Abstract
In this study we demonstrate, at an ultrastructural level, the in situ distribution of heterogeneous nuclear RNA transcription sites after microinjection of 5-bromo-UTP (BrUTP) into the cytoplasm of living cells and subsequent postembedding immunoelectron microscopic visualization after different labeling periods. Moreover, immunocytochemical localization of several pre-mRNA transcription and processing factors has been carried out in the same cells. This high-resolution approach allowed us to reveal perichromatin regions as the most important sites of nucleoplasmic RNA transcription and the perichromatin fibrils (PFs) as in situ forms of nascent transcripts. Furthermore, we show that transcription takes place in a rather diffuse pattern, without notable local accumulation of transcription sites. RNA polymerase II, heterogeneous nuclear ribonucleoprotein (hnRNP) core proteins, general transcription factor TFIIH, poly(A) polymerase, splicing factor SC-35, and Sm complex of small nuclear ribonucleoproteins (snRNPs) are associated with PFs. This strongly supports the idea that PFs are also sites of major pre-mRNA processing events. The absence of nascent transcripts, RNA polymerase II, poly(A) polymerase, and hnRNPs within the clusters of interchromatin granules rules out the possibility that this domain plays a role in pre-mRNA transcription and polyadenylation; however, interchromatin granule-associated zones contain RNA polymerase II, TFIIH, and Sm complex of snRNPs and, after longer periods of BrUTP incubation, also Br-labeled RNA. Their role in nuclear functions still remains enigmatic. In the nucleolus, transcription sites occur in the dense fibrillar component. Our fine structural results show that PFs represent the major nucleoplasmic structural domain involved in active pre-mRNA transcriptional and processing events.
INTRODUCTION
RNA transcription and processing take place in association with discrete subnuclear structures. The functional organization of these nuclear substructures still remains an incompletely explored area of cell biology, despite the fact that the first studies were performed years ago (e.g., Swift, 1962; Smetana et al., 1963; Monneron and Bernhard, 1969).
A number of earlier studies identified, using radioactively labeled RNA and high-resolution autoradiography, the border of condensed chromatin as the morphological substrate of transcription. Moreover, these investigations have demonstrated that perichromatin fibrils (PFs) are the in situ form of the nascent transcripts and further indicated a migration of a portion of PFs toward the interchromatin space while their RNA was undergoing processing (for reviews, see Fakan and Puvion, 1980; Fakan, 1986; Puvion and Moyne, 1981). Furthermore, immunocytochemical (Fakan et al., 1984; Puvion et al., 1984; Spector et al., 1991) and in situ hybridization (Visa et al., 1993b) analyses strongly support the idea that splicing and polyadenylation of pre-mRNA also occur in association with PFs (for reviews, see Spector, 1993; Fakan, 1994; van Driel et al., 1995; Puvion and Puvion-Dutilleul, 1996). These observations were later confirmed by means of nonradioactive RNA-labeling methods and immunofluorescence microscopy (Jackson et al., 1993; Wansink et al., 1993; Fay et al., 1997).
Interchromatin granules (IGs), coiled bodies, and the recently described compartment called IG-associated zone (Visa et al., 1993a) are considered to be initial sites for assembly of different splicing factors into processing complexes, which then move to the nuclear domains where RNA processing takes place (for reviews, see Fakan, 1994; Puvion and Puvion-Dutilleul, 1996; Spector, 1996). This favors previous indirect evidence showing cotranscriptional association of splicing factors (Fakan et al., 1986) and suggesting that splicing can be a cotranscriptional event (Beyer and Osheim, 1988) using splicing complexes probably assembled elsewhere in the nucleus (Amero et al., 1992). Whether IGs can act as recycling sites for some splicing factors remains unclear (Lamond and Carmo-Fonseca, 1993; Puvion and Puvion-Dutilleul, 1996).
Recent in vivo observations that focused on pre-mRNA processing events (Misteli et al., 1997) are in agreement with previous reports (e.g., Malatesta et al., 1994; Tamburini et al., 1996) suggesting that the interphase nucleus is a dynamic cell compartment. Moreover, it has been proposed that an interaction between mRNA and different mRNA-processing machinery factors depends on a transcriptional status of the nucleus (Huang and Spector, 1996; Misteli et al., 1997; Zeng et al., 1997).
To obtain new insights into the functional organization of the nucleus, we have investigated in this report nucleoplasmic RNA transcriptional events with relation to splicing and polyadenylation of pre-mRNA at the ultrastructural level. To localize the sites of transcription in situ by this high-resolution method, we microinjected 5-bromo-UTP (BrUTP) into living cells. Postembedding immunoelectron microscopy was used for visualizing both Br-labeled RNA (Br-RNA) and different factors involved in pre-mRNA transcription and processing. Moreover, we analyzed the possible colocalization of the nascent RNA with such factors within different nuclear subcompartments.
We demonstrate that PFs are the major in situ nucleoplasmic structural domains labeled after short BrUTP pulses and that important pre-mRNA transcription and processing factors are preferentially associated with these structural constituents. Our observations support the idea that PFs are not only the in situ form of nascent transcripts but they also represent sites of pre-mRNA processing.
MATERIALS AND METHODS
Cell Culture
Human bladder carcinoma cells T24 (ATCCHB4) were grown at 37°C under a 10% CO2 atmosphere in DMEM (Life Technologies, Breda, The Netherlands) supplemented with 10% heat-inactivated fetal calf serum (Boehringer Mannheim, Mannheim, Germany), 2 mM l-glutamine (Life Technologies), 100 IU/ml penicillin, and 100 μg/ml streptomycin (Life Technologies). Cells were cultured on Alcian Blue-coated (Brink et al., 1992) and microgridded Cellocate coverslips (Eppendorf, Hamburg, Germany); cells were used at 50–70% confluency.
Microinjection and Fixation
Microinjection of BrUTP (Sigma, St. Louis, MO) was performed as described previously (Wansink et al., 1993, 1994a). Briefly, cells were injected into the cytoplasm of living cells with 100 mM BrUTP in 140 mM KCl and 2 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.4. Approximately 5% cell volume was injected. After microinjection, cells were cultured for 4–90 min at 37°C and fixed with 4% paraformaldehyde in 0.1 M Sörensen phosphate buffer, pH 7.4, for 60 min on ice. For immunofluorescence, the microinjected cells were fixed with 4% paraformaldehyde in PBS for 10 min. To determine the exact time interval between injection and fixation, the injection time and the position of injected cells were recorded.
