migrates with unliganded trimer in BN-PAGE (Fig. 6B–E), the apparent trimer affinity may be reduced artificially. However, arguing against this possibility, unlike 2G12, the trimer-sized band at saturating concentrations of these IgGs is virtually absent. Nevertheless, to address this problem, we generated F(ab')2 fragments from IgGs by pepsin digestion. These smaller fragments should eliminate possible band co-migration, while retaining the valency of the parent IgGs. Pepsin digestion created functional F(ab')2 fragments of b12 and 4E10, as measured by ELISA (not shown). These were found to bind JR-FL trimers at precisely the same concentrations as the whole IgGs (not shown), suggesting that our affinity estimates are accurate.
Different consequences of 1:1 and 1:3 sCD4-trimer binding stoichiometry for infection
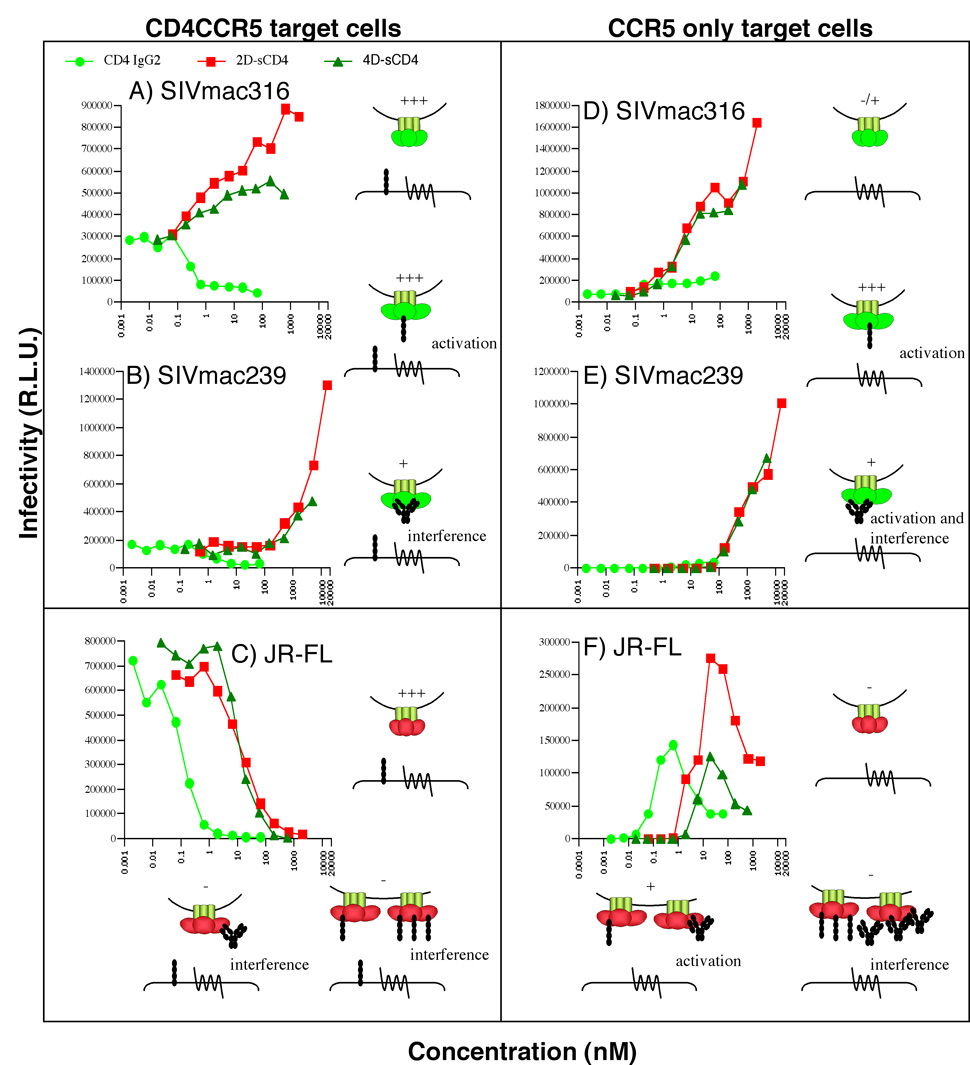
We next investigated the effects of CD4-based molecules of different sizes and valencies (2D-sCD4, 4D-sCD4 and CD4-IgG2) on the infection of HIV-1 and SIV bearing trimers that have differing CD4 binding stoichiometries (Fig. 7). We used cell lines expressing both CD4 and CCR5 receptors (Fig. 7, left panels) or CCR5 only (Fig. 7, right panels). The concentrations of the 3 CD4-based species are given in nM to allow direct comparisons, eliminating the effect of their differing molecular weights. The corresponding trimer binding IC50s in nM, where known, are given in Table 1 for JR-FL and in Fig. 5 for the two SIV isolates.
Fig. 7. Effect of CD4 binding stoichiometry on HIV-1 and SIV infection.
2D-sCD4, 4D-sCD4 and CD4-IgG2 were incubated with SIVmac316 (A, D), SIVmac239 (B, E) or JR-FL (C, F) gp160ΔCT viruses, and infection in relative light units (R.L.U.) was measured on CF2 target cells bearing both CD4 and CCR5 (left panels) or CCR5 only (right panels). Baseline infection of CCR5-expressing targets with no sCD4 (not shown) was identical to that at the lowest sCD4 concentrations shown. The effects of CD4-based ligands on SIV and HIV-1 infection are illustrated by cartoons. The levels of infection in each cartoon scenario is depicted by + signs. Soluble CD4 (2D- or 4D-) is depicted as 4 black circles, and CD4-IgG2 is depicted as a series of black circles in an IgG conformation.