Immunofluorescence Labeling and Confocal Microscopy
The fixed cells were permeabilized with 0.5% Triton X-100 (Sigma) in PBS and incubated with PBS containing 100 mM glycine (Sigma) for 10 min. Subsequently, cells were incubated overnight at 4°C with a rat anti-bromodeoxyuridine (BrdU) antibody (Table 1) diluted 1:500 in PBS. Anti-BrdU antibodies were shown to recognize bromouridine with high specificity and affinity (Vanderlaan and Thomas, 1985). After washing in PBS, cells were incubated with a secondary anti-rat immunoglobulin G antibody coupled to biotin (Jackson, West Grove, PA) for 1 h at room temperature and subsequently for 30 min with either Cy3- or FITC-conjugated streptavidin (Jackson) or FITC-conjugated donkey anti-rat antibody (Jackson), diluted in PBS. For double-immunolabeling experiments, cells were incubated overnight simultaneously with anti-BrdU and a mouse monoclonal antibody against the proliferation-associated nuclear antigen (PANA) considered as a marker of IG (Clevenger and Epstein, 1984). Anti-PANA was detected with either FITC- or Cy3-conjugated donkey anti-mouse immunoglobulin M (Jackson) diluted in PBS. When anti-PANA and anti-BrdU were incubated separately, the same localization results were obtained. The specificity of each of the second antibodies was tested by omitting one of the primary antibodies. Slides were mounted in Vectashield (Vector, Burlingame, CA); they were kept at 4°C until evaluation and were observed within 24 h.
Table 1.
Antibodies used for immunoelectron microscopy
| Primary antibody | Dilution | Origin | Gold marker (nm) |
|---|---|---|---|
| Mouse anti-BrdU | 1 :50 | Partec (Münster, Germany) | GAM 5, 6, 12, or 15 |
| Rat anti-BrdU | 1 :50 | Sera-Lab (Crawley Down, UK) | GARa 6 |
| Chicken anti-hnRNP core proteina | 1 :10000 | Jones et al. (1980) | GAR 15 |
| Rabbit anti-RNA polymerase IIa | 1 :2 | Kim and Dahmus (1986) | GAR 15 |
| Chicken anti-RNA polymerase IIa | 1 :10000 | Carroll and Stollar (1983) | GAR 15 |
| Rabbit anti-p80-coilina | 1 :50–200 | Andrade et al. (1991) | GAR 15 |
| Rabbit anti-poly(A)-polymerasea | 1 :3 | Martin and Keller (unpublished observations) | GAR 15 |
| Mouse anti-SC35 splicing factorb | 1 :50 | Fu and Maniatis (1990) | GAM 15 |
| Mouse anti-Sm complex snRNPsb | 1 :25 | Lerner et al. (1981) | GAM 15 |
| Mouse anti-transcription factor TFIIHb | 1 :50 | Schaeffer et al. (1994) | GAM 15 |
GAM, goat anti-mouse secondary antibody conjugated with colloidal gold (Aurion, Wageningen, The Netherlands); GAR, goat anti-rabbit secondary antibody conjugated with colloidal gold (Aurion); GARa, goat anti-rat secondary antibody conjugated with colloidal gold (Aurion).
Antibodies used for double-labeling with mouse anti-BrdU.
Antibodies used for double-labeling with rat anti-BrdU.
Images were recorded with a Leica confocal laser scanning microscope equipped with a 100×/1.23 NA oil immersion lens. A dual-wavelength argon ion laser was used to excite FITC and Cy3 fluorochromes simultaneously at 488 and 514 nm, respectively. Emitted fluorescence was detected using a 525 DF10 bandpass filter for FITC and a 550-nm longpass filter for Cy3. Pairs of images were collected simultaneously in the green and red channel. Three-dimensional images were scanned as 512 × 512 × 32 voxel images (sampling rate 49 nm lateral and 208 nm axial). Optical cross talk was quantified, and images were corrected (Manders et al., 1992). Image analysis was performed using SCIL-IMAGE software (Ten Kate et al., 1990; van Balen et al., 1994). Images were subjected to a restoration procedure to correct for diffraction-induced distortion using a measured point-spread function (van der Voort and Straster, 1995).
Immunoelectron Microscopy
After fixation, the cells were repeatedly washed in Sörensen phosphate buffer and PBS to remove free aldehydic groups as well as nonincorporated BrUTP. The cells were then dehydrated in ethanol and embedded in LRWhite resin that was polymerized by heat. The embedded cells were separated from the gridded coverslips after short treatment with liquid nitrogen. The injected cells were localized by the α-numeric imprint on the surface of the block and cut parallel to the substrate using a Leica Ultracut UCT ultramicrotome. The ultrathin sections were mounted on Formvar/carbon-coated nickel grids and processed, with minor changes, for postembedding immunogold labeling as described previously (Biggiogera et al., 1989; Malatesta et al., 1994). Antibodies used for immunoelectron microscopy are listed in Table 1. Briefly, the grids with sections were pretreated with 10% normal goat serum in PBS for 10 min and then reacted, for 17 h at 4°C, with a mixture of primary antibodies diluted in PBS containing 0.05% Tween 20 (Sigma) and 1% BSA (Fluka, Buchs, Switzerland). After washing with PBS-Tween and PBS alone, followed by a repeated treatment with normal goat serum for 10 min, the sections were reacted with a mixture of colloidal gold-conjugated secondary antibodies in PBS at room temperature for 30 min. When chicken primary antibodies were used, a rabbit anti-chicken probe (EY Labs, San Mateo, CA), diluted 1:100 in PBS/Tween/BSA, was used as a bridge before gold-complex labeling. Finally, all grids were thoroughly rinsed with PBS and ultrapure water and air-dried. The preparations were stained by the regressive technique, which is preferential for nuclear ribonucleoproteins (Bernhard, 1969): 4.7% aqueous uranyl acetate for 45 sec, 0.02 M EDTA for 3 min, and lead citrate for 45 sec.
As controls, noninjected cells were processed as above. Moreover, the injected cells treated without primary antibodies or without rabbit anti-chicken bridge probe were used. To confirm the RNA nature of Br-labeling, some sections were also submitted to RNA digestion with 0.2% ribonuclease (type IA, Sigma) in 1 mM triethanolamine–acetic acid buffer, pH 7.3, for 18 h at 37°C.
The grids were examined with a Philips (Eindhoven, The Netherlands) CM 100 electron microscope at 80 kV using a 30–40 μm objective aperture.
Evaluation of the Gold Grain Density
For semiquantitative evaluation, the estimation refers to the minimal and maximal presence of colloidal gold granules found on subnuclear compartments. Local density of gold grains was estimated separately for each antibody. The evaluation of the specific labeling was as follows: zero = very weak or no signal; single symbol = intensity of signal was moderate, but evidently above background level; double symbol = intensity of gold labeling is between minimal and maximal level; triple symbol = maximal labeling intensity. A similar method was also used for the estimation of colocalization between two antibodies reflected by codistribution of different size colloidal gold particles. Background staining was evaluated as a binding of antibodies to the resin over regions free of cells.
RESULTS
The results of this study show that microinjection of BrUTP into living cells is a very useful tool for in vivo labeling of RNA and its subsequent in situ localization by electron microscopical methods. The ability to reveal the incorporated precursor directly on ultrathin resin sections allows one to avoid permeabilization of cells and therefore prevents deleterious effects of detergents and makes possible a more satisfactory level of preservation of ultrastructural details. In addition, sectioning makes accessible antigenic sites in compact nuclear domains and thus prevents antibody penetration problems encountered in fluorescence or preembedding immunocytochemistry. The use of gridded coverslips allowed us to visualize, with good precision, the individual microinjected cells. In the ultrathin sections of cells processed in situ, the fine structural morphology of the cell nucleus was well preserved so that all the nuclear constituents could be identified easily.
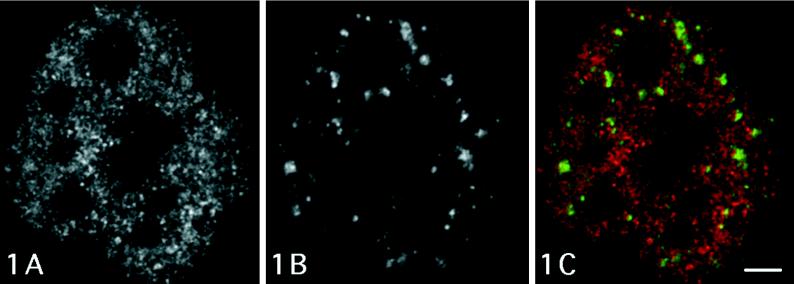
Figure 1 shows a confocal optical section of a cell nucleus visualizing BrUTP-labeled RNA after 10 min incubation and the distribution of anti-PANA labeling. Anti–Br-RNA signal exhibits the typical distribution pattern demonstrating diffuse labeling with a number of small spot-like areas, which represents transcriptionally active regions on the periphery of condensed chromatin clumps (Figure 1A). The nucleoli (large dark areas) remain virtually unlabeled because of an insufficient antibody penetration (Wansink et al., 1993). An immunofluorescence assay with the anti-PANA probe recognizing a marker protein of IG (Clevenger and Epstein, 1984) reveals several speckled regions corresponding to IG clusters (Figure 1B). When incubation with the anti-PANA antibody is carried out in a double-labeling experiment, the PANA-positive speckles occur in nucleoplasmic areas that do not exhibit significant anti-BrUTP labeling (Figure 1C).
Figure 1.
Confocal optical section of a microinjected T24 cell showing immunocytochemical signal for Br-RNA, after 10 min incubation, and PANA marker of clusters of IGs. Large unlabeled areas correspond to nucleoli. (A) The typical diffuse distribution of sites of nascent RNA is demonstrated. (B) Several anti-PANA–labeled speckled regions correspond to clusters of IGs. (C) Merged colocalization image shows very little overlapping between anti-PANA–labeled regions (green) and Br-RNA (red). Bar, 2.0 μm.
Tables 2 and 3 summarize results of ultrastructural immunolabeling of Br-RNA and of the distribution of antibodies recognizing different transcription and processing factors and the estimation of the extent of their colocalization, respectively.
Table 2.
Summary of immunoelectron microscopic distribution of individual antibodies
| Antibody | Nuclear area | 4–5 min | 6–7 min | 8–9 min | 10–11 min | 12–13 min | 14–15 min | 20–25 min | 30–35 min | 90 min |
|---|---|---|---|---|---|---|---|---|---|---|
| Anti-BrdU | PF | x | xx | xx | xx | xx | xx | xxx | xxx | xxx |
| IG | o | o | o | o | o | x | x | x | x | |
| IGAZ | o | o | o | o | x | x | xxx | xxx | xxx | |
| Anti-RNA pol II | PF | xx | xx | xx | xx | xx | xx | xx | xx | xx |
| IG | o | o | o | o | o | o | o | o | o | |
| IGAZ | x | x | x | x | x | x | x | x | x | |
| Anti-hnRNP | PF | xx | xx | xx | xx | xx | xx | xx | xx | − |
| IG | o | o | o | o | o | o | o | o | − | |
| IGAZ | o | o | o | o | o | o | o | o | − | |
| Anti-SC35 | PF | x | x | x | x | x | x | x | x | − |
| IG | xx | xx | xx | xx | xx | xx | xx | xx | − | |
| IGAZ | o | o | o | o | o | o | o | o | − | |
| Anti-Sm complex of snRNPs | PF | xx | xx | xx | xx | xx | xx | xx | xx | − |
| IG | xx | xx | xx | xx | xx | xx | xx | xx | − | |
| IGAZ | x | x | x | x | x | x | x | xx | − | |
| Anti-poly(A) polymerase | PF | xx | xx | xx | xx | xx | xx | xx | xx | − |
| IG | o | o | o | o | o | o | o | o | − | |
| IGAZ | o | o | o | o | o | o | o | o | − | |
| Anti-TFIIH | PF | xx | xx | xx | xx | xx | xx | xx | xx | − |
| IG | x | x | x | x | x | x | x | x | − | |
| IGAZ | x | x | x | x | x | x | x | x | − |
o, x, xx, xxx, Estimation of intensity of labeling; PF, perichromatin fibrils; IG, interchromatin granules; IGAZ, IG-associated zones; −, not done.
Table 3.
Summary of immunoelectron microscopic colocalization of Br-RNA and of transcription and processing factors
| Nuclear area | Antibody | 4–5 min | 6–7 min | 8–9 min | 10–11 min | 12–13 min | 14–15 min | 20–25 min | 30–35 min | 90 min |
|---|---|---|---|---|---|---|---|---|---|---|
| PF | Anti-pol II | xxx | xxx | xxx | xx | xx | xx | xx | xx | xx |
| Anti-hnRNP | x | x | xx | xx | xx | xx | xxx | xxx | − | |
| Anti-SC35 | x | xx | xx | xx | xx | xx | xx | xx | − | |
| Anti-Sm complex | x | xx | xx | xx | xx | xx | xx | xxx | − | |
| Anti-poly(A)pol | x | x | x | x | x | x | x | x | − | |
| Anti-TFIIH | xx | xx | xx | xx | xx | xx | xx | xx | − | |
| IG | Anti-pol II | o | o | o | o | o | o | o | o | − |
| Anti-hnRNP | o | o | o | o | o | o | o | o | − | |
| Anti-SC35 | o | o | o | o | o | x | x | xx | − | |
| Anti-Sm complex | o | o | o | o | o | x | x | xx | − | |
| Anti-poly(A)pol | o | o | o | o | o | o | o | o | − | |
| Anti-TFIIH | o | o | o | o | o | x | xx | xx | − | |
| IGAZ | Anti-pol II | o | o | o | o | x | x | x | x | − |
| Anti-hnRNP | o | o | o | o | o | o | o | o | − | |
| Anti-SC35 | o | o | o | o | o | o | o | o | − | |
| Anti-Sm complex | o | o | o | o | x | x | x | xx | − | |
| Anti-poly(A)pol | o | o | o | o | o | o | o | o | − | |
| Anti-TFIIH | o | o | o | o | x | x | x | x | − |
o, x, xx, xxx, Estimation of intensity of colocalization with anti-BrdU antibodies; PF, perichromatin fibrils; IG, interchromatin granules; IGAZ, IG-associated zones; −, not done.