In the absence of sCD4, SIVmac316 infection was detectable, consistent with its CD4-independent phenotype (Fig. 7D), in contrast to the CD4-dependent SIVmac239 virus (Fig. 7E) (Johnson et al., 2003; Puffer et al., 2002). Surprisingly, 2D- or 4D-sCD4 binding to SIV gp160ΔCT trimers dramatically enhanced infection of CD4/CCR5 cells (Fig. 7A, B). This suggests that one molecule of sCD4 binds each trimer, resulting in a conformational change that increases exposure of the CCR5 binding site (as illustrated by the cartoons adjacent to Fig. 6A and B). Though we were unable to ascertain the stoichiometry of sCD4 binding to SIVmac316 in native PAGE (Fig. 5), the similar enhancing effects of sCD4 on these two viruses implies that the 1:1 sCD4:trimer stoichiometry applies to both. The enhancement steadily increased over a broad concentration range, beginning at much lower concentrations of sCD4 for SIVmac316 (~0.1 nM) than for SIVmac239 (~100 nM) (compare Fig. 7A, B), consistent with the large sCD4 affinity differences observed in BN-PAGE (Fig. 5). The sCD4 concentration required for half maximal enhancement of SIVmac316 infection and trimer binding appeared to correspond (~20nM) (compare Fig. 5 and Fig. 7A). A similar consistency in enhancement and trimer binding appears likely for SIVmac239, though we have not determined the exact trimer binding IC50, which is between 4 and 80µM (Fig. 5 and Fig. 7B). In general, the smaller 2D-sCD4 resulted in more than 2 fold greater enhancement than the larger 4D-sCD4 (Fig. 7A, B). Thus, trimers complexed to the bulkier 4D-sCD4 have a somewhat sterically diminished CCR5 binding capacity compared to those complexed with the smaller CD4 molecule.
The influence of size appears to be further supported by the observation that CD4-IgG2 inhibited rather than enhanced SIV infection (Fig. 7A, B). Although CD4-IgG2 binding induces a conformational change in trimers, exposing CCR5 binding sites, its bulk appears to obscure CCR5 engagement, resulting in a net interfering effect (as illustrated in the cartoons associated with Figs. 7A, B).
When infection was assayed on cells expressing only CCR5 (no CD4), sCD4-induced enhancement was again observed (Fig. 7D and E), as illustrated in the associated cartoons. The degree of enhancement induced by 2D-sCD4 remained only slightly higher than 4D-CD4, contrasting with the more dramatic difference observed on CD4/CCR5 cells. CD4-IgG2 also moderately enhanced SIVmac316 and SIVmac239 infection, in contrast to its effect when infection was assayed on CD4/CCR5 cells (Fig. 7, compare parts A and B with parts D and E). Thus, in this low baseline infection scenario, CD4-IgG2-induced exposure of coreceptor binding sites was more important than its tendency to inhibit CCR5-engagement due to its bulk. In fact, all the CD4-based molecules ultimately lead to almost identical infection levels of both cell types and it is the baseline infection levels that differ (Fig. 7, compare parts A and B to parts D and E).
In contrast to their effects on SIV, CD4-based ligands all inhibited JR-FL infection of CD4/CCR5 target cells (Fig. 7C), presumably by blocking receptor and/or coreceptor binding. Concentrations of sCD4 that would be expected to lead to partial trimer occupation (~10 nM) did not enhance infection as they did for SIV. This may be because the remaining open CD4 binding sites on partially liganded JR-FL trimers can also interact with cellular CD4, leading to net interference (Fig. 7C, see cartoons). This scenario is not possible for SIV, because a maximum of only one sCD4 molecule can bind to each trimer (Fig. 7A, B).
In stark contrast to the effect on CD4/CCR5 cells, when CCR5 only cells were used to measure infection, we observed a remarkable enhancement of JR-FL by all CD4 ligands, peaking at concentrations roughly equivalent to the neutralization IC50s on CD4/CCR5 cells (compare Fig. 7C and F). The peaks correspond to concentrations sufficient partial CD4 binding, presumably causing a global conformational change that enhances trimer binding to cellular CCR5, most likely by the remaining open subunits of the trimer (see cartoons in Fig. 7F). When the trimer becomes fully saturated, the enhancement wanes, as sCD4 appears to interfere with cellular CCR5 binding by the same trimer subunit. The enhancement may occur on CCR5 only cells because non-productive interference between cellular and soluble forms of CD4 is eliminated (compare cartoons in Fig. 7C and F). As with SIV, the ultimate effects of the ligands are similar on both cell types and the differences observed in Fig. 7F versus Fig. 7C lies in the lower baseline infection level that reveals the activating effect of sCD4. As observed for SIV (Fig. 7A, C), 2D-sCD4 enhanced JR-FL infection to a somewhat greater extent than 4D-sCD4.
It is interesting to compare the IC50 titers of monovalent and multivalent CD4 molecules against JR-FL and SIV viruses. As mentioned earlier, the potency of CD4-IgG2 against JR-FL (enhancement or inhibition, depending on assay format) was much greater than that of sCD4 (IC50=~0.1 nM versus ~10 nM; Fig. 7C, Table 1). In contrast, the CD4-IgG2 IC50 against SIV trimers was exactly the same as monovalent sCD4 counterparts (compare Fig. 7A, B and C).
The enhancement of SIV isolates by sCD4 observed here contrasts with its neutralizing effect against SIV described previously (Pikora, Wittish, and Desrosiers, 2005; Yuste et al., 2005). The sCD4 IC50 concentrations required were comparable, however: SIVmac239 neutralization by 4D-sCD4 was previously reported to occur at an IC50 concentration similar to where enhancement occurs in our experiments (~500 nM; Fig. 7B). One possible explanation for this discrepancy was our use of truncated rather than full-length gp160 pseudovirions. Indeed, we found that sCD4 neutralized rather than enhanced equivalent full-length gp160 SIV pseudovirions (not shown), suggesting that the full-length gp160 allows a 3:1 sCD4: trimer binding stoichiometry similar to JR-FL.
DISCUSSION
Neutralization and trimer binding as probed by native PAGE
We showed here a direct, quantitative correlation between Fab trimer binding and neutralization, regardless of epitope, suggesting that affinity is a major factor in determining neutralization potency. Our data are consistent with neutralization beginning when one mAb binds to each spike (Schonning et al., 1999; Yang et al., 2005a; Yang et al., 2005b) and complete neutralization is reached when all spikes become saturated.