The anti-BrdU antibodies used in our experiments showed the strongest signal of several probes of different commercial origins; the antibody from Partec exhibits especially high affinity for Br-RNA. As to the specificity of anti-RNA polymerase II antibodies used in our experiments, the rabbit affinity-purified antibody (Kim and Dahmus, 1986) reacts predominantly with the phosphorylated C-terminal domain of the RNA polymerase II0 subunit (Cadena and Dahmus, 1987), whereas the chicken antibody (Carroll and Stollar, 1983) obviously recognizes several forms of the enzyme.
The distribution pattern of Br-RNA and of the different factors involved in pre-mRNA formation investigated in the present work were evaluated with regard to different nucleoplasmic domains previously reported as being involved in pre-mRNA metabolism. In the tables describing the degree of labeling in such nucleoplasmic domains, we analyzed PF as the major RNP constituent of the perichromatin region, whereas IG and IGλ-associated zones obviously represent the most important and remarkable domains in the interchromatin nucleoplasmic space.
Perichromatin Region
Intensive labeling of newly synthesized RNA, mainly occurring on the periphery of condensed chromatin areas where PFs were the major labeled nucleoplasmic constituents, was observed 20–35 min after BrUTP microinjection (Figures 2 and 3); however, evident signal was already identified at much shorter periods, from 4 min onward, after microinjection of cells. In this case, mainly individual gold particles were observed associated predominantly with PFs (Figures 4 and 5). No striking local accumulation of gold particles was observed, regardless of the duration of the labeling period. The labeling intensity within the perichromatin regions was increasing with the increasing labeling time. Some agregation of labeled PFs that had accumulated within these regions was observed after prolonged incubation periods (20–90 min); however, the general distribution pattern of Br-RNA within this region remained comparable for all labeling periods. Anti-RNA polymerase II antibodies colocalized frequently with Br-RNA, especially after shorter incubation periods (Figure 6). This frequency of colocalization, however, decreased with the increasing duration of BrUTP labeling.
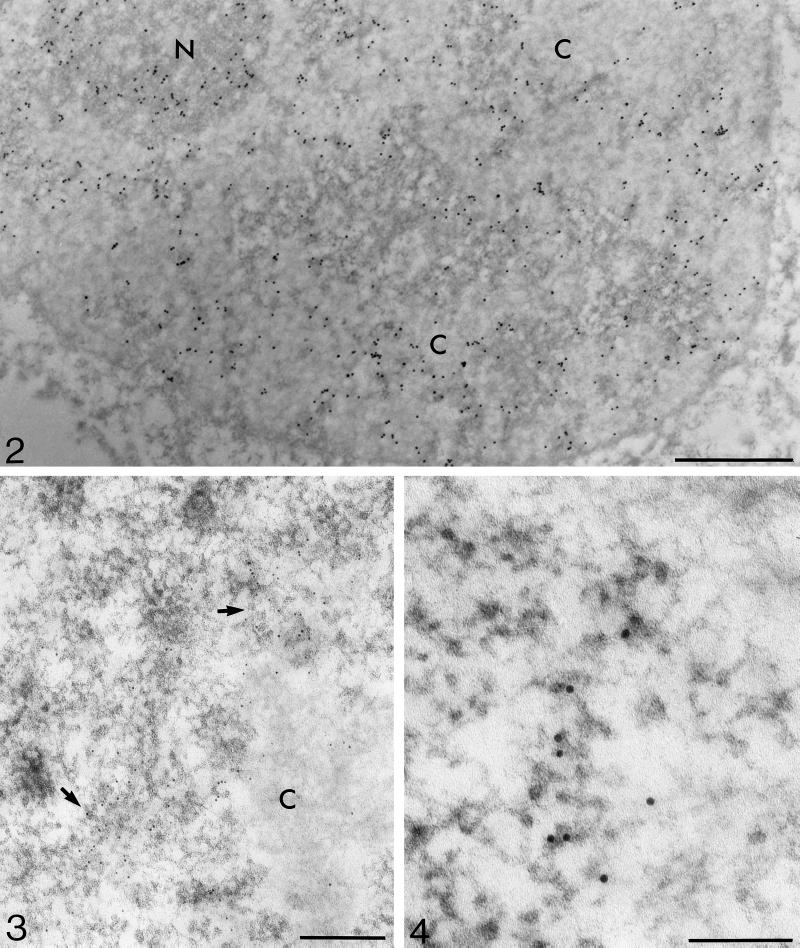
Figure 2.
Immunoelectron microscopic localization of BrU (mouse antibody) in BrUTP-microinjected T24 cells after 20-min incubation. Gold marker labels the perichromatin region in the nucleoplasm and the dense fibrillar component in the nucleolus (N). Condensed chromatin areas (C) and the interchromatin region (contrasted areas) are mostly devoid of label. Bar, 1.0 μm.
Figure 3.
Thirty minutes after microinjection of BrUTP, two regions located on condensed chromatin periphery, looking like transcriptional complexes with multiple transcripts, exhibit strongly anti-BrU–labeled (mouse anti-BrdU antibody) PFs (arrows). Bar, 0.2 μm.
Figure 4.
A high-power micrograph shows, after pulse of 7 min 40 sec, anti-BrU label (mouse anti-BrdU antibody) on PFs arranged in an array resembling a transcription complex. Bar, 0.1 μm.
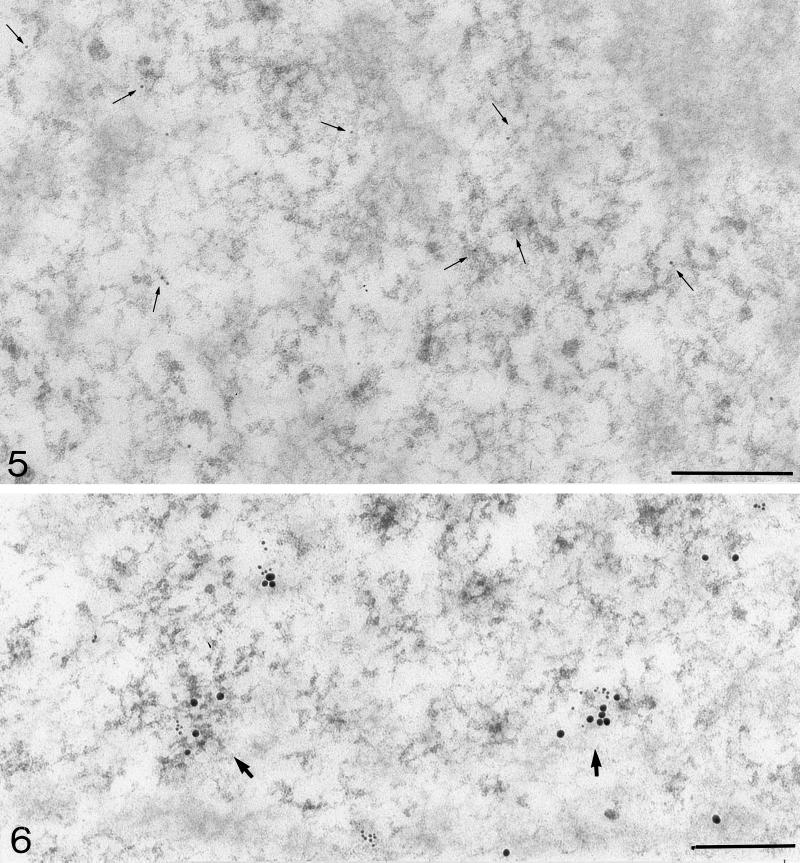
Figure 5.