MPER epitopes were found to be partially occluded in native trimers embedded in membranes. This constraint was lifted when trimers were liberated from particles. Similar findings were reported in a study that examined the neutralization sensitivities of viruses with the 2F5 epitope grafted into the SU or TM of MLV Env (Ou et al., 2006). The decreased sensitivity of the TM graft to neutralization matches the conformational occlusion we observed, suggesting that Abs directed to TM might have to overcome binding constraints not present for SU mAbs (Ou et al., 2006). However, our findings that detergent treatment exposes MPER epitopes on trimers appears to contrast with reports that membranes increase binding of MPER antibodies to Env proteins captured on the surfaces of proteoliposomes and other particles (Grundner et al., 2002; Phogat et al., 2008). Therefore, the form of Env captured could dictate the influence of phospholipid context on MPER Abs binding affinity.
Trimer binding stoichiometry
A report by Kim and coworkers showed that only one molecule of sCD4 bound to uncleaved soluble gp140 SIVmac32H trimers (Kim et al., 2001). We demonstrated here that this phenotype is preserved in functional cleaved SIV trimers with a truncated gp41 tail but not in full-length trimers. Several explanations were previously proposed to explain the 1:1 sCD4:trimer stoichiometry (Kim et al., 2001). We can now rule out inappropriate gp120-gp41 association, since our trimers are proteolytically mature and functional. The possibility that each gp120 comprises of a partial CD4 binding site that is only complete when gp120s are packed in the trimer structure can also be ruled out, because monomeric forms of SIV Env readily bind to sCD4, suggesting that 3 binding sites are potentially available in each trimer. Steric constraints were previously considered an unlikely explanation, since even the smallest 2D-sCD4 bound only once to SIV gp140 trimers (Kim et al., 2001). However, if the binding site is close to the trimer axis (Chen et al., 2005), perhaps only one sCD4 can gain access, regardless of its size, and therefore a single CD4 molecule occludes the remaining two sites. This partial exclusion of binding sites is reminiscent of findings with HIV-1 gp120 trimers generated by fusion with a C-terminal oligomerization motif, to which a maximum of only two molecules were shown to bind (Pancera et al., 2005). As with our SIV trimers, interactions between the gp120 monomers might restrict sCD4 access. Interestingly, removal of the V-loops relieved these constraints, allowing 3 molecules of sCD4 to bind.
Our data show that single sCD4 binding to trimers activates infection of CCR5-expressing cells but three sCD4 molecules interfere with infection (Fig. 7D, E, F). Thus, a single sCD4 molecule can induce a global structural change in all three gp120/gp41 subunits of trimers. The exposed coreceptor binding sites can then mediate infection of coreceptor-expressing cells. In particular, the open subunits of the trimer that are not bound to sCD4 appear to mediate CCR5 binding, as indicated by reduced enhancement of JR-FL at saturating sCD4 concentrations when infection is assayed on CCR5 cells (Fig. 7F). This is consistent with the idea that infection is optimally mediated by trimers bearing at least two wild type gp120/gp41 subunits, one responsible for binding CD4 and another for binding CCR5 (Yang et al., 2006a). Constraints on CCR5 binding are also suggested by our finding that only one X5 molecule appears able to bind trimers saturated by sCD4 (Crooks et al., 2005).
In contrast to JR-FL, SIV enhancement was observed even at very high sCD4 concentrations, as binding is saturated at a 1:1 stoichiometry, eliminating the possibility of coreceptor binding interference by further sCD4 molecules binding to the trimer. Soluble CD4: SIV trimer affinities measured by BN-PAGE corresponded well with the concentrations required to enhance infection (Fig. 5 and Fig. 7), just as Fab:trimer affinities corresponded to neutralization IC50s for JR-FL. Soluble CD4 bound SIVmac316 trimers incrementally over a broad concentration range, only showing signs of saturation at 1,000 nM, a 4 orders of magnitude higher concentration than that at which the enhancement began. The same slow saturation is probably true for SIVmac239, but insufficient quantities of sCD4 did not allow us to investigate. In contrast, JR-FL trimers are saturated over a narrow range. This is consistent with a global conformational change in trimers induced by a single sCD4 molecule binding, leading to the cooperative binding of other CD4 molecules (Fig. 3D; compare the effects of 2D-sCD4 in Figs. 7A and C). Clearly, this cooperative CD4 binding is not available for SIV trimers studied here.
Paradoxically, neutralizing mAbs can occasionally moderately enhance HIV-1 infection at limiting concentrations (Koch et al., 2003). It is possible that partial mAb binding induces slight conformational changes that result in enhanced infection, if the remaining unoccupied monomers function slightly more effectively than they do at ground state (albeit to a much lower extent than sCD4). Indeed, some nAbs are known to induce conformational changes upon binding to viral proteins (Emini et al., 1983). A study that showed entropic changes as a result of mAb-Env complexing provides further circumstantial evidence of this possibility (Kwong et al., 2002).
Gp41 truncation is a natural adaptation of some SIV isolates to growth in human T cell lines, with a stop codon appearing at residue 734 (Bonavia et al., 2005; Kodama et al., 1989; Puffer et al., 2004). SIVmac239 particles harboring this stop codon were recently analyzed by cryoelectron tomography, revealing that the majority of spikes reacted with only one sCD4 molecule, even when sCD4 was added in great molar excess (K.Roux and P.Zhu, personal communication). This agrees with the 1:1 trimer:sCD4 stoichiometry we showed here for SIVmac239 particles with a stop codon at 718, leaving a two amino acid gp41 tail. However, it appears that not all truncation mutants show this unexpected CD4 stoichiometry. SIVmac239 particles bearing trimers with a stop codon at residue 767 (Yuste et al., 2005) were neutralized rather than enhanced by sCD4, similar to the full length SIVmac239 trimer-bearing viruses, rather than our truncation mutant. Taken together, it appears that 1:1 CD4:trimer stoichiometry applies only to dramatically truncated SIV Env trimers and it will be worth investigating graded SIV gp41 tail truncations to determine where the switch from 3:1 to 1:1 stoichiometry occurs. We speculate that the 734 truncation mutant found in T cell cultures may be selected on the basis of increased growth capacity. It might be envisaged that trimers binding a single CD4 molecule rather than 3 leaves coreceptor binding sites unencumbered for more efficient coreceptor binding and entry.
It will be interesting to determine whether SIV trimers can also restrict mAb binding to a 1:1 stoichiometry. Kim et al. reported that, in contrast to sCD4, uncleaved SIV gp140 trimers can bind 3 Ab molecules (Kim et al., 2001). Despite a paucity of neutralizing reagents reactive with SIVmac239, this point could be investigated using viruses bearing the more sensitive SIVmac316 trimers. Moreover, it will be interesting to compare the structures of SIV trimers that exhibit differing CD4 stoichiometries to understand the basis and possible implications for infection and neutralization. The very large SIV oligomeric band observed in BN-PAGE that may be dimers of trimers (Fig. 5) also clearly warrants further investigation.
Possible explanations for the increased neutralization potency of IgGs compared to monovalents
The greater neutralization potencies of IgG1b12, CD4-IgG2 and 2F5 IgG compared to their monovalent counterparts might be attributable to any or all of several unique IgG properties. Size clearly seems to matter. Increased bulk has been suggested to account for the greater neutralizing potency of a dodecameric CD4 molecule, D1D2-IgP (Arthos et al., 2002). In the present study, the contribution of size is supported by a comparison of 2D- and 4D-sCD4, of which the bulkier of the two exhibited greater interference (Fig. 7). CD4-IgG2 neutralized rather than enhanced SIV infection, perhaps further suggesting an effect of size. The difference is probably not due to increased avidity, as the IC50 titer would be expected to be lower if more than one CD4-IgG2 Fab arm is engaged (as for JR-FL, compare Fig. 7A and C). Overall, increasing size appears to affect magnitude of the effect but not the titer, i.e. in Fig 7, increasing size lowers viral infectivity (vertical scale), but not IC50 (horizontal scale).
Avidity may also influence neutralization, increasing the potency of multivalent ligands. This may occur in 3 ways; crosslinking epitopes on a single spike, crosslinking neighboring spikes on the same particle or bridging spikes on different particle(s). It is likely that higher avidity would increase neutralization titer, shifting curves to the left in Fig. 7, contrasting the effect of ligand size. CD4-IgG2 had a lower IC50 against JR-FL trimers, suggesting that it crosslinks more than one gp120 subunit.
We first consider the possibility of multivalent binding to a single spike. It might be envisaged that one IgG Fab arm bound to trimer puts the other Fab arm in close vicinity of the 2 remaining targets on the same trimer. IgGs molecules are flexible at their hinge regions, allowing Fab arm “wag" in 3 dimensions (Burton, 1990; Hanson, Yguerabide, and Schumaker, 1981; Roux, Strelets, and Michaelsen, 1997; Zhu, Olson, and Roux, 2001). The Fab arms themselves are also able to rotate about their long axis and considerable flexing can occur at the elbow between the variable and constant domains. This could mean that IgGs can contort in conformations allowing simultaneous engagement of Fab arms. However, specificity may exert an important constraint on the possibility of multivalent binding to a single spike. While IgGs that bind epitopes near the trimer apex may be able to crosslink, those that bind radially or laterally to their epitopes may not, due to the geometric limits of Fab arm flexibility. Modeling studies suggest that 2G12 may fall into the first category, and IgG1b12, CD4-IgG2, 2F5 and 4E10 fall into the latter (Kwong et al., 2000; Zhu, Olson, and Roux, 2001), though definitive data are still lacking. 2G12 is an exceptional case since the locked Fabs have no flexibility with respect to one another, intra-spike crosslinking appears to be highly unlikely
Despite their theoretical inability to bind individual spikes more than once, IgGs that bind laterally may be able to bridge different spikes on the same virus. The difficulty in observing clear trimer-IgG complexes in BN-PAGE in many cases comparable to the clear trimer-Fab complexes (compare Fig. 3 and Fig. 6) could imply that both Fab arms engage different spikes, forming higher order complexes that are difficult to resolve. However, it is also possible that these smears stem simply from inherent IgG flexibility. Dodecameric D1D2-IgP has been shown to crosslink trimers on SIV particles that bare ~70–79 spikes (Bennett et al., 2007). Others have suggested that CD4-IgG2 is also capable of intravirion trimer crosslinking (Zhu, Olson, and Roux, 2001). The trimer affinities of IgG1b12, CD4-IgG2, and 2F5 IgG we measured in BN-PAGE appear to support this possibility. Multivalent binding to a single spike would be expected to increase the trimer affinity compared to the equivalent Fabs. If, on the other hand, increased IgG avidity emerges from crosslinking separate spikes, binding each only once, the affinity on a per trimer basis measured against individual spikes in BN-PAGE might be expected to match that of the respective Fabs, as observed (Fig. 3, Fig. 6, Table 1). In light of the lower spike density of our JR-FL VLPs (~27/particle (Moore et al., 2006)), this model would require that trimers are either clustered or mobile in the lipid bilayer (Sougrat et al., 2007; Zhu et al., 2006). The cytoplasmic tail truncated trimers used here may make the possibility of mobility more likely. It may therefore be worth comparing these viruses to those bearing full-length gp160 trimers that might be properly anchored and therefore immobile.
IgGs may also be able to bridge spikes on separate particles (Bennett et al., 2007). This would be expected to be less dependent of specificity, and more on sufficient particle concentrations to bring neighboring viruses into proximity. In theory, the number of spikes inactivated compared to the number of spikes actually bound by ligand may be increased if the spikes on closely apposed surfaces of crosslinked particles are rendered incapable of accessing the target cell surface and mediating entry (Bennett et al., 2007). A potential caveat for inter-particle crosslinking is that non-neutralizing Abs can bind to non-functional forms of Env such as gp41 stumps present on particle surfaces (Poignard et al., 2003) and might therefore conceivably be able to crosslink particles. However, these mAbs do not neutralize, perhaps because in practice the density of non-functional Env is too low for this effect to be significant.