Localization of brominated RNA (mouse anti-BrdU antibody) after incubation of 4 min 20 sec. Mostly individual 5-nm gold particles are localized on PFs (arrows). Bar, 0.2 μm.
Figure 6.
Double-labeling with mouse anti-BrdU antibody (small grains, 6 nm) and chicken anti-RNA polymerase II antibody (large grains, 15 nm) after incubation of 7 min 20 sec. Colocalization is visible on PFs (arrows). Bar, 0.2 μm.
The other specific probes colocalized to different extents during all BrUTP incubation periods (Figures 7 and 8 and Table 3). With increasing BrU incubation time, the fre-quency of BrU colocalization with antipoly(A) polymerase and with anti-TFIIH remained equal while the colocalization with anti-hnRNP and anti-SC35 was increasing.
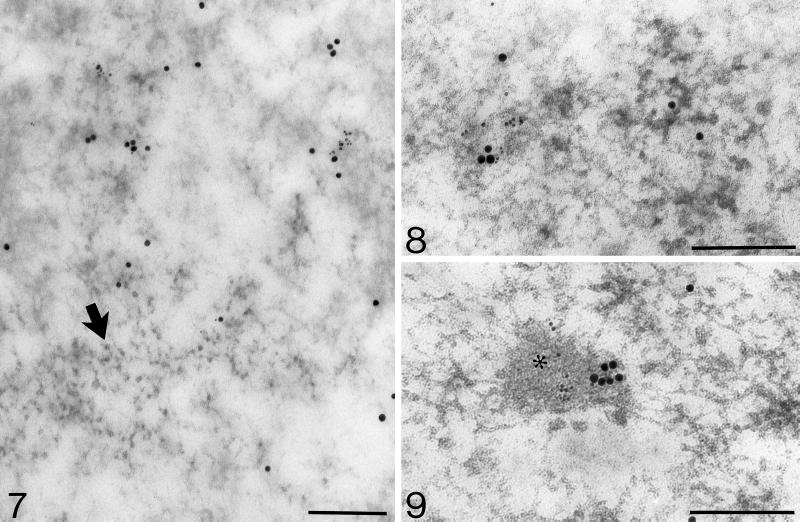
Figure 7.
Mouse anti-BrdU antibody (6-nm grains) and hnRNP (15-nm grains) colocalization on PFs after 8-min incubation. Note unlabeled IGs (arrow). Bar, 0.2 μm.
Figure 8.
Colocalization of Br-RNA (mouse anti-BrdU antibody; 6-nm particles) and poly(A)-polymerase (15-nm particles) on PFs after 5 min incubation. Bar, 0.2 μm.
Interchromatin Region
We have not observed major variations in the overall BrUTP labeling of the interchromatin space, which remained relatively low at all incubation periods. After longer BrUTP labeling (12 min onward), the signal was associated with recently described IG-associated zones (Visa et al., 1993a). A rather frequent colocalization with anti-RNA polymerase II was found here (Figure 9). In these areas, TFIIH and Sm complex of snRNPs were also accumulated and colocalized to some extent with anti-BrdU (Figure 10); however, anti–p80-coilin antibody was not associated with such zones (our unpublished observation).
Figure 9.
Colocalization of brominated RNA (mouse anti-BrdU antibody; 6-nm grains) and RNA polymerase II (chicken anti-pol II antibody; 15-nm grains) in the IG-associated zone (asterisk) after 30-min incubation. Bar, 0.2 μm.
Figure 10.
Double-labeling with rat anti-BrdU antibody (6-nm particles) and mouse anti-Sm antibody (15-nm particles) after pulse of 8 min 40 sec. Sm complex occurs in both the IGs and the IG-associated zone (asterisk), whereas Br-RNA is not detected in these areas. Bar, 0.2 μm.
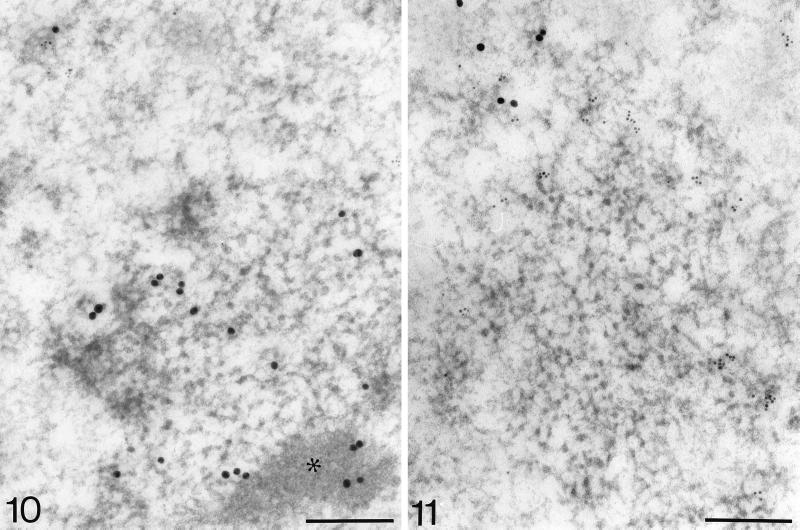
Anti-BrdU antibody recognized some BrU label that was also in association with clusters of IG, but only after longer incubation times (14–90 min); however, the centers of these clusters remained devoid of label (Figures 7 and 11). Incorporated BrUTP was partly colocalized with SC-35 and Sm complex of snRNPs (Figure 10). No significant signal for anti-RNA polymerase II (Figure 11) and antipoly(A) polymerase was revealed in IG clusters. A low signal for TFIIH was identified in both IG clusters and IG-associated zones.
Figure 11.