Overall, it is not yet clear which of the modes of multivalent binding is more important in neutralization, and it may depend on the specific features of the ligand. The balance of data in our study suggests that inter-spike crosslinking is predominant. The only apparent contradiction comes from an analysis of the effects of CD4-IgG2 and sCD4 against truncated SIV trimers. The SIV particles bear a far greater spike density than JR-FL (~73 versus 27), perhaps increasing the likelihood of inter-spike crosslinking. However, the observed lack of an increased CD4-IgG2 neutralization titer compared to sCD4 suggests that CD4-IgG2 binds only once and inter-spike crosslinking does not occur. By default, this might be taken to suggest that intra-spike crosslinking (not available for our SIV mutant) explains the increased titer of CD4-IgG2 against JR-FL (Fig. 7). Our counter-argument is that the different positioning of the single CD4 binding site on SIV trimers may limit spike crosslinking so that neutralization is reduced to a simple 1:1 trimer binding scenario. In contrast, the orientation of the 3 CD4 binding sites on JR-FL trimers renders them available for inter-spike cross-linking.
The observation that the 4E10 IgG (and probably Z13e1 IgG) trimer affinity matched its neutralization potency was unexpected, considering the differences observed for IgG1b12, CD4-IgG2, and 2F5 IgG. In fact, in this respect, 4E10's behavior better reflects that of the monovalent ligands. A similarity between 4E10 IgG, Z13e1 IgG and monovalent Fabs is further supported by the observation that they form clear complexes with trimers in BN-PAGE (Fig. 6), unlike the smears observed for IgG1b12, CD4-IgG2, and 2F5 IgG. Interestingly, molecular weight estimates indicated that only a single 4E10 or Z13e1 IgG molecule binds to trimers at saturation, in contrast to Fab Z13e1, of which 3 molecules can bind. This might indicate that steric constraints limit access of the larger IgGs to their membrane proximal epitopes. Overall, it is difficult at this stage to determine whether the lack of a marked increase in Z13e1 IgG neutralization compared to its Fab counterpart stems from limited access to its epitope or an inability to crosslink spikes, or indeed both.
A recent study reported that dodecameric D1D2-IgP induce particle “rupturing” (Bennett et al., 2007). Conceivably, this could be the result of the separation of previously crosslinked particles, leading to removal of a “plug” of Env-complex from one partner. Perhaps related to this notion, our consistent observation of significant amounts of unliganded gp120/gp41 monomer in native PAGE at saturating concentrations of IgG1b12 and CD4-IgG2 was unexpected (right lanes in Fig. 6B, C). Since trimers are fully liganded at this concentration, this free monomer is unlikely to indicate low mAb affinity, especially considering that saturating concentrations of monovalent Fab b12 and sCD4 did not leave unliganded monomer (Fig. 3B, D), as did a non-neutralizing IgG, b6 to a similar epitope (Fig. 6A). It might therefore be proposed that IgG binding destabilizes trimers, leading to free monomers and permanent spike inactivation. This provides another possible explanation for the difficulty in visualizing IgG-trimer complexes in Fig. 6. Since the SOS mutant we used here prevents gp120 shedding, the putative dissociation most likely occurs at the trimer axis, leading to gp120/gp41 monomers (Moore et al., 2006). Given the permanent neutralizing effect of this phenomenon, it may be worth investigating the cumulative effects of these IgGs in native PAGE and neutralization assays over time. Clearly, more work will be required to determine exactly how neutralizing IgGs interact with trimers and how this affects neutralization.
MATERIALS AND METHODS
MAbs, HIVIG, C34-Ig and sCD4
Anti-HIV-1 gp120 monoclonal antibodies (mAbs) included b12 and b6, directed to epitopes that overlap the CD4 binding site (CD4bs) (Burton et al., 1994); 2G12, directed to a unique glycan-dependent epitope on gp120 (Sanders et al., 2002; Scanlan et al., 2002); X5, directed to a CD4-inducible (CD4i) epitope (Darbha et al., 2004; Labrijn et al., 2003); 447D, 39F, LE311 and AH48, directed to the V3 loop (Crooks et al., 2005; Schulke et al., 2002); P7, directed to a conformational epitope of the C1 region of gp120 (Ditzel et al., 1997). Anti-gp41 mAbs 2F5, Z13 WT and its derivative Z13e1 are directed to the membrane proximal ectodomain region (Zwick et al., 2001). T2 and T3 are directed to clusters 1 and 2 of gp41, respectively (Binley et al., 1996). Anti-SIV mAbs included 5B11, 7D3, 171C2, and 8C7, 1.7A, and 3.10A (Cole et al., 2001; Edinger et al., 2000). A plasmid expressing the chimeric fusion of the C34 peptide and the Fc portion of IgG (C34-Ig) that recognizes the putative prehairpin gp41 fusion intermediate was expressed in 293T cells and purified over protein G (Si et al., 2004). HIVIG was purified IgG derived from pools of HIV-infected donor plasma. Soluble forms of CD4 (sCD4) included one consisting of all 4 ectodomains (4D-sCD4) and another consisting of domains 1 and 2 only (2D-sCD4).
Fab fragments were in most cases expressed in bacteria, using the pComb3 vector (Barbas et al., 1993). Fab b12 was prepared by papain digestion of IgG according to manufacturer's protocols (Pierce, Rickford, IL). IgGs were digested into monovalent F(ab')2 fragments using immobilized pepsin, respectively, also according to manufacturer's protocols (Pierce, Rickford, IL).
MAbs b12, 2G12, and 2F5 are broadly and potently neutralizing (Binley et al., 2004); mAb Z13 and its improved derivative are weak, but broadly neutralizing; mAb X5 neutralizes primary isolates of HIV-1 in the presence of sCD4 (Crooks et al., 2005; Moulard et al., 2002); the V3 loop-specific Abs neutralize a subset of primary isolates (Binley et al., 2004); HIVIG, T2, T3, b6 and P7 do not neutralize primary HIV-1 isolates.