BrUTP (mouse anti-BrdU antibody; 6-nm grains) and RNA polymerase II (rabbit anti-pol II antibody; 15-nm grains) double-labeling after 90-min incubation. Even after a prolonged incubation time, the center of the IG cluster remains devoid of label. Some BrU signal can be observed on the periphery of the cluster, whereas no RNA polymerase II is associated with the IG cluster. Bar, 0.2 μm.
Nucleolus
The nucleolus exhibited detectable labeling after all BrUTP incubation periods. Although short pulses gave rise to signal limited to the dense fibrillar component (Figure 2), the granular component became labeled only after prolonged incubation (90 min) with the precursor (our unpublished results). No detectable signal was observed in fibrillar centers, regardless of the labeling period duration.
Cytoplasm
After paraformaldehyde fixation, ultrastructural morphology of the cytoplasmic constituents is not very well preserved. Nevertheless, the occurrence of brominated RNA in the cytoplasm, mainly after 90 min incubation, was detected repeatedly. During such incubation periods, label was also observed close to some nuclear pores. No detectable signal was identified after short labeling pulses, showing at the same time that the intracellular pool of unincorporated BrUTP was removed during washing of cells and subsequent dehydration.
Controls
The results of control experiments, in which the primary or the bridge antibody was omitted, were negative. Also the background on the resin areas outside the cells was negligible. Moreover, noninjected cells observed in the sections were devoid of label.
Ribonuclease digestion of sections labeled with brominated precursor leads to a complete removal of labeled RNA and gives rise to a signal corresponding to background level. This assay therefore confirms that the observed labeling represents RNA. The overall contrast of RNA-containing nuclear constituents, in particular of PFs usually visualized after the EDTA staining, was also reduced after ribonuclease treatment (our unpublished observations).
DISCUSSION
Our immunoelectron microscopic observations of cells microinjected with BrUTP reveal the following points. 1) During all different incubation periods with BrUTP (4–90 min), PFs occurring mainly on the periphery of condensed chromatin regions represent the major labeled nucleoplasmic ribonucleoprotein constituents. All transcription or processing factors, which we probed for, colocalized to different extents on PFs. 2) After longer BrUTP incubation periods, interchromatin granule clusters become occasionally labeled only on their periphery, whereas they remain unlabeled after short pulses. They do not contain detectable amounts of hnRNP core proteins, RNA polymerase II, and poly(A) polymerase, but they accumulate splicing factors. 3) Interchromatin granule-associated zones contain significant amounts of Br-RNA after longer labeling periods. They also accumulate RNA polymerase II and transcription factor TFIIH as well as the Sm complex of snRNPs. 4) After short incubation periods, BrUTP is incorporated only into the dense fibrillar component of the nucleoli, whereas after 90 min the signal also occurs in the granular component. 5) After 90 min incubation with the brominated precursor, label can be observed in the proximity of nuclear pores as well as in the cytoplasm.
To avoid deleterious effects of cell permeabilization on nuclear fine structure, as well as a possible protein extraction and/or displacement attributable to detergent treatment, we have microinjected BrUTP into the cytoplasm of living cells. Visualization of the incorporated brominated nucleotide by means of postembedding immunoelectron microscopy on sections of cells cut in situ allowed us to follow, at high resolution, sites of transcription and the association of newly synthesized RNA with nuclear structural constituents. Moreover, the use of gridded coverslips as cell supports allows for identification of individual cells and recording of a precise labeling period after microinjection of RNA precursor. This labeling system was previously used in fluorescence digital imaging analyses of the intranuclear spatial distribution of newly synthesized RNA (Wansink et al., 1993; Fay et al., 1997). In our laboratories, we have not been able to obtain detectable labeling, at either light or electron microscopic level, using direct incorporation of Br-uridine by intact cells in culture. Because previous studies of Br-labeled RNA processing indicated alteration of RNA processing events attributable to the presence of bromine atoms within the pre-mRNA molecule (Wansink et al., 1994b; Fay et al., 1997), our analyses were mainly concentrated on the investigation of transcription sites and on topological relationships between localization of newly synthesized RNA and constituents of nuclear architecture.
The results of our experiments confirm earlier ultrastructural observations carried out by means of high-resolution autoradiography of 3H-uridine in vivo labeled cells (e.g., Bachellerie et al., 1975; Fakan et al., 1976) and clearly identify PF as the in situ forms of nascent transcripts. It is interesting to note that after short incubation periods following BrUTP microinjection (Figure 5), label associated with PFs containing Br-RNA is mostly represented by individual gold grains, thus suggesting that transcription takes place in individual transcription sites rather than within “transcription factories” (Iborra et al., 1996; Jackson et al., 1998). Moreover, similar to autoradiographic observations (for review, see Fakan, 1978), our present findings show that most transcription sites occur on the periphery of condensed chromatin areas.
In agreement with previous reports, antibodies recognizing all transcription and splicing factors probed so far associate with PFs (e.g., Fakan et al., 1984; Puvion et al., 1984; Spector et al., 1991, Spector et al., 1993). A high degree of colocalization between Br-RNA 10 min after BrUTP microinjection and RNA polymerase II was reported using confocal immunofluorescence microscopy (Grande et al., 1997), coinciding with our observations.
The decreasing colocalization of Br-RNA with RNA polymerase II with increasing BrUTP-labeling time further suggests that a portion of labeled RNA is separated from the polymerase complex after longer labeling periods. After longer BrUTP incubation periods, some label occurs on the periphery of clusters of interchromatin granules, in agreement with previous autoradiographic observations (e.g., Fakan and Bernhard, 1973), whereas virtually no label is identified after short labeling pulses. The data therefore strongly support the conclusion that significant transcription does not take place on the IG clusters and does not favor the idea that these nuclear domains represent “transcript domains” in the nucleus (Carter et al., 1993). Similarly, no accumulation of transcription sites on the periphery of IG clusters, suggesting a concentration of active genes within this region (Moen et al., 1995), was noticed. With regard to RNA polymerase II distribution in the nucleoplasm, our findings demonstrate most anti-RNA polymerase II labeling associated with PFs within perichromatin regions, whereas only a very weak signal is revealed in interchromatin granule clusters. This is in agreement with immunofluorescence microscopic observation showing that in cells exhibiting abundant BrUTP incorporation, Br-RNA, the large RNA polymerase II subunit, and splicing and polyadenylation factors are located mostly within the nucleoplasm and do not concentrate in speckles (Zeng et al., 1997).
The high-resolution observations of the present work are in agreement with findings of fluorescent microscopic experiments indicating a large number of BrUTP signal sites diffusely distributed in the nucleoplasm of various types of cells (Jackson et al., 1993; Wansink et al., 1993; Fay et al., 1997). Moreover, an elegant fluorescence microscopic quantitative digital analysis of nascent Br-RNA, SC-35, and poly(A) distribution demonstrated that for all probes examined, most of the signal was diffusely distributed in the nucleoplasm (Fay et al., 1997).