MAbs 2G12 and 2F5 were provided by Dr. H. Katinger (Polymun Scientific Inc., Vienna, Austria). Two and four domain sCD4 and CD4-IgG2 (Jacobson et al., 2004) were provided by Progenics Pharmaceuticals Inc (Tarrytown, NY). MAb 447D was provided by Dr. Susan Zolla-Pazner (NYU). HIVIG was provided by John Mascola (NIH VRC). The C34-Ig plasmid was provided by Drs. Joe Sodroski and Zhihai Si.
Viruses and plasmids
Pseudoviruses were produced by transient transfection of 293T cells with the plasmids pNL-LucR-E- and an Env expressing plasmid, pCAGGS to express JR-FL gp160.CT SOS, as described previously (Moore et al., 2006). Gp160.CT WT Env clones of SIVmac239, SIVmac316, KNH1144 ec1, 3988, 1196, REJO, TRJO, 92US715, THRO, RHPA, QHO515, CAAN, ZM214, ZM135, ZM249, DU422 and DU172 were constructed in pCAGGS in an analogous manner to JR-FL (Binley et al., 2003). The SIV Envs were cloned with a stop codon at residue 718, leaving a 2 amino acid gp41 tail. Env source plasmids were provided by Welkin Johnson (Harvard), David Montefiori (Duke University), Francine McCutchan and the NIH AIDS Repository. Concentrated preparations of inactivated HIV-1 MN and ADA and SIVmac239 viruses were provided by Drs. Larry Arthur and Jeff Lifson. The SIVmac239 virus has been grown in SupT1 cells and is known to have a stop codon at residue 734.
Neutralization assays
Pseudovirus neutralization assays were performed as described previously (Binley et al., 2003; Crooks et al., 2005). Briefly, in standard assays, virus was incubated for 1 hour with inhibitors, before adding the mixture to CF2.CD4.CCR5 target cells for 2 hours, followed by a wash and a further 2 day incubation. In post-CD4 neutralization assays, virus was incubated with a fixed concentration of 5 µg/ml 4D-sCD4 and graded concentrations of mAb. Subsequently, infection was assayed on CF2 cells bearing high concentrations of surface CCR5. All assays involved a 15 minute 1000 rpm spinoculation step with the remainder of the infection in an incubator at 37°C.
Native PAGE
BN-PAGE was performed as described previously (Crooks et al., 2007; Crooks et al., 2005; Moore et al., 2006). Briefly, concentrated VLPs or inactivated HIV particles were incubated with or without mAb or sCD4 for 5 minutes at RT. The concentration of sCD4 and/or Fab or IgG used in trimer shift experiments was recorded as that in the final mixture with VLPs. In experiments using IgG, VLPs pellets were washed with 1 ml PBS to remove possible interference by unbound IgG upon anti-Fc detection in Western blot. The mixture was then gently solubilized for 5 minutes in 2x solubilization buffer (1% Triton X-100/1% NP40 in 1 mM EDTA) in 1.5 M aminocaproic acid to liberate Env. An equal volume of 2x sample buffer (100 mM MOPS, 100 mM Tris-HCl, pH 7.7, 40 % glycerol, 0.1 % Coomassie blue) was then added prior to loading onto a 4–12% Bis-Tris NuPAGE gel (Invitrogen). Ferritin (Sigma) was used as a size standard. Samples were electrophoresed at 4°C for 3 h at 100 V with 50 mM MOPS/50 mM Tris pH 7.7 containing 0.002 % Coomassie blue as cathode buffer and the same buffer without Coomassie blue as the anode buffer. The gel was then blotted onto PVDF, destained, and probed using mAbs b12, 2G12, E51, 39F, 2F5 and 4E10 (anti-HIV mAb cocktail), a pool of plasmas from HIV+ donors (Crooks et al., 2007) or 5B11, 7D3, 171C2, 8C7, 1.7A and 3.10A (anti-SIV cocktail) (Cole et al., 2001; Edinger et al., 2000). Goat anti-human Fc alkaline phosphatase conjugates were then used to detect the primary Abs (Jackson). Trimer binding was determined by depletion of unliganded trimer in Western blots. Band densities were determined using UN-SCAN-IT software (Silk Scientific). The IC50 of trimer shifts were determined by plotting these data versus ligand concentration. Molecular weights were also estimated using this software according to software guidelines, assigning JR-FL or SIV gp160ΔCT trimers and monomers with molecular weights of 420 and 140, respectively, as markers for defining the masses of other bands.