Anti-poly(A) polymerase labeling observed in our specimens occurred rather frequently on PFs, although it moderately colocalized with newly synthesized RNA; however, no signal was observed on IG clusters, contrary to reports for poly(A)+ RNA (Visa et al., 1993b) and poly(A) binding protein II (Krause et al., 1994), which both were partly localized in this domain, in addition to a frequent occurrence on PFs. The absence of the polyadenylation enzyme in IG aggregates further indicates that polyadenylation of pre-mRNA does not take place within this nucleoplasmic domain. Moreover, in the absence of significant amounts of hnRNP core proteins within IG clusters, the nature of poly(A)+ RNA previously detected in this nucleoplasmic constituent remains to be elucidated.
IG-associated zones, a novel nucleoplasmic domain recently described in HeLa cells (Visa et al., 1993a), were also frequently observed in T24 cells. Originally reported as containing U1 but not U2 snRNP (Visa et al., 1993a), this domain begins to be labeled 10–15 min after BrUTP microinjection and is strongly labeled later. Although we also found some RNA polymerase II, Sm complex of snRNP, and TFIIH in the IG-associated zones, we have not been able to identify p80-coilin within this domain (Puvion-Dutilleul et al., 1995), although coiled bodies were labeled with anti–p80-coilin antibodies. Because IG-associated zones do not contain poly(A) polymerase, they do not seem to correspond to dots containing this enzyme and other pre-mRNA 3′ processing factors observed close to the periphery of “speckles” by laser confocal microscopy (Schul et al., 1998). The functional significance of this nucleoplasmic compartment, which has not been shown so far as a ubiquitous nuclear domain, remains to be elucidated.
Previous analysis of in vitro splicing of an adenovirus major late II pre-mRNA construct labeled with BrUTP has shown that splicing is strongly inhibited if all uridines were substituted for BrU, whereas it was restored to some extent at 50% substitution ratio and returned to an almost normal level when every tenth uridine was substituted (Wansink et al., 1994b). Although we cannot estimate the percentage of BrU substitution in our experiments, the fact that some label, especially after long periods after microinjection, is found in the vicinity of nuclear pores and in the cytoplasm is in favor of some migration of Br-RNA. Whether the molecules reaching the cytoplasm have been properly processed remains unclear; however, our observations show at the same time that there is much less intranuclear migration and export to the cytoplasm of Br-RNA compared with 3H-uridine-labeled RNA in cultured cells (e.g., Fakan and Bernhard, 1971; Fakan and Nobis, 1978). Whether accumulation of some Br-labeled PFs in small agregates observed within perichromatin regions after longer incubation periods can account for “transcription factories” reported after comparable labeling times with Br-RNA precursors (e.g., Iborra et al., 1996; Jackson et al., 1998) remains to be elucidated.
As for the distribution of label in the nucleolus, it is interesting to note that during 4–30 min post-injection periods, the signal is observed exclusively on the dense fibrillar component and reflects pre-rRNA transcription sites. This confirms some previously reported data (e.g., Granboulan and Granboulan, 1965; Fakan and Bernhard, 1971; Hozak, 1995). After a 90 min incubation, Br-RNA occurs also on the nucleolar granular component. Even if this RNA represents correctly processed molecules, its translocation from the fibrillar component is considerably slower than after radioactive labeling (Granboulan and Granboulan, 1965).
Together, these observations suggest that some RNA molecules might, with a considerable delay, follow their intracellular pathways, although the number of such molecules would be much lower than that observed for tritium-labeled RNA.
In conclusion, our high-resolution observations demonstrate perichromatin regions as being the major sites of nucleoplasmic RNA transcription. PFs, which represent the in situ forms of nascent transcripts, are also the nucleoplasmic structural constituents that bring together all transcription and processing factors analyzed so far. These in situ findings therefore support a previous suggestion that splicing could take place cotranscriptionally on PFs. Furthermore, most transcripts appear more or less individually without any striking accumulation of label even after longer incorporation periods. Finally, no accumulation of transcripts occurs within the regions of IG aggregates. IG clusters contain limited amounts of rather slowly labeled RNA. Further studies that are now in progress aim at defining labeling conditions allowing for high-resolution analyses of RNA kinetics and intranuclear trafficking under conditions of unaltered pre-mRNA processing.
ACKNOWLEDGMENTS
We thank Mrs. J. Fakan, V. Mamin, and F. Voinesco for excellent technical assistance, Miss. N. Ruchonnet and Mr. E. Bernardi for skillful photographic work, and Dr. W. Schul and Mrs. I. van der Kraan for help with microinjection. We are indebted to Dr. E.K. Chan for kindly providing anti–p80-coilin antibody, to Dr. J.-M. Egly for anti-TFIIH antibody, and to Dr. G. Martin and Dr. W. Keller, who raised the antipoly(A) polymerase antibody and prepared recombinant poly(A) polymerase. This work was supported by the Swiss National Science Foundation (grant 31-43333.95) and the Swiss Federal Office of Education and Science (OFES 95.0823) in the frame of the European Union Biomed II Program (project BMH4-CT 95-1139). P.J.V. acknowledges support from the same European Union Biomed II Program grant.
REFERENCES
- Amero SA, Raychaudhuri G, Cass CL, van Venrooij WJ, Habets WJ, Krainer AR, Beyer AL. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc Nat Acad Sci USA. 1992;89:8409–8413. doi: 10.1073/pnas.89.18.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J-P, Puvion E, Zalta J-P. Ultrastructural organization and biochemical characterization of chromatin-RNA-protein complexes isolated from mammalian cell nuclei. Eur J Biochem. 1975;58:327–337. doi: 10.1111/j.1432-1033.1975.tb02379.x. [DOI] [PubMed] [Google Scholar]
- Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection rate of splicing and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Biggiogera M, Fakan S, Kaufmann SH, Black A, Shaper JH, Busch H. Simultaneous immunoelectron microscopic visualization of protein B23 and C23 distribution in the HeLa cell nucleolus. J Histochem Cytochem. 1989;9:1371–1374. doi: 10.1177/37.9.2768807. [DOI] [PubMed] [Google Scholar]
- Brink M, Humbel BM, de Kloet ER, van Driel R. The unliganded glucocorticoid receptor is localized in the nucleus, not in the cytoplasm. Endocrinology. 1992;130:3575–3581. doi: 10.1210/endo.130.6.1597154. [DOI] [PubMed] [Google Scholar]
- Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- Carroll SB, Stollar BD. Antibodies to calf thymus RNA polymerase II from egg yolks of immunized hens. J Biol Chem. 1983;258:24–26. [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Clevenger C, Epstein A. Component of interchromatin granules using a monoclonal antibody and immunogold electron microscopy. Exp Cell Res. 1984;151:194–207. doi: 10.1016/0014-4827(84)90368-9. [DOI] [PubMed] [Google Scholar]
- Fakan S. The Cell Nucleus. Vol. 5. H. Busch, New York: Academic Press; 1978. High resolution autoradiography studies on chromatin functions; pp. 3–53. [Google Scholar]
- Fakan S. Structural support for RNA synthesis in the cell nucleus. In: Jasmin G, Simard R, editors. Nuclear Submicroscopy. Methods and Achievements in Experimental Pathology. Vol. 12. Basel: Karger; 1986. pp. 105–140. [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W. Localization of rapidly and slowly labeled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res. 1971;67:129–141. doi: 10.1016/0014-4827(71)90628-8. [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W. Nuclear labelling after prolonged 3H-uridine incorporation as visualized by high resolution autoradiography. Exp Cell Res. 1973;79:431–444. doi: 10.1016/0014-4827(73)90463-1. [DOI] [PubMed] [Google Scholar]
- Fakan S, Nobis P. Ultrastructural localization of transcription sites and of RNA distribution during the cell cycles of synchronized CHO cells. Exp Cell Res. 1978;113:327–337. doi: 10.1016/0014-4827(78)90373-7. [DOI] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. In: Bourne GH, Danielli JF, editors. International Review of Cytology. Vol. 65. San Diego: Academic Press; 1980. pp. 255–299. [DOI] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J Cell Biol. 1984;98:358–363. doi: 10.1083/jcb.98.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Immunoelectron microscope visualization of nuclear ribonucleoprotein antigens within spread transcription complexes. J Cell Biol. 1986;103:1153–1157. doi: 10.1083/jcb.103.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E, Spohr G. Localization and characterization of newly synthesized nuclear RNA in isolated rat hepatocytes. Exp Cell Res. 1976;99:155–164. doi: 10.1016/0014-4827(76)90690-x. [DOI] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distribution of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Granboulan N, Granboulan P. Cytochimie ultrastructurale du nucléole. II. Etude des sites de synthèse du RNA dans le nucléole et le noyau. Exp Cell Res. 1965;38:604–619. doi: 10.1016/0014-4827(65)90384-8. [DOI] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- Hozak P. Catching RNA polymerase I in flagranti: ribosomal genes are transcribed in the dense fibrillar component of the nucleolus. Exp Cell Res. 1995;216:285–289. doi: 10.1006/excr.1995.1036. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EMM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RE, Okamura CS, Martin TE. Immunofluorescent localization of the proteins of nuclear ribonucleoprotein complexes. J Cell Biol. 1980;86:235–243. doi: 10.1083/jcb.86.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Dahmus ME. Immunochemical analysis of mammalian RNA polymerase II subspecies. Stability and relative in vivo concentration. J Biol Chem. 1986;261:14219–14225. [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. Localisation of splicing snRNPs in mammalian cells. Mol Biol Rep. 1993;18:127–133. doi: 10.1007/BF00986767. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz J. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune diseases. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Amalric F, Lührmann R, Vogel P, Fakan S. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol. 1994;65:82–93. [PubMed] [Google Scholar]
- Manders EMM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factors in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Moen PT, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum Mol Gen. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Puvion E, Moyne G. The Cell Nucleus. Vol. 8. H. Busch, New York: Academic Press; 1981. In situ localization of RNA structures; pp. 59–115. [Google Scholar]
- Puvion E, Puvion-Dutilleul F. Ultrastructure of the nucleus in relation to transcription and splicing: roles of perichromatin fibrils and interchromatin granules. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- Puvion E, Viron A, Assens C, Leduc EH, Jeanteur P. Immunocytochemical identification of nuclear structures containing snRNPs in isolated rat liver cells. J Ultrastruct Res. 1984;87:180–189. doi: 10.1016/s0022-5320(84)80077-5. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Besse S, Chan EKL, Tan EM, Puvion E. p80-coilin: a component of coiled bodies and interchromatin granule-associated zones. J Cell Sci. 1995;108:1143–1153. doi: 10.1242/jcs.108.3.1143. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers JH, Egly JM. The ERCC2/DNA repair proteins associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp Cell Res. 1998;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- Smetana K, Steele WJ, Busch H. A nuclear ribonucleoprotein network. Exp Cell Res. 1963;182:521–533. [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear organization and gene expression. Exp Cell Res. 1996;229:189–197. doi: 10.1006/excr.1996.0358. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, O’Keefe RT, Jiménez-Garcia LF. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harb Symp Quant Biol. 1993;58:799–805. doi: 10.1101/sqb.1993.058.01.087. [DOI] [PubMed] [Google Scholar]
- Swift H. The Interpretation of Ultrastructure. Symposia of the International Society for Cell Biology. Vol. 1. R.J.C. Harris, New York: Academic Press; 1962. Nucleoprotein localization in electron micrographs: metal binding and radioautography; pp. 213–232. [Google Scholar]
- Tamburini M, Malatesta M, Zancanaro C, Martin TE, Fu X-D, Vogel P, Fakan S. Dense granular bodies: a novel nucleoplasmic structure in hibernating dormice. Histochem Cell Biol. 1996;106:581–586. doi: 10.1007/BF02473273. [DOI] [PubMed] [Google Scholar]
- Ten Kate R, van Balen R, Smeulders AWM, Groen FCA, den Boer G. SCILAIM, a multi-level interactive image processing environment. Pattern Recogn Lett. 1990;11:429–441. [Google Scholar]
- van Balen R, Ten Kate T, Koelma D, Mosterd B, Smeulders AWM. ScilImage: A multi-layered environment for use and envelopment of image processing software. In: Christensen HI, Crowley JL, editors. Experimental Environments for Computer Vision and Image Processing. Singapore: World Scientific Press; 1994. pp. 107–126. [Google Scholar]
- Vanderlaan M, Thomas CB. Characterization of monoclonal antibodies to bromodeoxyuridine. Cytometry. 1985;6:501–505. doi: 10.1002/cyto.990060603. [DOI] [PubMed] [Google Scholar]
- Van der Voort HTM, Straster K. Restoration of confocal images for quantitative image analysis. J Microsc. 1995;178:165–181. [Google Scholar]
- van Driel R, Wansink DG, van Steensel B, Grande MA, Schul W, de Jong L. Nuclear domains and the nuclear matrix. Int Rev Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Bachellerie J-P, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur J Cell Biol. 1993a;60:308–321. [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie J-P, Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993b;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Motley AM, van Driel R, de Jong L. Fluorescent labeling of nascent RNA in the cell nucleus using 5-bromouridine 5′-triphosphate. In: Celis JE, editor. Cell Biology—A Laboratory Handbook. Vol. 2. San Diego: Academic Press; 1994a. pp. 368–374. [Google Scholar]
- Wansink DG, Nelissen RLH, de Jong L. In vitro splicing of pre-mRNA containing bromouridine. Mol Biol Rep. 1994b;19:109–113. doi: 10.1007/BF00997156. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:282–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Kim E, Warren SL, Berget SM. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]