ACKNOWLEDGEMENTS
This study was supported by NIH RO1 AI58763 (JMB), Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery Vaccine Immune Monitoring Consortium grant #38619 (JMB, JER), the AIDS and Infectious Disease Science Center at the Torrey Pines Institute for Molecular Studies (JMB) and AI49784 (JAH). We thank Dr. Joseph Sodroski and Zhihai Si for providing the CF2-based cell lines and C34-Ig, Kenneth Kang and William Olson at Progenics Pharmaceuticals for providing sCD4, CD4-IgG2 and JR-FL gp120, Hermann Katinger for the 2G12, 2F5 and 4E10 mAbs, Susan Zolla-Pazner for 447-52D, Dennis Burton and Ralph Pantophlet for various Fabs, John Mascola for HIVIG, Francine McCutchan for the KNH1144 ec1 source plasmid, Larry Arthur and Jeff Lifson for live inactivated HIV and SIV preparations, Welkin Johnson for the SIVmac239 and SIVmac316 Env genes, the NIH AIDS Repository for panels of Env source plasmids, Michelle Davis and Ruth Christian for administrative support, and Norbert Schülke, Linda Harris, Ken Roux, Bill Schief and Quentin Sattentau for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arthos J, Cicala C, Steenbeke TD, Chun TW, Dela Cruz C, Hanback DB, Khazanie P, Nam D, Schuck P, Selig SM, Van Ryk D, Chaikin MA, Fauci AS. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J Biol Chem. 2002;277(13):11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230(3):812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- Bennett A, Liu J, Van Ryk D, Bliss D, Arthos J, Henderson RM, Subramaniam S. Cryoelectron tomographic analysis of an HIV-neutralizing protein and its complex with native viral gp120. J Biol Chem. 2007;282(38):27754–27759. doi: 10.1074/jbc.M702025200. [DOI] [PubMed] [Google Scholar]
- Binley JM, Cayanan CS, Wiley C, Schulke N, Olson WC, Burton DR. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J Virol. 2003;77(10):5678–5684. doi: 10.1128/JVI.77.10.5678-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ditzel HJ, Barbas CF, 3rd, Sullivan N, Sodroski J, Parren PW, Burton DR. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12(10):911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- Binley JM, Ngo-Abdalla S, Moore P, Bobardt M, Chatterji U, Gallay P, Burton DR, Wilson IA, Elder JH, de Parseval A. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology. 2006;3:39. doi: 10.1186/1742-4690-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A, Bullock BT, Gisselman KM, Margulies BJ, Clements JE. A single amino acid change and truncated TM are sufficient for simian immunodeficiency virus to enter cells using CCR5 in a CD4-independent pathway. Virology. 2005;341(1):12–23. doi: 10.1016/j.virol.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68(9):6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BK, Darden JM, Tovanabutra S, Oblander T, Frost J, Sanders-Buell E, de Souza MS, Birx DL, McCutchan FE, Polonis VR. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J Virol. 2005;799(10):6089–6101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR. Fc Receptors and the Action of Antibodies American Societry for Microbiology. Washington, D.C: 1990. The Conformation of Antibodies; pp. 31–54. [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Burton DR, Saphire EO, Parren PW. A model for neutralization of viruses based on antibody coating of the virion surface. Curr Top Microbiol Immunol. 2001;260:109–143. doi: 10.1007/978-3-662-05783-4_7. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Center RJ, Earl PL, Lebowitz J, Schuck P, Moss B. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J Virol. 2000;74(10):4448–4455. doi: 10.1128/jvi.74.10.4448-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Choudhry V, Zhang MY, Harris I, Sidorov IA, Vu B, Dimitrov AS, Fouts T, Dimitrov DS. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem Biophys Res Commun. 2006;348(3):1107–1115. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KS, Alvarez M, Elliott DH, Lam H, Martin E, Chau T, Micken K, Rowles JL, Clements JE, Murphey-Corb M, Montelaro RC, Robinson JE. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology. 2001;290(1):59–73. doi: 10.1006/viro.2001.1144. [DOI] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, de Vries RP, L WC, Zharkikh I, Schülke N, Roux KH, Montefiori DC, Burton DR, Binley JM. A comparative immunogenicity study of HIV-1 virus-like particles 3 bearing various forms of envelope proteins, particles bearing no 4 envelope and soluble monomeric gp120. Virology. 2007 doi: 10.1016/j.virol.2007.04.033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Richman D, Robinson J, Crooks JA, Franti M, Schulke N, Binley JM. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum Antibodies. 2005;14(3–4):101–113. [PMC free article] [PubMed] [Google Scholar]
- Darbha R, Phogat S, Labrijn AF, Shu Y, Gu Y, Andrykovitch M, Zhang MY, Pantophlet R, Martin L, Vita C, Burton DR, Dimitrov DS, Ji X. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry. 2004;43(6):1410–1417. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201(9):1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel HJ, Parren PW, Binley JM, Sodroski J, Moore JP, Barbas CF, 3rd, Burton DR. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267(3):684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- Earl PL, Doms RW, Moss B. Multimeric CD4 binding exhibited by human and simian immunodeficiency virus envelope protein dimers. J Virol. 1992;66(9):5610–5614. doi: 10.1128/jvi.66.9.5610-5614.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Ahuja M, Sung T, Baxter KC, Haggarty B, Doms RW, Hoxie JA. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J Virol. 74 17;:7922–7935. doi: 10.1128/jvi.74.17.7922-7935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini EA, Kao SY, Lewis AJ, Crainic R, Wimmer E. Functional basis of poliovirus neutralization determined with monospecific neutralizing antibodies. J Virol. 1983;46(2):466–474. doi: 10.1128/jvi.46.2.466-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71(4):2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2008;105(10):3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5(4):276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Grundner C, Mirzabekov T, Sodroski J, Wyatt R. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J Virol. 2002;76(7):3511–3521. doi: 10.1128/JVI.76.7.3511-3521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson DC, Yguerabide J, Schumaker VN. Segmental flexibility of immunoglobulin G antibody molecules in solution: a new interpretation. Biochemistry. 1981;20(24):6842–6852. doi: 10.1021/bi00527a016. [DOI] [PubMed] [Google Scholar]
- Herrera C, Spenlehauer C, Fung MS, Burton DR, Beddows S, Moore JP. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J Virol. 2003;77(2):1084–1091. doi: 10.1128/JVI.77.2.1084-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson JM, Israel RJ, Lowy I, Ostrow NA, Vassilatos LS, Barish M, Tran DN, Sullivan BM, Ketas TJ, O'Neill TJ, Nagashima KA, Huang W, Petropoulos CJ, Moore JP, Maddon PJ, Olson WC. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob Agents Chemother. 2004;48(2):423–429. doi: 10.1128/AAC.48.2.423-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Sanford H, Schwall L, Burton DR, Parren PW, Robinson JE, Desrosiers RC. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J Virol. 2003;77(18):9993–10003. doi: 10.1128/JVI.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Chen B, Hussey RE, Chishti Y, Montefiori D, Hoxie JA, Byron O, Campbell G, Harrison SC, Reinherz EL. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J Biol Chem. 2001;276(46):42667–42676. doi: 10.1074/jbc.M104166200. [DOI] [PubMed] [Google Scholar]
- Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83(Pt 9):2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313(2):387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- Kodama T, Wooley DP, Naidu YM, Kestler HW, 3rd, Daniel MD, Li Y, Desrosiers RC. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63(11):4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74(4):1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. Access of Antibody Molecules to the Conserved Coreceptor Binding Site on Glycoprotein gp120 Is Sterically Restricted on Primary Human Immunodeficiency Virus Type 1. J Virol. 2003;77(19):10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney TL, McLain L, Armstrong SJ, Dimmock NJ. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a postfusion event. Virology. 1997;233(2):313–326. doi: 10.1006/viro.1997.8547. [DOI] [PubMed] [Google Scholar]
- Mkrtchyan SR, Markosyan RM, Eadon MT, Moore JP, Melikyan GB, Cohen FS. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J Virol. 2005;79(17):11161–11169. doi: 10.1128/JVI.79.17.11161-11169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80(5):2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A. 2002;99(10):6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol. 1998;72(11):9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W, Lu N, Yu SS, Silver J. Effect of epitope position on neutralization by anti-human immunodeficiency virus monoclonal antibody 2F5. J Virol. 2006;80(5):2539–2547. doi: 10.1128/JVI.80.5.2539-2547.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Lebowitz J, Schon A, Zhu P, Freire E, Kwong PD, Roux KH, Sodroski J, Wyatt R. Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis. J Virol. 2005;79(15):9954–9969. doi: 10.1128/JVI.79.15.9954-9969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Mondor I, Naniche D, Ditzel HJ, Klasse PJ, Burton DR, Sattentau QJ. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72(5):3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PWHI, Burton DR. The anti-viral activity of antibodies in vitro and in vivo. Adv. Immunol. 2000 doi: 10.1016/S0065-2776(01)77018-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phogat S, Svehla K, Tang M, Spadaccini A, Muller J, Mascola J, Berkower I, Wyatt R. Analysis of the human immunodeficiency virus type 1 gp41 membrane proximal external region arrayed on hepatitis B surface antigen particles. Virology. 2008;373(1):72–84. doi: 10.1016/j.virol.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikora C, Wittish C, Desrosiers RC. Identification of two N-linked glycosylation sites within the core of the simian immunodeficiency virus glycoprotein whose removal enhances sensitivity to soluble CD4. J Virol. 2005;79(19):12575–12583. doi: 10.1128/JVI.79.19.12575-12583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P, Fouts T, Naniche D, Moore JP, Sattentau QJ. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1996;183(2):473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77(1):353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer BA, Altamura LA, Pierson TC, Doms RW. Determinants within gp120 and gp41 contribute to CD4 independence of SIV Envs. Virology. 2004;327(1):16–25. doi: 10.1016/j.virol.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol. 2002;76(6):2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol. 1997;159(7):3372–3382. [PubMed] [Google Scholar]
- Salzwedel K, Smith ED, Dey B, Berger EA. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74(1):326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76(14):7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293(5532):1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182(1):185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of a1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Saphire EO, Calarese D, Stanfield R, Wilson IA, Katinger H, Dwek RA, Burton DR, Rudd PM. The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv Exp Med Biol. 2003;535:205–218. doi: 10.1007/978-1-4615-0065-0_13. [DOI] [PubMed] [Google Scholar]
- Schonning K, Lund O, Lund OS, Hansen JE. Stoichiometry of monoclonal antibody neutralization of T-cell line-adapted human immunodeficiency virus type 1. J Virol. 1999;73(10):8364–8370. doi: 10.1128/jvi.73.10.8364-8370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76(15):7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, 3rd, Sodroski JG. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A. 2004;101(14):5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3(5):e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenlehauer C, Kim A, Aubertin AM, Moog C. Antibody-mediated neutralization of primary human immunodeficiency virus type 1 isolates: investigation of the mechanism of inhibition. J Virol. 2001;75(5):2235–2245. doi: 10.1128/JVI.75.5.2235-2245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Kan E, Sun Y, Sharma VA, Cisto J, Burke B, Lian Y, Hilt S, Biron Z, Hartog K, Stamatatos L, Cheng RH, Ulmer JB, Barnett SW. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2007 doi: 10.1016/j.virol.2007.10.022. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Legg H, Kan E, Fong A, Coates SR, Leung L, Wininger M, Donnelly JJ, Ulmer JB, Barnett SW. Purification and characterization of oligomeric envelope glycoprotein from a primary r5 subtype B human immunodeficiency virus. J Virol. 2002;76(6):2835–2847. doi: 10.1128/JVI.76.6.2835-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69(11):6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S-H, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-Induced Epitopes on the HIV-1 gp120 Envelope Glycoprotein Recognized by Neutralizing Human Monoclonal Antibodies. AIDS Research and Human Retroviruses. 2002 doi: 10.1089/08892220260387959. In Press. [DOI] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Lee S, Sodroski J. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J Virol. 2005a;79(6):3500–3508. doi: 10.1128/JVI.79.6.3500-3508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005b;79(19):12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J Virol. 2006a;80(9):4388–4395. doi: 10.1128/JVI.80.9.4388-4395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lipchina I, Cocklin S, Chaiken I, Sodroski J. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J Virol. 2006b;80(22):11404–11408. doi: 10.1128/JVI.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Craig S, Si Z, Farzan M, Sodroski J. CD4-induced T-20 binding to human immunodeficiency virus type 1 gp120 blocks interaction with the CXCR4 coreceptor. J Virol. 2004;78(10):5448–5457. doi: 10.1128/JVI.78.10.5448-5457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste E, Johnson W, Pavlakis GN, Desrosiers RC. Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J Virol. 2005;79(19):12455–12463. doi: 10.1128/JVI.79.19.12455-12463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex In Situ. PLoS Pathog. 2006;2(8) doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zhu P, Olson WC, Roux KH. Structural flexibility and functional valence of CD4-IgG2 (PRO 542): potential for cross-linking human immunodeficiency virus type 1 envelope spikes. J Virol. 2001;75(14):6682–6686. doi: 10.1128/JVI.75.14.6682-6686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]